Abstract
Although interactions between microalgae and bacteria are observed in both natural environment and the laboratory, the modalities of coexistence of bacteria inside microalgae phycospheres in laboratory cultures are mostly unknown. Here, we focused on well-controlled cultures of the model green picoalga Ostreococcus tauri and the most abundant member of its phycosphere, Marinobacter algicola. The prevalence of M. algicola in O. tauri cultures raises questions about how this bacterium maintains itself under laboratory conditions in the microalga culture. The results showed that M. algicola did not promote O. tauri growth in the absence of vitamin B12 while M. algicola depended on O. tauri to grow in synthetic medium, most likely to obtain organic carbon sources provided by the microalgae. M. algicola grew on a range of lipids, including triacylglycerols that are known to be produced by O. tauri in culture during abiotic stress. Genomic screening revealed the absence of genes of two particular modes of quorum-sensing in Marinobacter genomes which refutes the idea that these bacterial communication systems operate in this genus. To date, the ‘opportunistic’ behaviour of M. algicola in the laboratory is limited to several phytoplanktonic species including Chlorophyta such as O. tauri. This would indicate a preferential occurrence of M. algicola in association with these specific microalgae under optimum laboratory conditions.
1. Introduction
Phytoplankton and bacterial dynamics are closely linked in coastal marine environments [1,2]. Numerous findings suggest that these specific relationships result from a long coexistence in the oceans for more than 200 million years [3,4,5]. Some bacterial strains are frequently isolated from natural blooms and cultivated in the laboratory [6,7,8]. Phytoplankton exudates can be important carbon substrates for bacteria, especially in early phytoplankton bloom conditions [9,10] although other carbon sources might also be important for bacterial growth [9]. In turn, it is clear now that heterotrophic bacteria can improve or inhibit microalgal growth [10,11,12,13]. Some bacteria provide marine algae with essential vitamins and nutrients while others compete with microalgae for nutrients or produce algicidal components [10], but very often the compounds involved in the partnership have not been fully characterised [11]. In addition, the coexistence of many bacteria in microalgae laboratory cultures is still not completely understood [12].
Ostreococcus is a widely distributed marine phytoplankton genus detected around the world [13] and contributes significantly to primary production in oligotrophic waters. Ostreococcus tauri was discovered in the Mediterranean Thau Lagoon in 1994 and was described as the smallest free-living eukaryotic cell [14]. This species can be observed as a dominant species during phytoplanktonic blooms in coastal seas [15]. O. tauri picoplanktonic cells (<2 µm) are auxotrophic for vitamin B1 (thiamine) and vitamin B12 (cobalamin). They depend on exogenous sources of vitamin or vitamin precursors for growth [16,17,18,19,20]. O. tauri possesses the metH gene coding for vitamin B12 methionine synthase but lacks the metE gene coding for B12-independent methionine synthase which allows growth in the absence of vitamin B12 [21]. It has been previously demonstrated that in the absence of vitamin B12, diatoms lacking the functional metE gene cannot survive [22]. In B12 depleted environments, microalgae which lack methionine synthase activity would need available sources of B12 produced exclusively by prokaryotes in the environment [23]. The biosynthetic pathway of vitamin B12, a complex tetrapyrrole, appears to be confined to prokaryotes [23]. It is intriguing that many oligotrophic species such as O. tauri are nonetheless vitamin auxotrophs [20,24] and so must be able to obtain a ready source of these organic micronutrients from their surroundings. Specific biotic interactions between algae and bacteria have been shown to take place for B12 acquisition to the algae from the bacteria [18,19] that in exchange benefit from algal exudates [18,22]. Recent studies have demonstrated that heterotrophic bacteria can satisfy microalgal requirements for B12 via mutualistic interactions [6,18]. It has been shown recently that cross-exchange of B-vitamins underpins a mutualistic interaction between O. tauri and the alpha-proteobacterium Dinoroseobacter shiba [20]. To survive in oligotrophic environments depleted in cobalamin, we expect that O. tauri would take up cobalamin produced by bacteria.
The bacterial diversity of the phycosphere of the green algae Ostreococcus tauri RCC4221 cultivated in the laboratory revealed a large dominance of M. algicola [25], hereafter noticed as M. algicola OT. The M. algicola-type strain was isolated from a dinoflagellate culture and was then frequently detected in dinoflagellate and coccolithophorid cultures [26,27]. Bacteria from the Marinobacter genus are found in numerous marine habitats including the deep ocean, costal seawater, sediments and sea-ice, and in associated animal or algal hosts. Many Marinobacter strains can adopt two life-styles, living planktonically or as biofilms [28]. Marinobacter species are frequently present in diverse microalgae phycospheres in laboratory cultures. Strains of M. adhaerens are frequently detected in diatom cultures [26,27,29,30]. They interact with diatom cells and are involved in the aggregation of microalgae [27,31]. An important feature is the high prevalence of M. algicola OT in O. tauri cultures [26] and it raises the question of how these bacteria could communicate to rapidly grow and spread in their environment. Relatedly, colonisation of the phycosphere by bacterial populations can occur through quorum sensing (QS) [32]. QS is a widespread cell to cell bacterial communication method that regulates and synchronises numerous bacterial activities through the diffusion of autoinducer (AI) molecules [33]. However, to date, only one study investigating QS in Marinobacter has been previously published [34].
In this work, we explored several hypotheses to investigate whether any positive, neutral or negative interaction occurred between the algae O. tauri RCC4221 and the bacterial strain M. algicola OT. Since O. tauri was auxotrophic for vitamin B12, the possibility that M. algicola provided the vitamin to the algae was tested. After M. algicola OT was isolated from a culture of O. tauri RCC4221 and maintained under laboratory conditions, coculture experiments with inoculation of different M. algicola OT concentrations were conducted to check whether O. tauri RCC4221 needs M. algicola OT for its growth under different conditions. We combined the searches in genomes for genes coding for proteins possibly involved in such interactions with experiments aimed at supporting the genomic data. Then, given the ability of Marinobacter species to degrade lipids [31] and the diversity of fatty acids and triacylglycerols (TAGs) produced by O. tauri [35], the capacity of M. algicola OT to use lipids for growth was tested. We again developed a double approach to combine bioinformatics screening of Marinobacter genomes and experiments on M. algicola OT to find insights of QS capacity in Marinobacter. Lastly, a phylogenetic reconstruction including the M. algicola strain from the O. tauri RCC4221 culture and all available Marinobacter 16S sequences was first performed to detect whether there was any evidence of a common evolutionary trend between Marinobacter strains found in microalgae cultures and if they share similar phylogenetic traits.
2. Material and Methods
2.1. Phylogenetic Analysis
To comment on the phylogenetic position of our M. algicola OT strain with respect to the other strains, a molecular phylogeny of 16S rRNA gene sequences was performed using sequences from different Marinobacter species. The dataset comprises 56 nucleotide sequences from GenBank including the M. algicola OT sequence (MT023716), for a total of 1415 positions after multiple alignment using the ClustalW algorithm in MEGA X version 10.1.7 software (available at https://www.megasoftware.net/, accessed on 9 August 2021) [36]. A tree was constructed using a GTR + I (invariable sites proportion) + G (discrete gamma distribution) model selected using the ‘Find Best DNA Models’ option in MEGA X version 10.1.7 software (available at https://www.megasoftware.net/, accessed on 9 August 2021) [36] and the maximum likelihood (ML) method using 100 bootstrap replicates (BP). All accession numbers are indicated in the tree (Figure 1). Particular attention was focused on the M. algicola clade with a comparison of sequences and the identification of specifically shared nucleic acid characteristics.

Figure 1.
Phylogenetic and morphological features of M. algicola OT MT023716. (A) ML tree from a 1415-position 16S alignment analysed with GTR + G + I model and 100 bootstrap replicates. These 56 nucleotide sequences constitute the totality of Marinobacter species. The M. algicola OT MT023716 sequence (black arrow) obtained from the O. tauri RCC4221 culture (in the ‘green microalgae, dinoflagellate and haptophyte-associated M. algicola clade’), the sequence from the unique Marinobacter strain previously tested for AI production (in the ‘diatom-associated Marinobacter clade’) and the OTB1 sequence from a Marinobacter strain previously observed in another O. tauri strain (RCC745) (grey arrow) are shown in bold. Branch lengths are proportional to the number of substitutions per site. (B) Partial alignment showing position 762 with the T nucleotide shared by sequences from only the M. algicola clade. (C) Transmission electron micrograph of O. tauri culture supernatant cells showing O. tauri cells (large cells approximately 0.8 µm in size) and rod-shaped bacteria (black arrows) with size characteristics corresponding to M. algicola DG893 [26]. Black arrows show bacterial cells in different plane slices (probably M. algicola).
2.2. Transmission Electron Microscopy
We used transmission electron microscopy as routinely performed in the laboratory [37], to check the possibility of a close physical interaction between microalgae and bacteria. A pellet of five hundred million microalgae cells in exponential growth phase was prefixed in L1 culture medium with glutaraldehyde 1% (#16320, Electron Microscopy Sciences, Hatfield, PA, USA) for 30 min at room temperature. Then, the cells were centrifuged at 2500× g (Beckman Avanti™ centrifuge J-20XP, Beckman Coulter, Fullerton, CA, USA) for 25 min at 8 °C. Pellets were then resuspended in 40 µL of 1% low melting point agar (#A9414, Sigma, St. Louis, MO, USA) in L1 culture medium. After solidification, the plug was fixed for 2 h at 4 °C with 2.5% glutaraldehyde in one volume of 0.4 M sodium cacodylate buffer (#12300, Electron Microscopy Sciences, Hatfield, PA, USA) and two volumes of culture medium. The plug was then washed three times for 15 min in 0.2 M sodium cacodylate buffer, postfixed in 2% osmium tetroxide (#19150, Electron Microscopy Sciences, Hatfield, PA, USA) for 1 h and finally washed three times again as previously described. Samples were dehydrated in successive baths with increasing ethanol concentrations (70%, 95% and 100%) and propylene oxide (#O4332, Fisher Scientifics, Hampton, NH, USA). Samples were progressively impregnated and embedded in Epon 812 (#14120, Electron Microscopy Sciences, Hatfield, PA, USA). Thin sections were prepared with an ultramicrotome (Leica Ultracut R, Leica Biosystems, Nussloch, Germany) and stained with 2% uranyl acetate in 50% ethanol (ETOH) solution and 0.5% lead citrate in aqueous solution before examination on a transmission electron microscope (Hitachi H-7500, Hitachi, Tokyo, Japan).
2.3. O. tauri RCC4221 and M. algicola OT Cultures and Growth
2.3.1. O. tauri RCC4221 Culture
Ostreococcus tauri strain RCC4221 was isolated in 1994 from the North-West MediterraneanThau lagoon [14] and maintained in the laboratory (cultures and cryopreservation). The O. tauri RCC4221 strain was grown in liquid medium in 50 mL aerated flasks (Sarstedt) at 20 °C under white light at approximately 100 µmol photons·m−2·s−1 using an LD 12:12 photoperiod. Cell growth was followed by measuring the cell concentrations in small aliquots (50 µL) of culture every two or three days with a flow cytometer (CytoFlex, Beckman Coulter, Fullerton, CA, USA). O. tauri cells were detected using the red fluorescence emission (FL3 acquisition at 670 nm) of chlorophyll pigments.
2.3.2. M. algicola OT Culture
The M. algicola OT strain, used for the experiments in this study, is the same that the one initially isolated from O. tauri RCC4221 in 2016 [25]. More precisely, numerous M. algicola OTUs with partial 16S sequence (obtained by Illumina sequencing) were evidenced in this previous study [25]. After cultivation of the O. tauri RCC4221 phycosphere in solid medium, only one M. algicola strain could be successfully isolated, the M. algicola OT strain used in this study. Cell growth was followed either by optical density (OD) or by measuring the cell concentrations with a flow cytometer as above. In this latter case, for enumeration of bacteria, small aliquots (50 µL) of culture were used for measurement by flow cytometry every two or three days. Bacterial nucleic acids were labelled with SYBR® Green I (#50513, Lonza, Basel, Switzerland) and detected by green fluorescence (FL1 acquisition at 530 nm) [38].
2.3.3. Culture Media
For all O. tauri cultures and coculture experiments with M. algicola OT hereafter, ESAW-F/2 culture media [39] were prepared with artificial seawater and supplemented with nitrogen (NaNO3, 8.8 × 10−5 mol·L−1), phosphorus (NaH2PO4, 3.62 × 10−5 mol·L−1), vitamins (B1, 2.96 × 10−7 mol·L−1, B12, 1.48 × 10−9 mol·L−1 and H, 4.09 × 10−9 mol·L−1) and trace metals. A culture medium without vitamin B12 was also prepared for B12 depletion experiments. In addition, one growth experiment of M. algicola OT monoculture (positive control) was performed using a highly enriched medium, the marine broth (MB 2216) medium, typically used for bacteria growth.
2.4. O. tauri RCC4221 and M. algicola OT Coculture Experiments
O. tauri RCC4221 cultures used for coculture experiments were not axenic despite the use of antibiotic treatments. At the lowest possible bacterial contamination (approximately 3%) at the stationary phase, the O. tauri RCC4221 culture was diluted to 106 cells·mL−1 in ESAW-F/2 medium and distributed into several 50 mL sterile flasks. To test the effects of M. algicola OT on O. tauri RCC4221, different final bacterial concentrations (5 × 106 and 2 × 107 M. algicola OT cells·mL−1) were added to the O. tauri culture RCC4221. A control (without the addition of M. algicola OT to the O. tauri RCC4221 culture) allowed us to visualise the growth pattern of the few remaining bacteria (fewer than 3 × 105 cells·mL−1) after axenisation was not totally efficient. M. algicola OT cells were distinguished easily from other bacterial communities in O. tauri RCC4221 cultures owing to the fluorescence signal for M. algicola OT after SYBR Green labelling being lower than those for other bacteria. The same experiment was carried out in a medium without vitamin B12. As before, different final concentrations (5 × 106 and 2 × 107 M. algicola OT cells·mL−1) were added to the O. tauri culture. All coculture experiments were carried out in triplicates experiments and were performed for a total duration of 30 days.
2.5. M. algicola OT Growth Tests on Lipids
M. algicola OT (MT023716) was tested for its capacity to grow on palmitic acid (16:0), tricaprylin (TAG 8:0/8:0), tripalmitin (TAG 16:0/16:0/16:0), triolein (TAG 18:1/18:1/18:1), triarachidin (TAG 20:0/20:0/20:0) and hexadecyl palmitate (wax ester). Lipids were from Tokyo Chemical Industry Co. LTD, Tokyo, Japan. The strain was grown in marine broth (MB) overnight at 30 °C and resuspended at an optical density at 600 nm (OD600nm) of 0.1 in MB minimum [40]. Substrates were added at 0.1% (w/v) to the cell suspension in glass tubes and incubated at 30 °C under agitation at 150 rpm. Growth was monitored at 20, 44, 68 and 164 h by reading the OD600nm.
2.6. In Silico Genomic Screening of O. tauri Phycosphere Sequences and Complete Marinobacter Genomes
Whole-genome sequencing data of 19 O. tauri mutation accumulation line sets were used for genomic screening. These 19 O. tauri non-axenic culture lines (containing O. tauri and all bacterial communities from its phycosphere) were the result of a previous extinction-dilution experiment of 378 days, which corresponded to approximately 512 generations per line with 27 points of extinction dilution to one cell [41]. These lines were shown to exclusively exhibit M. algicola cultivable species at the end point [25]. To detect whether the genomes of the remaining bacteria present in the 19 non-axenic O. tauri line cultures possessed genes of interest, we aligned the raw reads against the target gene sequences (detailed in the following sections) using Burrows-Wheeler Aligner (BWA, http://bio-bwa.sourceforge.net, accessed on 26 July 2018) with standard parameters [42] and treated the resulting bam files with SAMtools (http://www.htslib.org, accessed on 26 July 2018) to obtain the coverage of these genes [43].
We performed BLASTP of translated sequences of interest against the reference proteomes of the 21 available and complete reference genomes and proteomes of Marinobacter species, including the M. algicola DG893 strain, the closest strain to date to M. algicola OT, which were also used for screening (all accession numbers are noticed in Table 2).
2.6.1. Searching for Methionine Synthases Genes
We searched for evidence of methionine synthesis by M. algicola by screening the different genomic datasets. We focused on the metH methionine synthase gene (locus_tag = ‘MDG893_RS02235’, NZ_ABCP01000002) and the metE methionine synthase gene (locus_tag = ‘MDG893_RS14865’, NZ_ABCP01000027) identified from the M. algicola DG893 annotated genome (PRJNA19321).
2.6.2. Searching for Lipid Degradation Genes
We searched evidence for lipid degradation genes. We focused on 2 genes already identified as important for lipid degradation [31]: fadB (Fatty acid degradation alpha subunit, TIGR02437) and fadA (3-ketoacyl-CoA thiolase, TIGR02445) from the fatty oxidation complex.
2.6.3. Searching for Quorum-Sensing Genes
We sought evidence of QS-associated genes in the different databases. We focused on the reference QS genes from Vibrio species including luxI (AY275714) from V. fischeri [44], luxR (DQ108980) from V. harveyi [45], luxS (AY391122) from V. alginolyticus, luxM (DQ987706) from V. splendidus [46], vanM (KT258634) and ainS (AY277634) from V. fischeri.
2.7. In Silico Protein Analysis
In addition to genomic analysis, the presence of potential AI synthase and LuxR-like receptors in the species M. algicola was examined in the InterPro Database v69.0 (https://www.ebi.ac.uk/interpro/, accessed on 9 August 2021) [47] with the queries ‘IPR001690—autoinducer synthase’, ‘IPR000792—Transcription regulator LuxR, C-terminal’ and ‘IPR005143 Transcription factor LuxR-like, autoinducer-binding domain’. Once protein sequences were retrieved, protein domains were then identified using SMART 7 software (online resource, http://smart.embl.de/, accessed on 9 August 2021) [48] and aligned using the ClustalW algorithm under MEGA X version 10.1.7 software (available at https://www.megasoftware.net/, accessed on 9 August 2021) [36] with Vibrio fischeri LuxR and Agrobacterium TraR sequences to check for potential conserved amino-acids. A prediction of ligand binding on LuxR proteins was performed to test whether the identified LuxR proteins of M. algicola could effectively be involved in QS. The ligands were identified using COACH-D software (online resource, https://yanglab.nankai.edu.cn/COACH-D/, accessed on 9 August 2021) available in the I-TASSER version 5.1 package (online resource, https://zhanglab.dcmb.med.umich.edu/I-TASSER/download/, accessed on 9 August 2021) [49].
2.8. Evaluation of M. algicola OT Quorum-Sensing Capacities
We investigated in this study AI-1 (AHL-driven) and AI-2 (Furanosyl diester borate-driven) modes of QS. Functional experiments with different types of biosensors were conducted to evaluate the QS capacities of M. algicola OT through AI-1 and AI-2 production, using protocols based on previously published research conducted in our group [50]. Briefly, M. algicola OT culture supernatants were tested using Pseudomonas putida and Escherichia coli-based biosensors. P. putida F117 (pRK-C12; Kmr; ppuI:mpt) was used to detect long-chain (>8 carbons in the acyl side chain) N-acyl homoserine lactones (AHLs) [51,52,53], and E. coli MT102 (pJBA132) was used to evaluate the emission of short-chain (≤8 carbons in the acyl side chain) AHLs [53,54]. A mutant strain of Vibrio harveyi, BB170, was cultivated to detect the presence of AI-2 in the culture supernatants [55,56]. Protocols are precisely described in Text S1 (Supplementary Materials).
3. Results
3.1. Phylogenetic Position of M. algicola OT in the Global Marinobacter Species Tree
A phylogenetic reconstruction was performed to more precisely describe the M. algicola OT phylogenetic position and to determine if all Marinobacter strains found in phytoplankton cultures tend to form a cluster. M. algicola OT, isolated from a culture of O. tauri RCC4221 [25], belongs to the M. algicola phylogenetic group (BP = 99) (black arrow, Figure 1A). This clade also includes M. algicola strains from the dinoflagellates Alexandrium AJ294359, Gymnodinium AY258116, AY258110 and Lingulodinium DQ486497, the chlorophytes Ostreococcus tauri and Dunaliella KJ573108 and the haptophytes Emiliana EF140750 and Coccolithus EF140753. All sequences from this clade share a diagnostic nucleotide: a T is present at position 762 in all sequences (Figure 1B) while in all other sequences, a C is present at this position. While all sequences belonging to this clade share 97.4% sequence identity (1378 identical nucleotide sites over the 1415 base pairs in the total alignment), it is not possible to state if M. algicola strains found in dinoflagellate cultures are more closely related to each other than to the other strains given the relatively low variability level inside the clade. Transmission electron microscopy observations of O. tauri RCC4221 cultures revealed numerous bacterial cells that exhibited a morphology similar to M. algicola DG893 (Figure 1C) [56]. The bacterial cells and the microalgae did not physically interact (Figure 1C).
3.2. Phylogenetic Features of M. algicola Relative to Other Marinobacter Species Found in Phytoplankton Cultures
The closest phylogenetically related Marinobacter species to M. algicola is M. salsuginis which appeared as a sister group of M. algicola (BP = 47, considered weak support, Figure 1A) with two diagnostic characters: A at position 618 (instead of the G in other sequences) and T at position 688 (instead of the C in other sequences) (personnal observation). Other Marinobacter strains observed in phytoplanktonic cultures, such as M. adhaerens known to be associated with diatoms, do not belong to the M. algicola and M. salsuginis clades (Figure 1A). Marinobacter sp. OTB1 (grey arrow, Figure 1A), did not cluster in the M. algicola clade but appeared to be related to a more distant lineage (M. alkaliphilus). The 16S sequences from Marinobacter sp. OTB1 and M. algicola OT strains exhibited 44 differences over the 829 homologous sites, representing 5.3% of the differences over these partial 16S sequences, which made these two strains evolutionarily distant from each other.
3.3. Effect of M. algicola OT on O. tauri RCC4221 Growth in the Absence of Vitamin B12
Coculture experiments were compared after inoculation of M. algicola OT at different concentrations and in presence and absence of vitamin B12. The absence of vitamin B12 in the culture medium affected the growth curve of O. tauri RCC4221. In the presence of vitamin B12 (complete ESAW-F/2 medium), the maximal cell abundance was 2.5-fold higher than that in the vitamin B12-depleted medium. Moreover, the decay phase was much faster in the absence of vitamin B12 (Figure 2). Interestingly, when M. algicola OT was added to the cultures (at 5 × 106 and 2 × 107 cells·mL−1 in ESAW-F/2 complete medium), O. tauri was maintained at the highest density from day 26 to the end of the experiment (Figure 2). However, the presence of M. algicola OT in the culture of O. tauri RCC4221 in vitamin B12-depleted medium did not restore the growth pattern of the algae observed in the presence of vitamin B12. As an evidence of the bacterial capacity to synthesise methionine (if genes are transcribed and translated), we retrieved that metH and metE methionine synthase genes from several O. tauri mutation accumulation lines containing reads from bacterial communities from its phycosphere (Table 1) and also from in the genome of M. algicola DG893, the closest sequenced strain to M. algicola OT (Table 2).
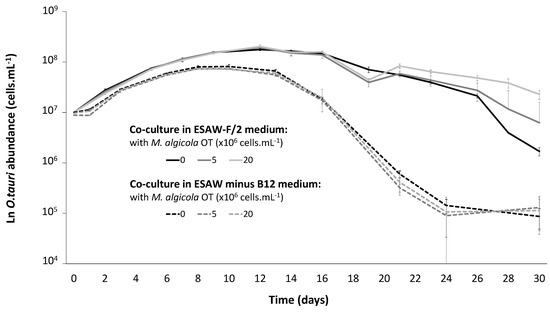
Figure 2.
O. tauri RCC4221 growth in coculture with M. algicola OT MT023716. Cocultures were performed in complete ESAW- F/2 medium (solid curves) and in ESAW medium depleted in vitamin B12 (ESAW minus B12) medium (dotted curves).

Table 1.
BWA alignment results of vitamin B12 synthase genes with raw genomics reads from 19 O. tauri (and its phycosphere) line cultures. From these 19 independent lines, which were endpoints diluted through serial subcultures [41], all the cultivable bacteria in solid media were determined to be M. algicola OT MT023716 [25].

Table 2.
Screening of methionine synthase genes and lipid degradation genes in 21 Marinobacter complete genomes (accession numbers in the NCBI). Vitamin B12 synthase genes are methionine synthase genes (metH and metE). Lipid degradation genes are fatty acid degradation alpha subunit genes (fadB) and 3-ketoacyl-CoA thiolase genes (fadA).
3.4. M. algicola OT Growth Behaviour in Coculture with O. tauri RCC4221
A bacterial monoculture experiment revealed that M. algicola OT did not grow in ESAW-F/2 medium contrary to what was observed in the positive control culture in MB medium (Figure 3A). However, M. algicola OT could grow in ESAW vitamin B12-depleted medium in coculture with O. tauri RCC4221 (ESAW minus B12, Figure 3B). M. algicola OT started to grow at day 8 and reached its maximal cell abundance at day 16, while O. tauri RCC4221 growth began at day 3 to reach maximal cell abundance at day 8 and then declined until complete cell disappearance at day 21 (Figure 3B). The M. algicola exponential growth phase was strongly delayed compared to the O. tauri one (10 days for M. algicola OT versus 1 day for O. tauri, Figure 3B). The two species exhibited shifted growth curves.
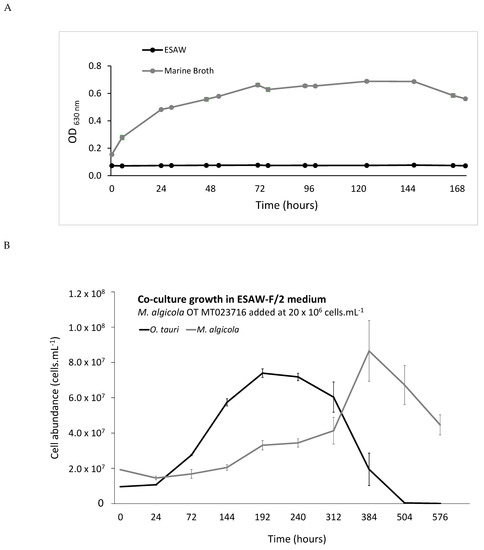
Figure 3.
M. algicola OT MT023716 growth under different experimental conditions. (A) M. algicola OT MT023716 monoculture growth in ESAW-F/2 and marine broth media; (B) O. tauri RCC4221 and M. algicola OT MT023716 coculture growth in ESAW minus vitamin B12, showing that a sufficient number of O. tauri cells in stationary phase induces M. algicola OT MT023716 growth.
3.5. M. algicola OT Growth on Lipids
For each compound tested, the M. algicola OT growth (until OD = 1.34 for the highest, Figure 4) was obtained at four different tested times, ranging from 20 to 164 h (Figure 4). Figure 4 shows that M. algicola OT was able to grow on four TAGs (tricaprylin, tripalmitin, triolein and triarachidin), a wax ester (hexadecyl palmitate) and a fatty acid (palmitic acid). These results indicated that M. algicola OT was able to assimilate the tested lipids. However, it was not possible to draw any conclusions from these data regarding the efficiency of the assimilation of the different lipids as they were nearly insoluble in water and their assimilation rate depended on the area of the water-lipid interface. The ability of M. algicola OT to assimilate a variety of lipids was also suggested by the occurrence of lipids and degradation genes in the M. algicola DG893 genome reference (if transcribed and translated). The two genes fadA and fadB target genes coding for proteins involved in lipid degradation, were found with relatively high coverage in genomics data of O. tauri phycosphere cultures (Table 3). Complete screening of available Marinobacter genomes also revealed that these two genes were retrieved in the M. algicola DB893 genome reference (Table 2) (with p-values = 0 after BLASTP searching). The occurrence of fadB and fadA indicates the probable presence of a fatty acid oxidation pathway in M. algicola DG893, and thus probably in M. algicola OT.
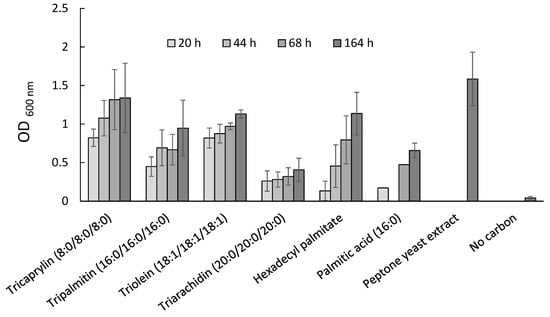
Figure 4.
M. algicola OT MT023716 growth on lipids and other sources.

Table 3.
BWA alignment results of lipid degradation genes with raw genomics reads from 19 O. tauri (and its phycosphere) line cultures. From these 19 independent lines, which were endpoints diluted through serial subcultures [41], all the cultivable bacteria in solid media were determined to be M. algicola OT MT023716 [25]. Here, the most abundant lipid degradation genes found from the genomic screening of the culture data are presented.
3.6. Attempts to Detect AI-1 or AI-2 from M. algicola OT by Experimentation or Genomic Screening
We evaluated the production of AHLs by M. algicola OT using sensor strains. The M. algicola OT strain did not produce type 1 (Figure S1A) or type 2 (Figure S1B) auto-inducers. However, genes coding for proteins involved in QS, mainly luxI, luxS and hdtS, were present with relatively high coverage in O. tauri phycosphere cultures. These genes belong to taxonomically distinct bacteria (Table S1) and were all present in O. tauri RCC4221 culture at very low abundance rate [25]. However, complete screening of available Marinobacter genomes did not identify any homologues of the luxI, luxS, luxM, vanM, ainS or hdtS QS genes involved in AI synthesis. Nevertheless, LuxR receptor homologues were retrieved in 13 out of 21 Marinobacter genomes, including M. algicola DG893 (Table S2). INTERPRO and SMART analysis of the Marinobacter LuxR protein sequences confirmed that all of them harbour the typical LuxR DNA-binding helix-turn-helix (HTH) domain at their C-terminal region. In the N-terminal region, eight sequences presented a signal transduction response regulator domain (IPR001789), three presented a Malt-like TPR region (IPR041617) and two presented N terminal domains that were not affiliated with any known type of protein domain. However, none of them presented the AI-binding domain (IPR005143), which was confirmed by a COACH-based analysis and alignment of LuxR sequences with the Vibrio fischeri and Agrobacterium LuxR sequences (personnal observation).
4. Discussion
4.1. Contrasting Behaviours of Marinobacter Strains in Microalgae Cultures
Bacterial growth can be substantially favoured because of organic nutrients in the surrounding environment. During blooms in Mediterranean coast, Trombetta and collaborators noticed dominant positive relationships between bacteria and phytoplankton [7] and it is known that bacterial cells can satisfy part of their own carbon demand by consuming phytoplankton exudates [57,58]. In coculture with O. tauri, organic matter quantity strongly depends on the growth stage of the microalgae, being particularly low until the middle of the exponential phase and high once the O. tauri cells reach the stationary phase. From our results, we hypothesise that bacteria largely benefit from O. tauri dead cells directly or from the exudates of accumulating living cells during exponential and decline phases. The behaviours of O. tauri RCC4221 and M. algicola in coculture contrast. Ostreococcus tauri is a known B12 auxotroph [21]. The maximal cell abundance of O. tauri was 2.5-fold lower in the ESAW medium without vitamin B12 than in the ESAW-F/2 complete medium in the control cultures, thus confirming that O. tauri RCC4221 needs vitamin B12 for optimal growth. The presence of vitamin B12 biosynthetic genes in many Marinobacter species suggested that M. algicola OT could provide vitamin B12 to O. tauri RCC4221. However, here, we did not demonstrate any impact of M. algicola OT in favour of O. tauri RCC4221 growth in coculture when the culture medium was depleted in vitamin B12. From our experiments, M. algicola OT could not provide vitamin B12 to O. tauri RCC4221. In contrast, the decline in O. tauri cells obviously matched the beginning of M. algicola OT growth, arguing that bacteria largely benefit from O. tauri (directly from dead cells or from the exudates of accumulating living cells) to sustain their growth. However, O. tauri was maintained at the highest density from day 21 to the end of the experiment when the initial density of M. algicola OT was twice that of the microalgae. This finding could indicate that O. tauri RCC4221 survived longer in the presence of the bacteria, probably benefiting from some decomposition of organic matter during the decline phase. Nevertheless, in a recent study, another Marinobacter behaviour, one that favoured O. tauri growth in culture, was evidenced [59]. In that study, the occurrence of a Marinobacter sp. OTB1 strain was reported in cultures of the O. tauri RCC745 strain, phylogenetically close to our O. tauri RCC4221 strain. The authors showed that Marinobacter sp. OTB1 favours the O. tauri RCC745 growth. They showed that in presence of Marinobacter sp. OTB1, the growth of O. tauri was stimulated following the addition of a precursor component in vitamin B1-limited cultures. In contrast, no growth stimulation was observed when antibiotics were added to prevent bacterial growth [59], therefore demonstrating the beneficial presence of bacteria for microalgae growth. In our case, even in the presence of vitamin B12, the presence of M. algicola OT did not enhance O. tauri RCC4221 growth in culture. It can be a little paradoxical that phylogenetically closed O. tauri strains (RCC4221 and RCC745) harbouring Marinobacter strains in their laboratory culture phycospheres, are not impacted the same way depending on the Marinobacter strain. In a previous study, the growth of two Prochlorococcus strains (MIT9313 and MED4) was differentially affected when they were cultivated with the same collection of heterotrophic bacteria [60]. The authors highlighted that the kind of interaction was dependent on the phylogenetic signature. From this point of view, closely related bacteria (whose 16S gene sequences differed by 1–2%) show more similar effects in cultures [60]. As pointed out previously from our phylogenetic reconstruction, M. algicola OT and Marinobacter OTB1 are two independent species that are not closely related, showing 5.3% differences in a 16S partial alignment (corresponding to the length of the 16S partial sequence of Marinobacter OTB1), and thus probably exhibit different metabolisms in microalgae cultures. Last, in our phylogenetic reconstruction, the clustering of the green microalgae, dinoflagellate and haptophyte Marinobacter clade (M. algicola) and diatom-associated Marinobacter clade (M. adhaerens—M. manganoxydans—M. flavimaris) together with M. salsuginis provided a highly bootstrap support (BP = 83). Contrary to all Marinobacter species mentioned above, which include bacteria with neutral or positive effects on microalgae to our knowledge, the M. salsuginis BS2 strain is the only species that does not seem to exhibit positive behaviour towards microalgae. The M. salsuginis BS2 strain was discovered from environmental sediments only two years after M. algicola was discovered. Although it appears closely phylogenetically related to M. algicola in our phylogenetic reconstruction, the M. salsuginis BS2 is an algicidal bacterium showing killing activity towards the harmful bloom-forming dinoflagellate Noctiluca scintillans [10].
4.2. M. algicola OT Metabolises Different Lipids
The growth defect of M. algicola OT in ESAW-F/2 without O. tauri RCC4221 was most likely due to the absence of a carbon and energy source in this medium. O. tauri RCC4221 is known to produce a wide range of lipids including TAGs that represent approximately 15% of the total lipids under nutrient-replete conditions and that accumulate up to 10 times more in nitrogen-depleted conditions [35]. Many Marinobacter species are known to degrade a large variety of hydrophobic substrates, including TAGs, wax esters, fatty acids, fatty alcohols and alkanes [31,61]. The ability of M. algicola OT MT023716 to grow in the presence of O. tauri RCC4221 in ESAW-F/2 medium, while it did not grow alone, suggested that the bacterium benefited from some exudates of O. tauri. From our experiments, M. algicola OT grew in the presence of different lipids, including four different TAGs. We can hypothesise that M. algicola OT might take advantage of TAGs produced by O. tauri, which are some of its major lipids [35]. In M. algicola OT and O. tauri coculture experiments, the co-occurrence of the onsets of the growth of the bacterium with the stationary phase of the algae is consistent with a hypothetical release of lipids or other substrates by O. tauri during the stationary phase, possibly by cell lysis. Even if it does not reveal they are actively transcribed and translated, the presence of different genes involved in different fatty acid degradation pathways in M. algicola DG893 would be a strong indicator to corroborate this hypothesis. In addition, M. algicola OT might also degrade starch from the microalga if an alpha-amylase (glycosidase) present in the M. algicola DG893 genome (personal observation) was also actively synthesised. Further investigations will be needed to directly assess the growth of M. algicola OT on O. tauri lipidic extracts and determine which microalga lipids are particularly metabolised.
4.3. M. algicola OT Does Not Produce AI-1 or AI-2 QS Autoinducers
The capacity of Marinobacter species to communicate by QS should be an essential feature of the genus. M. hydrocarbonoclasticus, an oil-degrading bacterium, forms biofilms at the substrate–water interface [62], when carbon sources such as lipids are condensed in nonaqueous phases in marine waters. Producing biofilms is a particular bacterial activity that is commonly regulated by QS in numerous bacterial families. Our first global screening for genes coding for proteins involved in QS found many QS genes involved in AI synthesis in the global phycosphere of O. tauri RCC4221 laboratory cultures. However, M. algicola and all other Marinobacter species screened do not possess any of the classical QS genes. Contrary to what we expected given the high prevalence of M. algicola OT in O. tauri RCC4221 cultures, the M. algicola OT MT023716 strain did not produce either AI-1 or AI-2 according to our various detection tests. In contrast, a Marinobacter strain recovered from snow particles in a marine environment (Marinobacter sp. PCOB-2 HP36, AJ000647) produce AHLs [34]. In our phylogenetic reconstruction, Marinobacter sp. PCOB-2 was not related to the M. algicola clade but rather to the diatom-associated Marinobacter clade, which included M. adhaerens known to live closely attached to diatoms. However, our genomic screening found no evidence of the QS genes involved in AI-1 and AI-2 synthesis in any Marinobacter genome available, including M. adhaerens. Nevertheless, other types of QS modes are possible in Gram-negative bacteria, but their study requires complex and in-depth multidisciplinary studies that are out of the context of this current work. In addition, except in a study from Gram and coworkers [34], experimental evidence of QS capacity in any Marinobacter strain has never been demonstrated. False positives are regularly observed in this kind of experiment [63], and thus, we hypothesise that this could probably be the case in the Gram and coworkers study unless new experiments demonstrate otherwise. Meanwhile, the encoding of a Lux-R receptor (assuming it is correctly translated and functional) in the majority of the Marinobacter genomes available, including M. algicola DG893, suggests that M. algicola senses AI from other bacterial communities and benefits from them to synchronise its own activities. However, for the thirteen retrieved M. algicola LuxR protein sequences, in silico protein analyses revealed the lack of an AI-binding domain (and of other features such as conserved amino acids involved in AHL binding and the presence of InterPro domains) in the LuxR protein sequences, thus eliminating this kind of ‘cheater’ behaviour [64]. In our study, we more specifically focused on AI-1 (AHL-driven) and AI-2 (furanosyl diester borate-driven) modes of QS. However, other types of QS have been depicted in the literature [65]. As their study requires intense experimental work, we did not consider including them in the context of this study. Nevertheless, their inclusion in future research could provide interesting insights into Marinobacter physiology, especially in the context of microalgae-bacteria interactions [32].
4.4. M. algicola OT Is a Commensal Species in O. tauri Cultures and Is Widespread in the Natural Environment
It appears clear that the colonisation of different phycospheres by Marinobacter sp. strains under laboratory conditions not only occurs in dinoflagellate cultures in which it was initially described [56] but can be extended to coccolithophorid, diatom and Chlorophyta microalgae cultures, including cultures with O. tauri RCC4221. From our partial 18S phylogenetic reconstruction, these strains all cluster together. It seems that bacterial species that best behave throughout rapid host growth have been selected from the diversity of bacteria observed in phytoplanktonic cultures, as previously suggested [12]. We wonder if this kind of selection over generations is a general phenomenon that also extends to other microalgae cultures to result in M. algicola OT MT023716 predominance in O. tauri RCC4221 cultures. The M. algicola OT growth phase matched the decline in O. tauri cells, suggesting that the bacteria fed on the exudates or dead cells of the algae. A study by Trombetta and collaborators during bloom period in mediterranean coast [7] revealed evidence of positive interactions between phytoplankton and bacteria that suggest that vitamin-synthesising bacteria would provide vitamins to phytoplankton in exchange for organic carbon [19,66,67]. Our results show that without an adequate carbon source, M. algicola is difficult to cultivate and grows particularly well in phytoplanktonic cultures in which it probably finds additional nutrients provided by living and/or dead algal cells. Given the relatively low variability level inside the M. algicola clade, it is not possible to state if M. algicola strains found in dinoflagellate cultures are more closely related to each other than to the other M. algicola strains inside the clade, which would indicate whether the bacteria are specific to some kinds of microalgae. In addition, a recent study reported the occurrence of the Marinobacter sp. OTB1 strain [59] in cultures of O. tauri RCC745. Another recent study suggests that algal-associated bacterial communities are controlled by algal hosts, culture conditions and the initial inoculum composition of the microalgae phycosphere [68].
The range of lipid compounds allowing M. algicola growth that were highlighted in this study could be the key elements explaining the occurrence of this bacterial species in so many different microalgae laboratory cultures. Interestingly, the majority of Marinobacter strains found in diverse Chlorophyta, Dinoflagellates and Haptophyte cultures are all in the M. algicola clade in our phylogenetic reconstruction, demonstrating a particular acclimation of this opportunistic species in different phycospheres in laboratory cultures. In the presence of organic carbon, Marinobacter can grow rapidly, out-competing other bacteria in enrichment cultures [69] and easily dominates other communities in natural marine aggregates [70]. This r-strategist (or copiotrophic) behaviour renders them very easy to cultivate, even compared with other heterotrophic marine bacteria [71]. Marinobacter species probably use other kinds of communication system to optimise their growth, and these bacteria might form biofilms, a central mechanism in the natural environment [72], to colonise any substrates that are not well dissolved [61], including phytoplanktonic blooms. Preliminary exploration of public datasets from Tara Oceans reveals probable colocalisation of M. algicola and O. tauri in coastal environments. It is highly probable that O. tauri, as a bloom-forming phytoplanktonic species, can be an important nutritive interface for numerous bacterial strains in the natural environment.
5. Conclusions
The persistence of M. algicola OT MT023716 bacteria in O. tauri RCC4221 cultures would result from opportunistic behaviour. Our experiments suggest that the M. algicola OT MT023716 strain needs O. tauri microalgae to grow in synthetic medium. Although neither genomic evidence nor experimental work argued in favour of QS activity by M. algicola OT MT023716, the strain grows at high density on a wide range of lipids, including triglycerides, known to be produced by O. tauri and numerous marine microalgae in culture during abiotic stress. Marinobacter algicola strains are frequently found in different microalgae cultures in the laboratory, which reveals their plasticity in colonising different laboratory culture phycospheres. Understanding why this colonisation is limited to certain phytoplanktonic species in the laboratory will bring new insights into the optimum activity of M. algicola in cultures, and perhaps also in marine environments.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms9081777/s1, Figure S1: No AI-1 (Figure S1A) or AI-2 (Figure S1B) production by M. algicola OT, Table S1: BWA alignment results of reference QS genes with raw genomics reads from 19 O. tauri (and its phycosphere) line cultures. Table S2: Quorum-sensing genes screening in 21 Marinobacter complete genomes (accession numbers in the NCBI). Text S1: Material and Methods for the evaluation of M. algicola OT quorum-sensing capacities.
Author Contributions
Conceptualization, S.S.-B.; methodology, J.P., R.L., M.K., J.L., R.G., L.U., M.-L.E., F.S. and S.S.-B.; software, J.P., M.K. and S.S.-B.; validation, R.L., M.K., J.L., R.G.; formal analysis, R.L., M.K., J.L., R.G. and S.S.-B.; investigation, J.P., R.L., M.K., J.L., R.G., L.U., M.-L.E. and S.S.-B.; resources, M.K., L.I., N.G. and G.P.; data curation, S.S.-B.; writing—original draft preparation, S.S.-B.; writing—review and editing, R.L., R.G., J.L., M.K., L.U., M.-L.E. and S.S.-B.; visualization, S.S.-B.; supervision, S.S.-B.; project administration, S.S.-B.; funding acquisition, S.S.-B., N.G. and G.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the PHYTNESS project ‘ANR-13-JSV6-0005’ and supported by the federal action ‘Biotic interactions’ of the Banyuls-sur-Mer Observatory (OOB).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
This work benefited from access to the BioPIC platform (Biology Platform of Imaging and flow Cytometry, OOB), an EMBRC-France and EMBRC-ERIC Site.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Fuhrman, J.; Ammerman, J.; Azam, F. Bacterioplankton in the coastal euphotic zone: Distribution, activity, and possible rela-tionships with phytoplankton. Mar Biol. 1980, 60, 201–207. [Google Scholar] [CrossRef]
- Rooney-Varga, J.; Giewat, M.; Savin, M.; Sood, S.; LeGresley, M.; Martin, J. Links between Phytoplankton and Bacterial Community Dynamics in a Coastal Marine Environment. Microb. Ecol. 2005, 49, 163–175. [Google Scholar] [CrossRef]
- Buchan, A.; LeCleir, G.R.; Gulvik, C.A.; Gonzalez, J. Master recyclers: Features and functions of bacteria associated with phytoplankton blooms. Nat. Rev. Genet. 2014, 12, 686–698. [Google Scholar] [CrossRef]
- Luo, H.; Csűros, M.; Hughes, A.L.; Moran, M.A. Evolution of Divergent Life History Strategies in Marine Alphaproteobacteria. mBio 2013, 4, e00373-13. [Google Scholar] [CrossRef] [Green Version]
- Grossart, H.-P.; Levold, F.; Allgaier, M.; Simon, M.; Brinkhoff, T. Marine diatom species harbour distinct bacterial communities. Environ. Microbiol. 2005, 7, 860–873. [Google Scholar] [CrossRef] [PubMed]
- Wagner-Döbler, I.; Ballhausen, B.; Berger, M.; Brinkhoff, T.; Buchholz, I.; Bunk, B.; Cypionka, H.; Daniel, R.; Drepper, T.; Gerdts, G.; et al. The complete genome sequence of the algal symbiont Dinoroseobacter shibae: A hitchhiker’s guide to life in the sea. ISME J. 2009, 4, 61–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trombetta, T.; Vidussi, F.; Roques, C.; Scotti, M.; Mostajir, B. Marine Microbial Food Web Networks During Phytoplankton Bloom and Non-bloom Periods: Warming Favors Smaller Organism Interactions and Intensifies Trophic Cascade. Front. Microbiol. 2020, 11, 502336. [Google Scholar] [CrossRef]
- Fouilland, E.; Tolosa, I.; Bonnet, D.; Bouvier, C.; Bouvier, T.; Bouvy, M.; Got, P.; Le Floc’H, E.; Mostajir, B.; Roques, C.; et al. Bacterial carbon dependence on freshly produced phytoplankton exudates under different nutrient availability and grazing pressure conditions in coastal marine waters. FEMS Microbiol. Ecol. 2013, 87, 757–769. [Google Scholar] [CrossRef] [Green Version]
- Fouilland, E.; Mostajir, B. Revisited phytoplanktonic carbon dependency of heterotrophic bacteria in freshwaters, transitional, coastal and oceanic waters. FEMS Microbiol. Ecol. 2010, 73, 419–429. [Google Scholar] [CrossRef] [Green Version]
- Keawtawee, T.; Fukami, K.; Songsangjinda, P. Use of a Noctiluca-killing bacterium Marinobacter salsuginis strain BS2 to reduce shrimp mortality caused by Noctiluca scintillans. Fish. Sci. 2012, 78, 641–646. [Google Scholar] [CrossRef]
- Sapp, M.; Schwaderer, A.S.; Wiltshire, K.; Hoppe, H.-G.; Gerdts, G.; Wichels, A. Species-Specific Bacterial Communities in the Phycosphere of Microalgae? Microb. Ecol. 2007, 53, 683–699. [Google Scholar] [CrossRef] [PubMed]
- Abby, S.; Touchon, M.; De Jode, A.; Grimsley, N.; Piganeau, G. Bacteria in Ostreococcus tauri cultures–friends, foes or hitchhikers? Front. Microbiol. 2014, 5, 505. [Google Scholar] [CrossRef] [PubMed]
- Countway, P.D.; Caron, D.A. Abundance and Distribution of Ostreococcus sp. in the San Pedro Channel, California, as Revealed by Quantitative PCR. Appl. Environ. Microbiol. 2006, 72, 2496–2506. [Google Scholar] [CrossRef] [Green Version]
- Courties, C.; Vaquer, A.; Troussellier, M.; Lautier, J.; Chrétiennot-Dinet, M.J.; Neveux, J.; Machado, C.; Claustre, H.; Chr, M.J. Smallest eukaryotic organism. Nat. Cell Biol. 1994, 370, 255. [Google Scholar] [CrossRef]
- O’Kelly, C.; Sieracki, M.; Their, A.; Ic, H. A transient bloom of Ostreococcus (Chlorophyta, Prasinophyceae) in West Neck Bay, Long Island New York. J. Phycol. 2003, 39, 850–854. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Simon, M. Interactions of planktonic algae and bacteria: Effects on algal growth and organic matter dynamics. Aquat. Microb. Ecol. 2007, 47, 163–176. [Google Scholar] [CrossRef] [Green Version]
- Worden, A.Z.; Lee, J.-H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green Evolution and Dynamic Adaptations Revealed by Genomes of the Marine Picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef] [Green Version]
- Croft, M.T.; Lawrence, A.D.; Raux-Deery, E.; Warren, M.; Smith, A. Algae acquire vitamin B12 through a symbiotic relationship with bacteria. Nat. Cell Biol. 2005, 438, 90–93. [Google Scholar] [CrossRef]
- Kazamia, E.; Czesnick, H.; Van Nguyen, T.T.; Croft, M.T.; Sherwood, E.; Sasso, S.; Hodson, S.J.; Warren, M.J.; Smith, A. Mutualistic interactions between vitamin B12-dependent algae and heterotrophic bacteria exhibit regulation. Environ. Microbiol. 2012, 14, 1466–1476. [Google Scholar] [CrossRef]
- Cooper, M.B.; Kazamia, E.; Helliwell, K.; Kudahl, U.J.; Sayer, A.; Wheeler, G.L.; Smith, A.G. Cross-exchange of B-vitamins underpins a mutualistic interaction between Ostreococcus tauri and Dinoroseobacter shibae. ISME J. 2019, 13, 334–345. [Google Scholar] [CrossRef] [Green Version]
- Helliwell, K.; Wheeler, G.L.; Leptos, K.; Goldstein, R.; Smith, A. Insights into the Evolution of Vitamin B12 Auxotrophy from Sequenced Algal Genomes. Mol. Biol. Evol. 2011, 28, 2921–2933. [Google Scholar] [CrossRef] [Green Version]
- Ellis, K.A.; Cohen, N.R.; Moreno, C.; Marchetti, A. Cobalamin-independent Methionine Synthase Distribution and Influence on Vitamin B12 Growth Requirements in Marine Diatoms. Protist 2017, 168, 32–47. [Google Scholar] [CrossRef]
- Warren, M.; Raux, E.; Schubert, H.; Escalante-Semerena, J. The biosynthesis of adenosylcobalamin (vitamin B12). Nat. Prod. Rep. 2002, 19, 390–412. [Google Scholar] [CrossRef] [PubMed]
- Green, D.; Hart, M.; Blackburn, S.; Bolch, C. Bacterial diversity of Gymnodinium catenatum and its relationship to dinoflagellate toxicity. Aquat. Microb. Ecol. 2010, 61, 73–87. [Google Scholar] [CrossRef]
- Lupette, J.; Lami, R.; Krasovec, M.; Grimsley, N.; Moreau, H.; Piganeau, G.; Sanchez-Ferandin, S. Marinobacter Dominates the Bacterial Community of the Ostreococcus tauri Phycosphere in Culture. Front. Microbiol. 2016, 7, 1414. [Google Scholar] [CrossRef] [Green Version]
- Green, D.H.; Echavarri-Bravo, V.; Brennan, D.; Hart, M.C. Bacterial diversity associated with the coccolithophorid algae Emiliania huxleyi and Coccolithus pelagicus f. braarudii. BioMed Res. Int. 2015, 2015, 194540. [Google Scholar] [PubMed] [Green Version]
- Kaeppel, E.C.; Gärdes, A.; Seebah, S.; Grossart, H.-P.; Ullrich, M.S. Marinobacter adhaerens sp. nov., isolated from marine aggregates formed with the diatom Thalassiosira weissflogii. Int. J. Syst. Evol. Microbiol. 2012, 62, 124–128. [Google Scholar] [CrossRef] [Green Version]
- Handley, K.M.; Lloyd, J.R. Biogeochemical implications of the ubiquitous colonization of marine habitats and redox gradients by Marinobacter species. Front. Microbiol. 2013, 4, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hold, G.; Smith, E.A.; Rappë, M.S.; Maas, E.W.; Moore, E.R.; Stroempl, C.; Stephen, J.R.; Prosser, J.I.; Birkbeck, T.; Gallacher, S. Characterisation of bacterial communities associated with toxic and non-toxic dinoflagellates: Alexandrium spp. and Scrippsiella trochoidea. FEMS Microbiol. Ecol. 2001, 37, 161–173. [Google Scholar] [CrossRef]
- Sonnenschein, E.C.; Syit, D.A.; Grossart, H.-P.; Ullrich, M.S. Chemotaxis of Marinobacter adhaerens and Its Impact on Attachment to the Diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 2012, 78, 6900–6907. [Google Scholar] [CrossRef] [Green Version]
- Bonin, P.; Vieira, C.; Grimaud, R.; Militon, C.; Cuny, P.; Lima, O.; Guasco, S.; Brussaard, C.P.D.; Michotey, V. Substrates specialization in lipid compounds and hydrocarbons of Marinobacter genus. Environ. Sci. Pollut. Res. 2015, 22, 15347–15359. [Google Scholar] [CrossRef] [PubMed]
- Rolland, J.L.; Stien, D.; Sanchez-Ferandin, S.; Lami, R. Quorum Sensing and Quorum Quenching in the Phycosphere of Phytoplankton: A Case of Chemical Interactions in Ecology. J. Chem. Ecol. 2016, 42, 1201–1211. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, H.B.; Greenberg, E.P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985, 163, 1210–1214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gram, L.; Grossart, H.; Schlingloff, A.; Kiørboe, T. Possible Quorum Sensing in Marine Snow Bacteria: Production of Acylated Homoserine Lactones by Roseobacter Strains Isolated from Marine Snow Possible Quorum Sensing in Marine Snow Bacteria: Production of Acylated Homoserine Lactones by Roseobacter Strai. Appl. Environ. Microbiol. 2002, 68, 4111–4116. [Google Scholar] [CrossRef] [Green Version]
- Degraeve-Guilbault, C.; Bréhélin, C.; Haslam, R.; Sayanova, O.; Marie-Luce, G.; Jouhet, J.; Corellou, F. Glycerolipid Characterization and Nutrient Deprivation-Associated Changes in the Green Picoalga Ostreococcus tauri. Plant Physiol. 2017, 173, 2060–2080. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Yau, S.; Krasovec, M.; Benites, L.F.; Rombauts, S.; Groussin, M.; Vancaester, E.; Aury, J.-M.; Derelle, E.; Desdevises, Y.; Escande, M.-L.; et al. Virus-host coexistence in phytoplankton through the genomic lens. Sci. Adv. 2020, 6, eaay2587. [Google Scholar] [CrossRef] [Green Version]
- Marie, D.; Partensky, F.; Jacquet, S.; Vaulot, D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid strain SYBRGreenI. Appl. Environ. Microbiol. 1997, 63, 186–193. [Google Scholar] [CrossRef] [Green Version]
- Harrison, P.J.; Waters, R.E.; Taylor, F.J.R. A broad spectrum artificial sea water medium for coastal and open ocean phytoplankton. J. Phycol. 1980, 16, 28–35. [Google Scholar] [CrossRef]
- Barnier, C.; Clerissi, C.; Lami, R.; Intertaglia, L.; Lebaron, P.; Grimaud, R.; Urios, L. Description of Palleronia rufa sp. nov., a biofilm-forming and AHL-producing Rhodobacteraceae, reclassification of Hwanghaeicola aestuarii as Palleronia aestuarii comb. nov., Maribius pontilimi as Palleronia pontilimi comb. nov., Maribius salinus as Palleronia salina comb. nov., Maribius pelagius as Palleronia pelagia comb. nov. and emended description of the genus Palleronia. Syst. Appl. Microbiol. 2020, 43, 126018. [Google Scholar] [CrossRef] [PubMed]
- Krasovec, M.; Eyre-Walker, A.; Grimsley, N.; Salmeron, C.; Pecqueur, D.; Piganeau, G.; Sanchez-Ferandin, S. Fitness Effects of Spontaneous Mutations in Picoeukaryotic Marine Green Algae. G3 Genes Genomes Genet. 2016, 6, 2063–2071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 genome project data processing subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, L.L.; Bright, N.G.; Carroll, R.J.J.; Scott, M.C.; Allen, M.S.; Applegate, B.M. Molecular characterization of autoinduction of biolu-minescence in the Microtox indicator strain Vibrio fischeri ATCC 49387. Can. J. Microbiol. 2005, 51, 549–557. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, C.M.; Chatterjee, J.; Swartzman, E.; Szittner, R.; Meighen, E.A. The role of the lux autoinducer in regulating luminescence in Vibrio harveyi; control of luxR expression. Mol. Microbiol. 1996, 19, 767–775. [Google Scholar] [CrossRef]
- Le Roux, F.; Binesse, J.; Saulnier, D.; Mazel, D. Construction of a Vibrio splendidus mutant lacking the metalloprotease gene vsm by use of a novel ounterselectable suicide vector. Appl. Environ. Microbiol. 2007, 73, 777–784. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, A.; Chang, H.Y.; Daugherty, L.; Fraser, M.; Hunter, S.; López, R.; McAnulla, C.; McMenamin, C.; Nuka, G.; Pesseat, S.; et al. The InterPro protein families database: The classification resource after 15 years. Nucleic Acids Res. 2015, 43, D213–D221. [Google Scholar] [CrossRef]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Wu, Q.; Peng, Z.; Zhang, Y.; Yang, J. COACH-D: Improved protein–ligand binding sites prediction with refined ligand-binding poses through molecular docking. Nucleic Acids Res. 2018, 46, W438–W442. [Google Scholar] [CrossRef] [Green Version]
- Blanchet, E.; Prado, S.; Stien, D.; Da Silva, J.O.; Ferandin, Y.; Batailler, N.; Intertaglia, L.; Escargueil, A.; Lami, R. Quorum Sensing and Quorum Quenching in the Mediterranean Seagrass Posidonia oceanica Microbiota. Front. Mar. Sci. 2017, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Andersen, J.B.; Heydorn, A.; Hentzer, M.; Eberl, L.; Geisenberger, O.; Christensen, B.B.; Molin, S.; Givskov, M. gfp-Based N-Acyl Homoserine-Lactone Sensor Systems for Detection of Bacterial Communication. Appl. Environ. Microbiol. 2001, 67, 575–585. [Google Scholar] [CrossRef] [Green Version]
- Girard, L.; Blanchet, É.; Intertaglia, L.; Baudart, J.; Stien, D.; Suzuki, M.; LeBaron, P.; Lami, R. Characterization of N-Acyl Homoserine Lactones in Vibrio tasmaniensis LGP32 by a Biosensor-Based UHPLC-HRMS/MS Method. Sensors 2017, 17, 906. [Google Scholar] [CrossRef] [Green Version]
- Riedel, K.; Steidle, A.; Eberl, L.; Wu, H.; Geisenberger, O.; Molin, S.; Huber, B.; Hentzer, M.; Høiby, N.; Givskov, M. N-Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology 2001, 147, 3249–3262. [Google Scholar] [CrossRef] [Green Version]
- Bassler, B.L.; Greenberg, E.P.; Stevens, A.M. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J. Bacteriol. 1997, 179, 4043–4045. [Google Scholar] [CrossRef] [Green Version]
- Taga, M.E. Methods for Analysis of Bacterial Autoinducer-2 Production. Curr. Protoc. Microbiol. 2005, 23, 1C.1.1–1C.1.8. [Google Scholar] [CrossRef]
- Green, D.H.; Bowman, J.; Smith, E.A.; Gutierrez, T.; Bolch, C.J.S. Marinobacter algicola sp. nov., isolated from laboratory cultures of paralytic shellfish toxin-producing dinoflagellates. Int. J. Syst. Evol. Microbiol. 2006, 56, 523–527. [Google Scholar] [CrossRef] [Green Version]
- Bratbak, G.; Thingstad, T.F. Phytoplankton-bacteria interactions: An apparant paradox? Analysis of a model system with both competition and commensalism. Mar. Ecol. Prog. Ser. 1985, 25, 23–30. [Google Scholar] [CrossRef]
- Gurung, T.B.; Urabe, J.; Nakanishi, M. Regulation of the relationship between phytoplankton Scenedesmus acutus and hetero-trophic bacteria by the balance of light and nutrients. Aquat. Microb. Ecol. 1999, 17, 27–35. [Google Scholar] [CrossRef]
- Paerl, R.; Bouget, F.-Y.; Lozano, J.-C.; Vergé, V.; Schatt, P.; E Allen, E.; Palenik, B.; Azam, F. Use of plankton-derived vitamin B1 precursors, especially thiazole-related precursor, by key marine picoeukaryotic phytoplankton. ISME J. 2017, 11, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Sher, D.; Thompson, J.W.; Kashtan, N.; Croal, L.; Chisholm, S.W. Response of Prochlorococcus ecotypes to co-culture with diverse marine bacteria. ISME J. 2011, 5, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Mounier, J.; Camus, A.; Mitteau, I.; Vaysse, P.-J.; Goulas, P.; Grimaud, R.; Sivadon, P. The marine bacteriumMarinobacter hydrocarbonoclasticusSP17 degrades a wide range of lipids and hydrocarbons through the formation of oleolytic biofilms with distinct gene expression profiles. FEMS Microbiol. Ecol. 2014, 90, 816–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ennouri, H.; D’Abzac, P.; Hakil, F.; Branchu, P.; Naïtali, M.; Lomenech, A.; Oueslati, R.; Desbrières, J.; Sivadon, P.; Grimaud, R. The extracellular matrix of the oleolytic biofilms ofMarinobacter hydrocarbonoclasticuscomprises cytoplasmic proteins and T2SS effectors that promote growth on hydrocarbons and lipids. Environ. Microbiol. 2016, 19, 159–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tourneroche, A.; Lami, R.; Hubas, C.; Blanchet, E.; Vallet, M.; Escoubeyrou, K.; Paris, A.; Prado, S. Bacterial–Fungal Interactions in the Kelp Endomicrobiota Drive Autoinducer-2 Quorum Sensing. Front. Microbiol. 2019, 10, 1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollak, S.; Omer-Bendori, S.; Even-Tov, E.; Lipsman, V.; Bareia, T.; Ben-Zion, I.; Eldar, A. Facultative cheating supports the coexistence of diverse quorum-sensing alleles. Proc. Natl. Acad. Sci. USA 2016, 113, 2152–2157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papenfort, K.; Bassler, B.L. Quorum sensing signal–response systems in Gram-negative bacteria. Nat. Rev. Microbiol. 2016, 14, 576–588. [Google Scholar] [CrossRef] [PubMed]
- Grant, M.A.A.; Kazamia, E.; Cicuta, P.; Smith, A.G. Direct exchange of vitamin B12 is demonstrated by modelling the growth dy-namics of algal-bacterial cocultures. ISME J. 2014, 8, 1418–1427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mayali, X. Editorial: Metabolic Interactions Between Bacteria and Phytoplankton. Front. Microbiol. 2018, 9, 727. [Google Scholar] [CrossRef]
- Kimbrel, J.A.; Samo, T.; Ward, C.; Nilson, D.; Thelen, M.P.; Siccardi, A.; Zimba, P.; Lane, T.W.; Mayali, X. Host selection and stochastic effects influence bacterial community assembly on the microalgal phycosphere. Algal Res. 2019, 40, 101489. [Google Scholar] [CrossRef]
- Handley, K.M.; Boothman, C.; Mills, R.A.; Pancost, R.D.; Lloyd, J.R. Functional diversity of bacteria in a ferruginous hy-drothermal sediment. ISME J. 2010, 4, 1193–1205. [Google Scholar] [CrossRef] [Green Version]
- Balzano, S.; Statham, P.J.; Pancost, R.D.; Lloyd, J.R. Role of microbial populations in the release of reduced iron to the water column from marine aggregates. Aquat. Microb. Ecol. 2009, 54, 291–303. [Google Scholar] [CrossRef]
- Kaye, J.Z.; Baross, J.A. High incidence of halotolerant bacteria in Pacific hydrothermal-vent and pelagic environments. FEMS Microbiol. Ecol. 2000, 32, 249–260. [Google Scholar] [CrossRef] [PubMed]
- Sivadon, P.; Barnier, C.; Urios, L.; Grimaud, R. Biofilm formation as a microbial strategy to assimilate particulate substrates. Environ. Microbiol. Rep. 2019, 11, 749–764. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).