Uptake of Dimethylsulfoniopropionate (DMSP) by Natural Microbial Communities of the Great Barrier Reef (GBR), Australia
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Experimental Design
2.2. Experiment 1: Quantification of DMSP Uptake by Size Fractions of the Microbial Community
2.3. Experiment 2: Effect of DMSP on Community Structure and Gene Regulation
2.4. Quantification of Biogenic Sulfur Compounds
2.5. Quantification of Microbial Composition
2.6. Nutrients
2.7. Microbial Community Composition and DMSP-Related Genes
2.8. DMSP Degradation Gene Profiling
2.9. Data Analysis
3. Results
3.1. Characteristics of Initial Water Masses
3.2. Experiment 1: Quantification of DMSP Uptake by Size Fractions of the Microbial Community
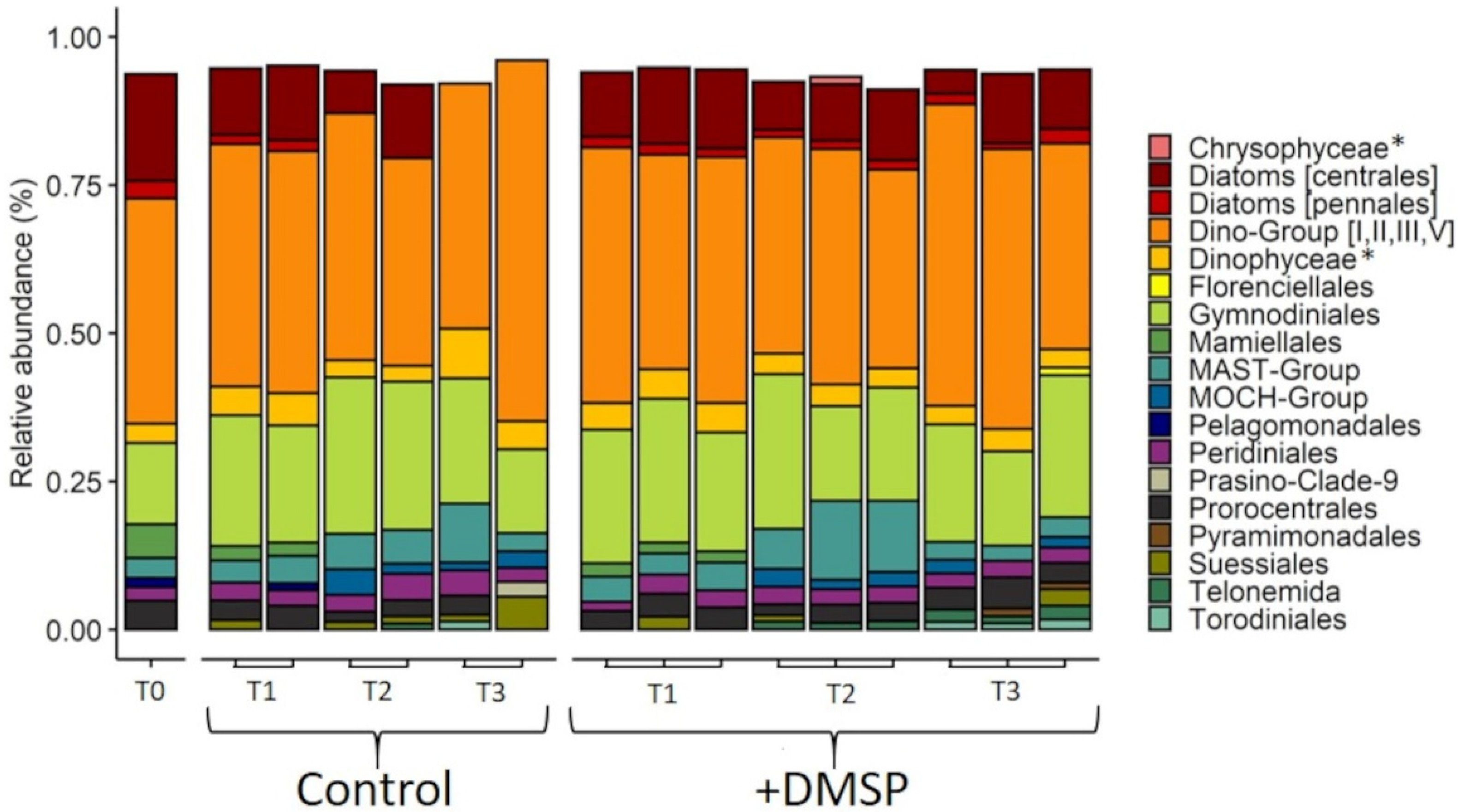
3.3. Experiment 2: Effect of DMSP on Community Structure and Gene Regulation
4. Discussion
4.1. Quantification of DMSP Uptake by Size Fractions of the Microbial Community—Experiment 1
4.2. Dynamic Changes in Microbial Community Composition and DMSP Degradation Gene Abundance in Reef Waters—Experiment 2
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simó, R. From cells to globe: Approaching the dynamics of DMS(P) in the ocean at multiple scales. Can. J. Fish. Aquat. Sci. 2004, 61, 673–684. [Google Scholar] [CrossRef] [Green Version]
- Keller, M.M.D.; Bellows, W.W.K.; Guillard, R.R.R.L. Dimethyl Sulfide Production in Marine Phytoplankton. In Biogenic Sulfur in the Environment; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 1989; Volume 393, pp. 167–182. [Google Scholar]
- Matrai, P.A.; Keller, M.D. Total organic sulfur and dimethylsulfoniopropionate in marine phytoplankton: Intracellular variations. Mar. Biol. 1994, 119, 61–68. [Google Scholar] [CrossRef]
- Caruana, A.M.N.; Steinke, M.; Turner, S.M.; Malin, G. Concentrations of dimethylsulphoniopropionate and activities of dimethylsulphide-producing enzymes in batch cultures of nine dinoflagellate species. Biogeochemistry 2012, 110, 87–107. [Google Scholar] [CrossRef]
- Keller, M.D.; Kiene, R.P.R.; Matrai, P.A.P.; Bellows, W.K. Production of glycine betaine and dimethylsulfoniopropionate in marine phytoplankton. II. N-limited chemostat cultures. Mar. Biol. 1999, 135, 249–257. [Google Scholar] [CrossRef]
- Simó, R.; Archer, S.D.; Pedrós-Alió, C.; Gilpin, L.; Stelfox-Widdicombe, C.E. Coupled dynamics of dimethylsulfoniopropionate and dimethylsulfide cycling and the microbial food web in surface waters of the North Atlantic. Limnol. Oceanogr. 2002, 47, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Zubkov, M.; Fuchs, B.; Archer, S.; Kiene, R.P.; Amann, R.; Burkill, P.H. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 2001, 3, 304–311. [Google Scholar] [CrossRef]
- Karsten, U.; Kück, K.; Vogt, C.; Kirst, G. Dimethylsulfoniopropionate production in phototrophic organisms and its physiological functions as a cryoprotectant. In Biological and Environmental Chemistry of DMSP and Related Sulfonium Compounds; Kiene, R.P., Visscher, P.T., Keller, M.D., Kirst, G.O., Visscher, P.T., Eds.; Plenum Press: New York, NY, USA, 1996; pp. 143–153. [Google Scholar]
- Dickson, D.; Kirst, G. The role of β-dimethylsulphoniopropionate, glycine betaine and homarine in the osmoacclimation of Platymonas subcordiformis. Planta 1986, 167, 536–543. [Google Scholar] [CrossRef]
- Sunda, W.; Kieber, D.J.; Kiene, R.P.; Huntsman, S. An antioxidant function for DMSP and DMS in marine algae. Nature 2002, 418, 317–320. [Google Scholar] [CrossRef]
- Saha, M.; Rempt, M.; Gebser, B.; Grueneberg, J.; Pohnert, G.; Weinberger, F. Dimethylsulphopropionate (DMSP) and proline from the surface of the brown alga Fucus vesiculosus inhibit bacterial attachment. Biofouling 2012, 28, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Strom, S.; Wolfe, G.; Slajer, A.; Lambert, S.; Clough, J. Chemical defense in the microplankton II: Inhibition of protist feeding by β-dimethylsulfoniopropionate (DMSP). Limnol. Oceanogr. 2003, 48, 230–237. [Google Scholar] [CrossRef]
- Wolfe, G.V.; Steinke, M.; Kirst, G.O. Grazing-activated chemical defence in a unicellular marine alga. Nature 1997, 387, 894–897. [Google Scholar] [CrossRef]
- Hill, R.; White, B.; Cottrell, M.; Dacey, J. Virus-mediated total release of dimethylsulfoniopropionate from marine phytoplankton: A potential climate process. Aquat. Microb. Ecol. 1998, 14, 1–6. [Google Scholar] [CrossRef]
- Matrai, P.A.; Keller, M.D. Dimethylsulfide in a large-scale coccolithophore bloom in the Gulf of Maine. Cont. Shelf Res. 1993, 13, 831–843. [Google Scholar] [CrossRef]
- Dacey, J.W.H.; Wakeham, S.G. Oceanic Dimethylsulfide: Production During Zooplankton Grazing on Phytoplankton. Science 1986, 233, 1314–1316. [Google Scholar] [CrossRef] [PubMed]
- Malmstrom, R.R.; Kiene, R.P.; Kirchman, D.L. Identification and enumeration of bacteria assimilating dimethylsulfoniopropionate (DMSP) in the North Atlantic and Gulf of Mexico. Limnol. Oceanogr. 2004, 49, 597–606. [Google Scholar] [CrossRef]
- Ruiz-González, C.; Galí, M.; Sintes, E.; Herndl, G.G.J.; Gasol, J.J.M.; Simó, R.; Neilson, A.; Lewin, R.; Amblard, C.; Wright, R.; et al. Sunlight Effects on the Osmotrophic Uptake of DMSP-Sulfur and Leucine by Polar Phytoplankton. PLoS ONE 2012, 7, e45545. [Google Scholar] [CrossRef] [Green Version]
- Curson, A.R.J.; Todd, J.D.; Sullivan, M.J.; Johnston, A.W.B. Catabolism of dimethylsulphoniopropionate: Microorganisms, enzymes and genes. Nat. Rev. Microbiol. 2011, 9, 849–859. [Google Scholar] [CrossRef]
- Reisch, C.R.; Stoudemayer, M.J.; Varaljay, V.A.; Amster, I.J.; Moran, M.A.; Whitman, W.B. Novel pathway for assimilation of dimethylsulphoniopropionate widespread in marine bacteria. Nature 2011, 473, 208–211. [Google Scholar] [CrossRef]
- Kiene, R.P.; Linn, L.J.; Bruton, J.A. New and important roles for DMSP in marine microbial communities. J. Sea Res. 2000, 43, 209–224. [Google Scholar] [CrossRef]
- Kiene, R.P.; Linn, L.J.; González, J.; Moran, M.A.; Bruton, J.A. Dimethylsulfoniopropionate and methanethiol are important precursors of methionine and protein-sulfur in marine bacterioplankton. Appl. Environ. Microbiol. 1999, 65, 4549–4558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiene, R.P.; Linn, L.J. The fate of dissolved dimethylsulfoniopropionate (DMSP) in seawater: Tracer studies using 35S-DMSP. Geochim. Cosmochim. Acta 2000, 64, 2797–2810. [Google Scholar] [CrossRef]
- Yoch, D.C. Dimethylsulfoniopropionate: Its Sources, Role in the Marine Food Web, and Biological Degradation to Dimethylsulfide. Appl. Environ. Microbiol. 2002, 68, 5804–5815. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moran, M.A.; Reisch, C.R.; Kiene, R.P.; Whitman, W.B. Genomic Insights into Bacterial DMSP Transformations. Ann. Rev. Mar. Sci. 2012, 4, 523–542. [Google Scholar] [CrossRef]
- Simó, R. Production of atmospheric sulfur by oceanic plankton: Biogeochemical, ecological and evolutionary links. Trends Ecol. Evol. 2001, 16, 287–294. [Google Scholar] [CrossRef]
- Gao, C.; Fernandez, V.I.; Lee, K.S.; Fenizia, S.; Pohnert, G.; Seymour, J.R.; Raina, J.-B.; Stocker, R. Single-cell bacterial transcription measurements reveal the importance of dimethylsulfoniopropionate (DMSP) hotspots in ocean sulfur cycling. Nat. Commun. 2020, 11, 1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, E.C.; Henriksen, J.R.; Buchan, A.; Reisch, C.R.; Burgmann, H.; Welsh, R.; Ye, W.; Gonzalez, J.M.; Mace, K.; Joye, S.B.; et al. Bacterial Taxa That Limit Sulfur Flux from the Ocean. Science 2006, 314, 649–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Howard, E.C.; Sun, S.; Biers, E.J.; Moran, M.A. Abundant and diverse bacteria involved in DMSP degradation in marine surface waters. Environ. Microbiol. 2008, 10, 2397–2410. [Google Scholar] [CrossRef]
- Todd, J.D.; Curson, A.R.J.; Dupont, C.L.; Nicholson, P.; Johnston, A.W.B. The dddP gene, encoding a novel enzyme that converts dimethylsulfoniopropionate into dimethyl sulfide, is widespread in ocean metagenomes and marine bacteria and also occurs in some Ascomycete fungi. Environ. Microbiol. 2009, 11, 1624–1625. [Google Scholar] [CrossRef]
- Choi, D.H.; Park, K.-T.; An, S.M.; Lee, K.; Cho, J.-C.; Lee, J.-H.; Kim, D.; Jeon, D.; Noh, J.H. Pyrosequencing revealed SAR116 clade as dominant dddP-containing bacteria in oligotrophic NW Pacific Ocean. PLoS ONE 2015, 10, e0116271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Todd, J.D.; Thrash, J.C.; Qian, Y.; Qian, M.C.; Temperton, B.; Guo, J.; Fowler, E.K.; Aldrich, J.T.; Nicora, C.D.; et al. The abundant marine bacterium Pelagibacter simultaneously catabolizes dimethylsulfoniopropionate to the gases dimethyl sulfide and methanethiol. Nat. Microbiol. 2016, 1, 16065. [Google Scholar] [CrossRef]
- Vila-Costa, M.; Simó, R.; Harada, H.; Gasol, J.M.; Slezak, D.; Kiene, R.P. Dimethylsulfoniopropionate uptake by marine phytoplankton. Science 2006, 314, 652–654. [Google Scholar] [CrossRef] [PubMed]
- Petrou, K.; Nielsen, D.A. Uptake of dimethylsulphoniopropionate (DMSP) by the diatom Thalassiosira weissflogii: A model to investigate the cellular function of DMSP. Biogeochemistry 2018, 141, 265–271. [Google Scholar] [CrossRef]
- Spielmeyer, A.; Gebser, B.; Pohnert, G. Investigations of the uptake of dimethylsulfoniopropionate by phytoplankton. ChemBioChem 2011, 12, 2276–2279. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-González, C.; Simó, R.; Vila-Costa, M.; Sommaruga, R.; Gasol, J.M. Sunlight modulates the relative importance of heterotrophic bacteria and picophytoplankton in DMSP-sulphur uptake. ISME J. 2012, 6, 650–659. [Google Scholar] [CrossRef] [Green Version]
- Theseira, A.M.; Nielsen, D.A.; Petrou, K. Uptake of dimethylsulphoniopropionate (DMSP) reduces free reactive oxygen species (ROS) during late exponential growth in the diatom Thalassiosira weissflogii grown under three salinities. Mar. Biol. 2020, 167, 1–7. [Google Scholar] [CrossRef]
- Alcolombri, U.; Ben-Dor, S.; Feldmesser, E.; Levin, Y.; Tawfik, D.S.; Vardi, A. Marine sulfur cycle. Identification of the algal dimethyl sulfide-releasing enzyme: A missing link in the marine sulfur cycle. Science 2015, 348, 1466–1469. [Google Scholar] [CrossRef]
- Malin, G. New pieces for the Marine Sulfur Cycle Jigsaw. Science 2006, 314, 607–608. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Maltrud, M.E.; Burrows, S.M.; Elliott, S.M.; Cameron-Smith, P. Impacts of Shifts in Phytoplankton Community on Clouds and Climate via the Sulfur Cycle. Glob. Biogeochem. Cycles 2018, 32, 1005–1026. [Google Scholar] [CrossRef]
- McCoy, D.T.; Burrows, S.M.; Wood, R.; Grosvenor, D.P.; Elliott, S.M.; Ma, P.-L.; Rasch, P.J.; Hartmann, D.L. Natural aerosols explain seasonal and spatial patterns of Southern Ocean cloud albedo. Sci. Adv. 2015, 1, e1500157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blondeau-Patissier, D.; Ernesto Brando, V.; Lønborg, C.; Leahy, S.M.; Dekker, A.G. Phenology of trichodesmium spp. Blooms in the great barrier reef lagoon, Australia, from the ESA-MERIS 10-year mission. PLoS ONE 2018, 13, e0208010. [Google Scholar] [CrossRef] [Green Version]
- Revelante, N.; Williams, W.T.; Bunt, J.S. Temporal and spatial distribution of diatoms, dinoflagellates and trichodesmium in waters of the Great Barrier Reef. J. Exp. Mar. Bio. Ecol. 1982, 63, 27–45. [Google Scholar] [CrossRef]
- Westberry, T.K.; Siegel, D.A. Spatial and temporal distribution of Trichodesmium blooms in the world’s oceans. Glob. Biogeochem. Cycles 2006, 20, 1–13. [Google Scholar] [CrossRef]
- Partensky, F.; Blanchot, J.; Vaulot, D. Differential distribution and ecology of Prochlorococcus and Synechococcus in oceanic waters: A review. In Marine Cyanobacteria; Charpy, L., Larkum, A., Eds.; Musée Océanographique: Monaco, Monaco, 1999; pp. 457–475. ISBN 2726002102. [Google Scholar]
- Broadbent, A.; Jones, G. Seasonal and Diurnal Cycles of Dimethylsulfide, Dimethylsulfoniopropionate and Dimethylsulfoxide at One Tree Reef Lagoon. Environ. Chem. 2006, 3, 260. [Google Scholar] [CrossRef]
- Broadbent, A.D.; Jones, G.B.; Jones, R.J. DMSP in Corals and Benthic Algae from the Great Barrier Reef. Estuar. Coast. Shelf Sci. 2002, 55, 547–555. [Google Scholar] [CrossRef]
- Jones, G.; Curran, M.; Broadbent, A.; King, S.; Fischer, E. Factors affecting the cycling of dimethylsulfide and dimethylsulfoniopropionate in coral reef waters of the Great Barrier Reef. Environ. Chem. 2007, 4, 310–322. [Google Scholar] [CrossRef]
- Jones, G.; Curran, M.; Broadbent, A. Dimethylsulphide in the South Pacific. In Recent Advances in Marine Science and Technology 1994; Bellwood, O., Choat, H., Saxena, N., Eds.; PACON International and James Cook University of North Queensland: Townsville, Australia, 1994; pp. 183–190. [Google Scholar]
- Del Valle, D.A.; Slezak, D.; Smith, C.M.; Rellinger, A.N.; Kieber, D.J.; Kiene, R.P. Effect of acidification on preservation of DMSP in seawater and phytoplankton cultures: Evidence for rapid loss and cleavage of DMSP in samples containing Phaeocystis sp. Mar. Chem. 2011, 124, 57–67. [Google Scholar] [CrossRef]
- Harada, H.; Rouse, M.-A.; Sunda, W.; Kiene, R.P. Latitudinal and vertical distributions of particle-associated dimethylsulfoniopropionate (DMSP) lyase activity in the western North Atlantic Ocean. Can. J. Fish. Aquat. Sci. 2004, 61, 700–711. [Google Scholar] [CrossRef]
- Marie, D.; Partensky, F.; Jacquet, S.; Vaulot, D. Enumeration and Cell Cycle Analysis of Natural Populations of Marine Picoplankton by Flow Cytometry Using the Nucleic Acid Stain SYBR Green I. Appl. Environ. Microbiol. 1997, 63, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Gasol, J.M.; Del Giorgio, P.A.; Giorgio, P.A. del Using flow cytometry for counting natural planktonic bacteria and understanding the structure of planktonic bacterial communities. Sci. Mar. 2000, 64, 197–224. [Google Scholar] [CrossRef] [Green Version]
- Seymour, J.R.; Seuront, L.; Mitchell, J.G. Microscale gradients of planktonic microbial communities above the sediment surface in a mangrove estuary. Estuar. Coast. Shelf Sci. 2007, 73, 651–666. [Google Scholar] [CrossRef]
- Cowley, R.; Critchley, G.; Eriksen, R.; Latham, V.; Plaschke, R.; Rayner, M.; Terhell, D. CSIRO Research Publications Repository—Hydrochemistry Operations Manual; CSIRO Marine Laboratories: Hobart, Australia, 1999. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Levine, N.M.; Varaljay, V.A.; Toole, D.A.; Dacey, J.W.H.; Doney, S.C.; Moran, M.A. Environmental, biochemical and genetic drivers of DMSP degradation and DMS production in the Sargasso Sea. Environ. Microbiol. 2012, 14, 1210–1223. [Google Scholar] [CrossRef] [Green Version]
- Varaljay, V.A.; Howard, E.C.; Sun, S.; Moran, M.A. Deep sequencing of a dimethylsulfoniopropionate-degrading gene (dmdA) by using PCR primer pairs designed on the basis of marine metagenomic data. Appl. Environ. Microbiol. 2010, 76, 609–617. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, M.T.; Taylor, L.T.; DeLong, E.F. Quantitative analysis of small-subunit rRNA genes in mixed microbial populations via 5′-nuclease assays. Appl. Environ. Microbiol. 2000, 66, 4605–4614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clarke, K.R.; Gorley, R.N. PRIMER V6 Software; Primer-e: Auckland, New Zealand.
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.; Guillaume Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, R.P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package 2020. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 10 April 2021).
- Kassambara, A. Ggpubr: “Ggplot2” Based Publication Ready Plots 2020. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 10 April 2021).
- Kiene, R.P.; Linn, L.J. Distribution and turnover of dissolved DMSP and its relationship with bacterial production and dimethylsulfide in the Gulf of Mexico. Limnol. Oceanogr. 2000, 45, 849–861. [Google Scholar] [CrossRef]
- Andrews, J.C. Water masses, nutrient levels and seasonal drift on the outer central queensland shelf (great barrier reef). Mar. Freshw. Res. 1983, 34, 821–834. [Google Scholar] [CrossRef]
- Crosbie, N.D.; Furnas, M.J. Abundance, distribution and flow-cytometric characterization of picophytoprokaryote populations in central (17°S) and southern (20°S) shelf waters of the Great Barrier Reef. J. Plankton Res. 2001, 23, 809–828. [Google Scholar] [CrossRef] [Green Version]
- Muslim, I.; Jones, G. The seasonal variation of dissolved nutrients, chlorophyll a and suspended sediments at Nelly Bay, Magnetic Island. Estuar. Coast. Shelf Sci. 2003, 57, 445–455. [Google Scholar] [CrossRef]
- Brimblecombe, P.; Shooter, D. Photo-oxidation of dimethylsulphide in aqueous solution. Mar. Chem. 1986, 19, 343–353. [Google Scholar] [CrossRef]
- Visscher, P.; Diaz, M.; Taylor, B. Enumeration of bacteria which cleave or demethylate dimethylsulfoniopropionate in the Caribbean Sea. Mar. Ecol. Prog. Ser. 1992, 89, 293–296. [Google Scholar] [CrossRef]
- Zeyer, J.; Eicher, P.; Wakeham, S.G.; Schwarzenbach, R.P. Oxidation of Dimethyl Sulfide to Dimethyl Sulfoxide by Phototrophic Purple Bacteria. Appl. Environ. Microbiol. 1987, 53, 2026–2032. [Google Scholar] [CrossRef] [Green Version]
- Malmstrom, R.R.; Kiene, R.P.; Vila, M.; Kirchman, D.L. Dimethylsulfoniopropionate (DMSP) assimilation by Synechococcus in the Gulf of Mexico and northwest Atlantic Ocean. Limnol. Oceanogr. 2005, 50, 1924–1931. [Google Scholar] [CrossRef]
- Campbell, L.; Nolla, H.A.; Vaulot, D. The importance of Prochlorococcus to community structure in the central North Pacific Ocean. Limnol. Oceanogr. 1994, 39, 954–961. [Google Scholar] [CrossRef]
- Partensky, F.; Hess, W.R.; Vaulot, D. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 1999, 63, 106–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vila-Costa, M.; del Valle, D.A.; González, J.M.; Slezak, D.; Kiene, R.P.; Sánchez, O.; Simó, R. Phylogenetic identification and metabolism of marine dimethylsulfide-consuming bacteria. Environ. Microbiol. 2006, 8, 2189–2200. [Google Scholar] [CrossRef]
- Lei, L.; Alcolombri, U.; Tawfik, D.S. DddY is a bacterial dimethylsulfoniopropionate lyase representing a new cupin enzyme superfamily with unknown primary function. bioRxiv 2017. [Google Scholar] [CrossRef] [Green Version]
- Varaljay, V.A.; Gifford, S.M.; Wilson, S.T.; Sharma, S.; Karl, D.M.; Moran, M.A. Bacterial dimethylsulfoniopropionate degradation genes in the oligotrophic north pacific subtropical gyre. Appl. Environ. Microbiol. 2012, 78, 2775–2782. [Google Scholar] [CrossRef] [Green Version]
- Malmstrom, R.R.; Kiene, R.P.; Cottrell, M.T.; Kirchman, D.L. Contribution of SAR11 Bacteria to Dissolved Dimethylsulfoniopropionate and Amino Acid Uptake in the North Atlantic Ocean. Appl. Environ. Microbiol. 2004, 70, 4129–4135. [Google Scholar] [CrossRef] [Green Version]
- Cui, Y.; Suzuki, S.; Omori, Y.; Wong, S.-K.; Ijichi, M.; Kaneko, R.; Kameyama, S.; Tanimoto, H.; Hamasaki, K. Abundance and distribution of dimethylsulfoniopropionate degradation genes and the corresponding bacterial community structure at dimethyl sulfide hot spots in the tropical and subtropical pacific ocean. Appl. Environ. Microbiol. 2015, 81, 4184–4194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Liu, J.; Zhang, S.-H.; Liang, J.; Lin, H.; Song, D.; Yang, G.-P.; Todd, J.D.; Zhang, X.-H. Novel Insights Into Bacterial Dimethylsulfoniopropionate Catabolism in the East China Sea. Front. Microbiol. 2018, 9, 3206. [Google Scholar] [CrossRef] [PubMed]
- Motard-Côté, J.; Kiene, R. Osmoprotective role of dimethylsulfoniopropionate (DMSP) for estuarine bacterioplankton. Aquat. Microb. Ecol. 2015, 76, 133–147. [Google Scholar] [CrossRef]
- Simó, R.; Pedrós-Alió, C.; Malin, G.; Grimalt, J. Biological turnover of DMS, DMSP and DMSO in contrasting open-sea waters. Mar. Ecol. Prog. Ser. 2000, 203, 1–11. [Google Scholar] [CrossRef]
- Simó, R.; Pedrós-Alió, C. Short-term variability in the open ocean cycle of dimethylsulfide. Glob. Biogeochem. Cycles 1999, 13, 1173–1181. [Google Scholar] [CrossRef] [Green Version]
- Wolfe, G.; Steinke, M. Grazing-activated production of dimethyl sulfide (DMS) by two clones of Emiliania huxleyi. Limnol. Oceanogr. 1996, 41, 1151–1160. [Google Scholar] [CrossRef] [Green Version]
- Van Boekel, J.; Stefels, W. Production of DMS from dissolved DMSP in axenic cultures of the marine phytoplankton species Phaeocystis sp. Mar. Ecol. Prog. Ser. 1993, 97, 11–18. [Google Scholar] [CrossRef]
- Yost, D.M.; Mitchelmore, C.L. Dimethylsulfoniopropionate (DMSP) lyase activity in different strains of the symbiotic alga symbiodinium microadriaticum. Mar. Ecol. Prog. Ser. 2009, 386, 61–70. [Google Scholar] [CrossRef]
- Yost, D.M.; Mitchelmore, C.L. Substrate kinetics of DMSP-lyases in various cultured symbiodinium strains. Bull. Mar. Sci. 2012, 88, 267–282. [Google Scholar] [CrossRef]
- Nowinski, B.; Motard-Côté, J.; Landa, M.; Preston, C.M.; Scholin, C.A.; Birch, J.M.; Kiene, R.P.; Moran, M.A. Microdiversity and temporal dynamics of marine bacterial dimethylsulfoniopropionate genes. Environ. Microbiol. 2019, 21, 1687–1701. [Google Scholar] [CrossRef]
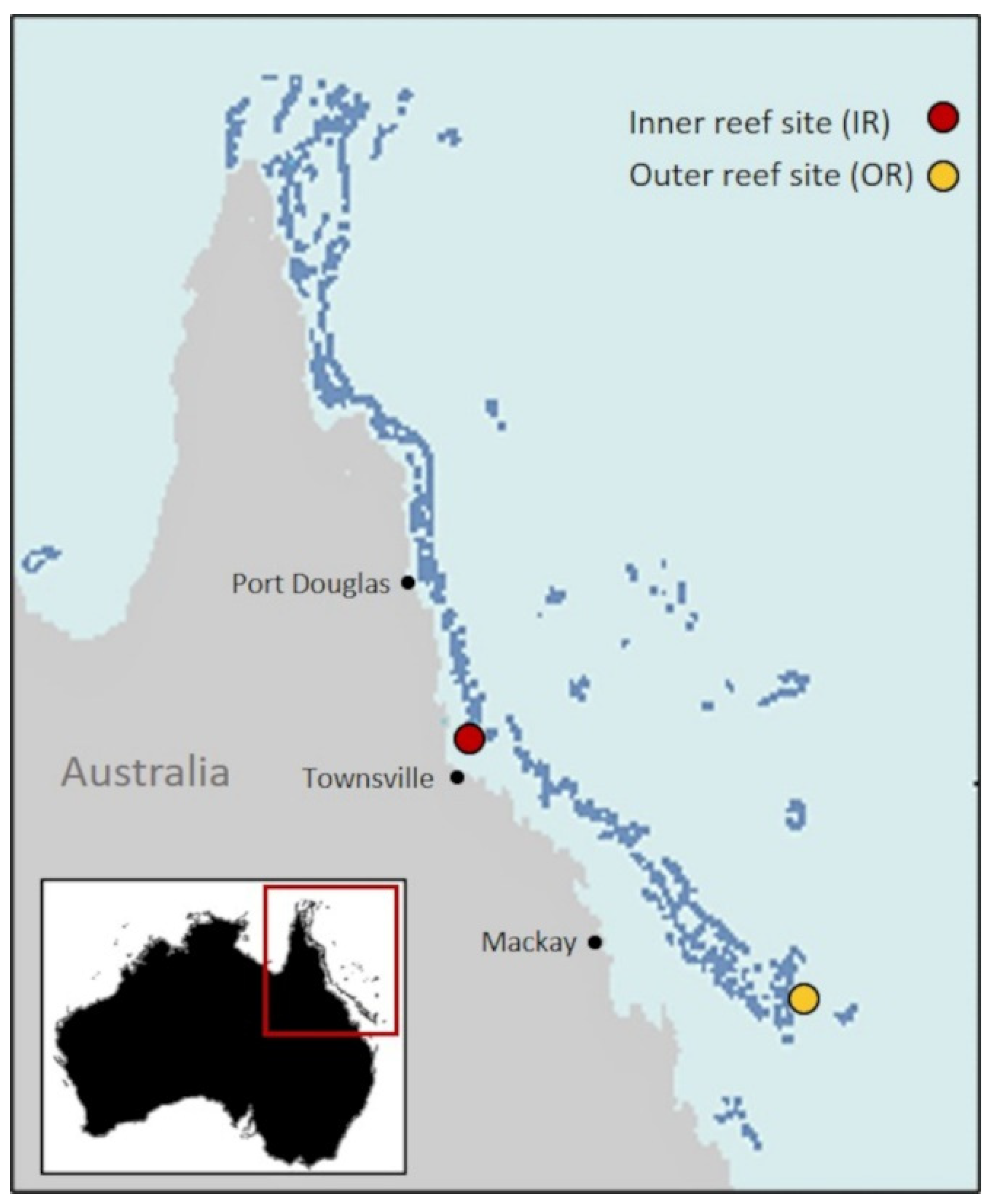
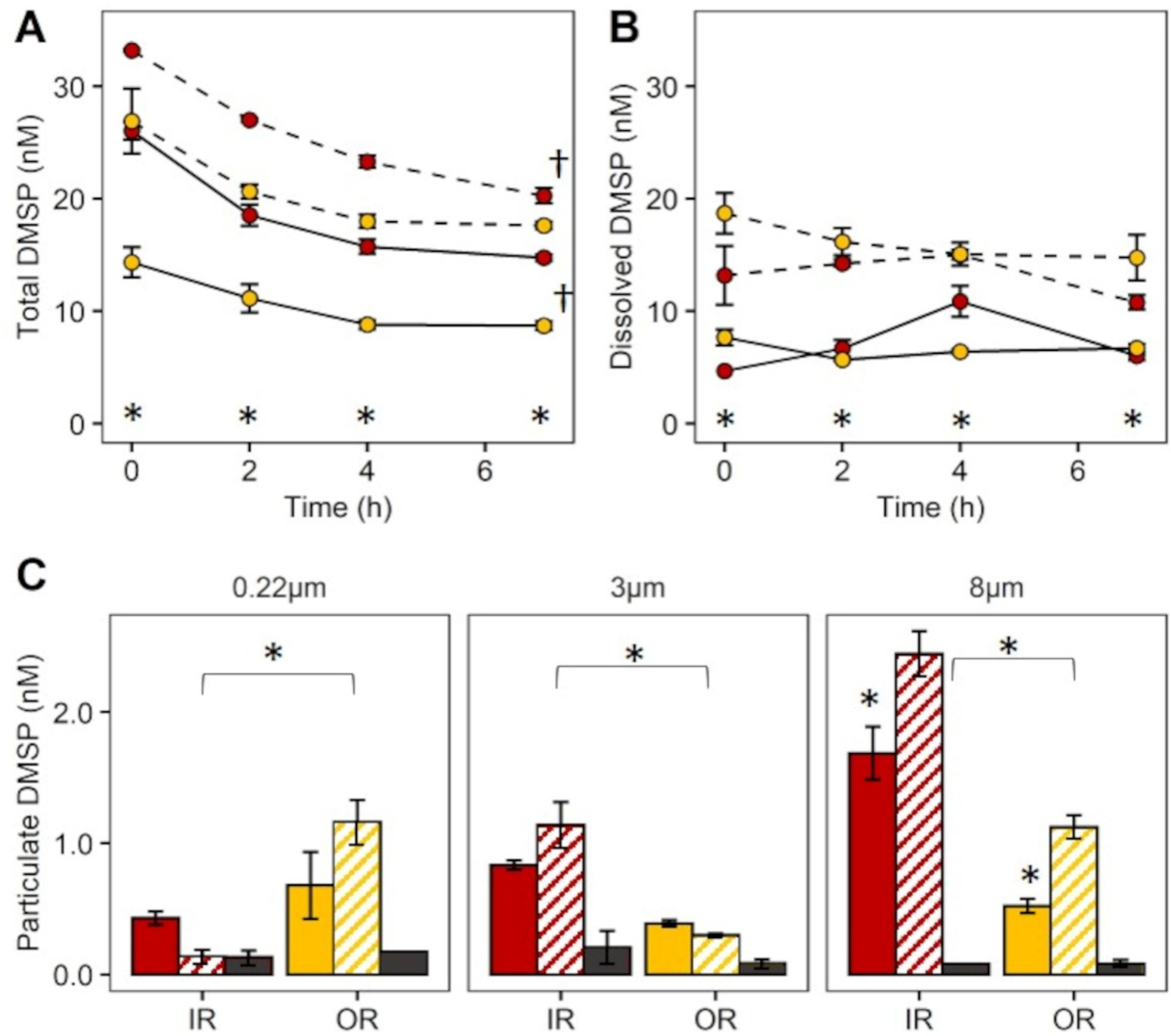
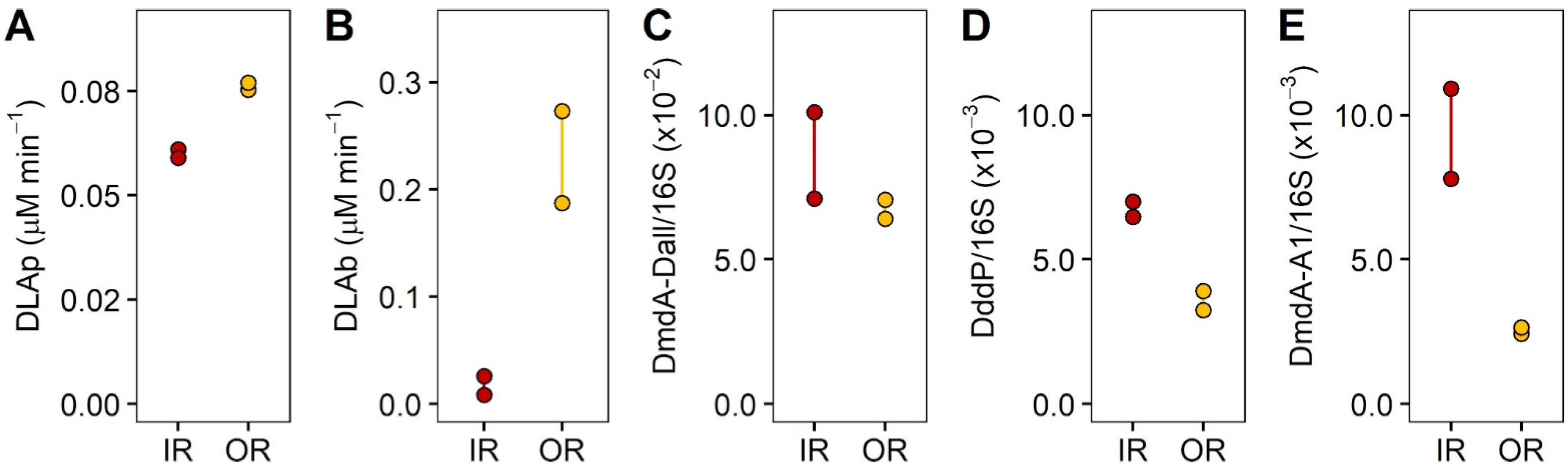

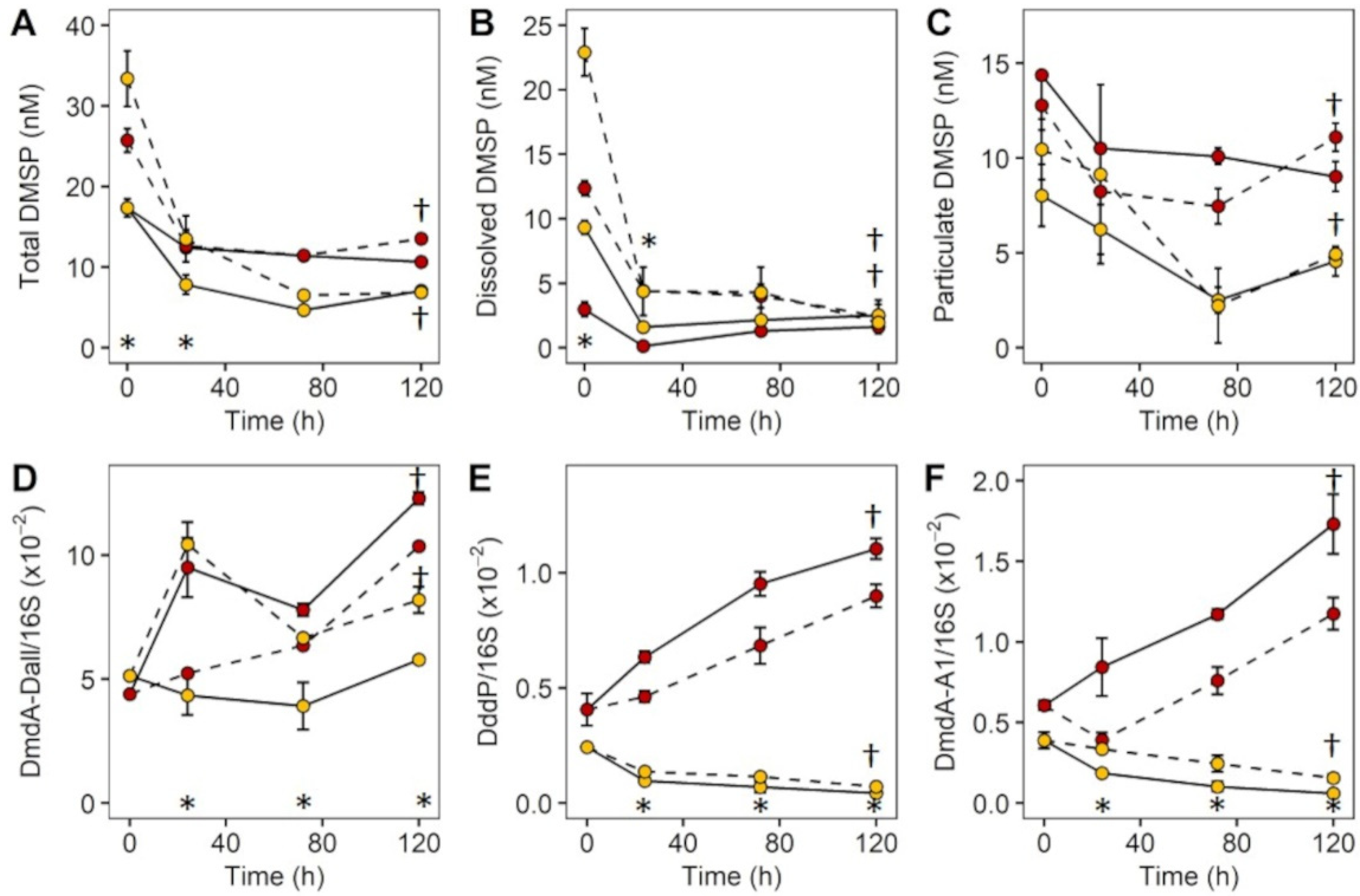




| Inner Reef | Outer Reef | |||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 1 | Experiment 2 | |
| Sample depth (m) | 4.0 | 3.4 | 5.0 | 5.0 |
| Temperature (°C) | 26.3 | 27.1 | 26.7 | 26.5 |
| Salinity (psu) | 35.4 | 35.4 | 35.4 | 35.5 |
| DMS (nM) | BDL | 0.90 | 0.86 | 1.0 |
| NOX (µM) | 0.014 | 0.020 | 0.020 | BDL |
| NO2− (µM) | 0.006 | 0.007 | 0.004 | 0.020 |
| PO43− (µM) | 0.038 | 0.050 | 0.049 | 0.066 |
| SiO4− (µM) | 0.623 | 0.500 | 0.798 | 0.882 |
| NH4+ (µM) | BDL | 0.010 | 0.021 | 0.004 |
| Inner Reef | Outer Reef | |||
|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 1 | Experiment 2 | |
| Synechococcus (cells × 103 mL−1) | 2.65 ± 0.71 a | 1.73 ± 0.03 ac | 0.20 ± 0.14 b | 0.93 ± 0.02 bc |
| Prochlorococcus (cells × 105 mL−1) | 6.56 ±2.7 ac | 2.11 ± 0.15 b | 2.29 ± 0.06 b | 5.59 ± 0.53 bc |
| Heterotrophic Bacteria (cells × 105 mL−1) | 9.30 ± 0.05 a | 6.50 ± 0.02 b | 3.24 ± 0.02 c | 2.67 ± 0.05 c |
| Picoeukaryotes (cells mL−1) | 1566 ± 361 a | 1416 ± 57.0 a | 616 ± 246 b | 716 ± 144 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernandez, E.; Ostrowski, M.; Siboni, N.; Seymour, J.R.; Petrou, K. Uptake of Dimethylsulfoniopropionate (DMSP) by Natural Microbial Communities of the Great Barrier Reef (GBR), Australia. Microorganisms 2021, 9, 1891. https://doi.org/10.3390/microorganisms9091891
Fernandez E, Ostrowski M, Siboni N, Seymour JR, Petrou K. Uptake of Dimethylsulfoniopropionate (DMSP) by Natural Microbial Communities of the Great Barrier Reef (GBR), Australia. Microorganisms. 2021; 9(9):1891. https://doi.org/10.3390/microorganisms9091891
Chicago/Turabian StyleFernandez, Eva, Martin Ostrowski, Nachshon Siboni, Justin R. Seymour, and Katherina Petrou. 2021. "Uptake of Dimethylsulfoniopropionate (DMSP) by Natural Microbial Communities of the Great Barrier Reef (GBR), Australia" Microorganisms 9, no. 9: 1891. https://doi.org/10.3390/microorganisms9091891
APA StyleFernandez, E., Ostrowski, M., Siboni, N., Seymour, J. R., & Petrou, K. (2021). Uptake of Dimethylsulfoniopropionate (DMSP) by Natural Microbial Communities of the Great Barrier Reef (GBR), Australia. Microorganisms, 9(9), 1891. https://doi.org/10.3390/microorganisms9091891







