Sympatric Yaks and Plateau Pikas Promote Microbial Diversity and Similarity by the Mutual Utilization of Gut Microbiota
Abstract
:1. Introduction
2. Materials and Methods
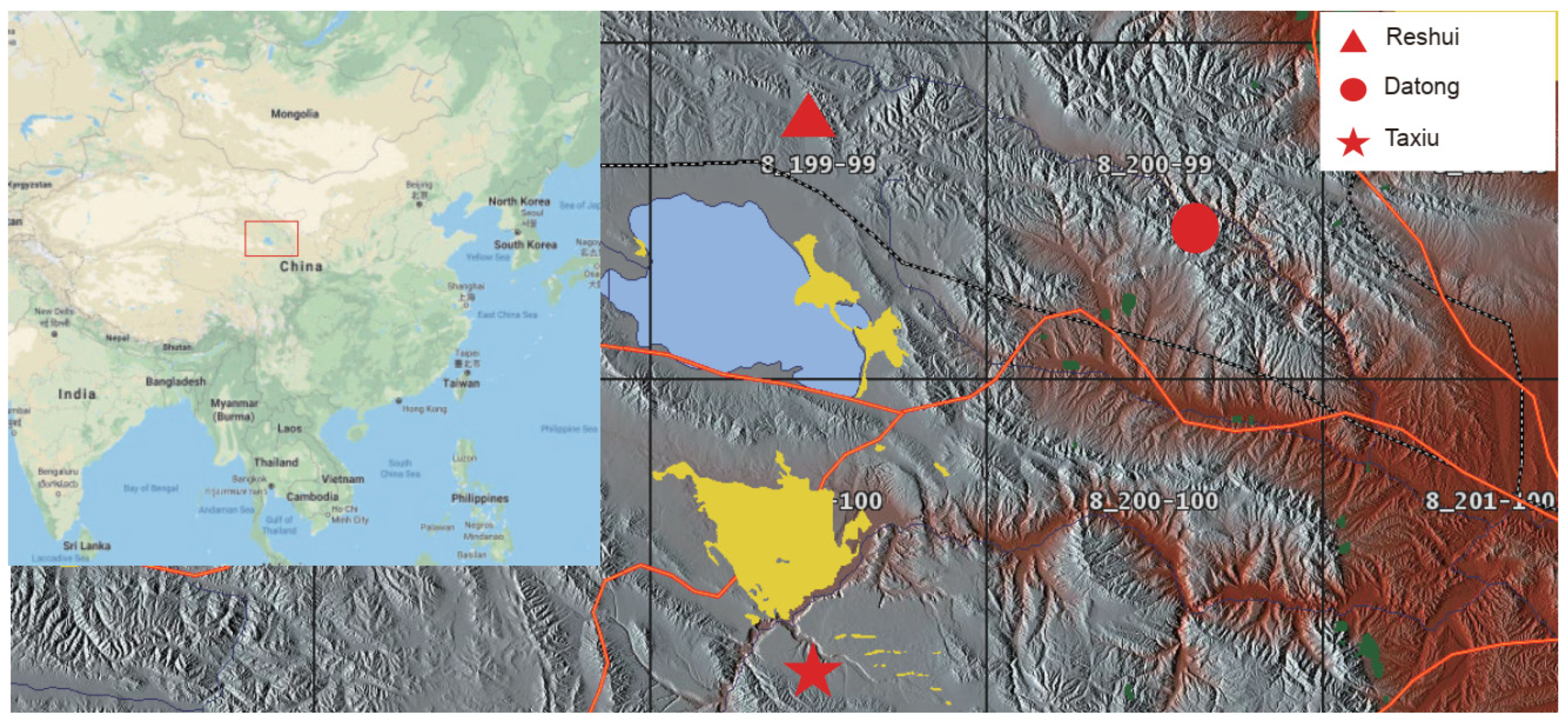
2.1. Experimental Procedures
2.2. Sample Collection
2.3. DNA Extraction and Sequencing
2.4. 16S rDNA Data Analysis
2.5. Identification of Horizontally Transmitted Microbiota
2.6. Function Prediction and Comparison
3. Results
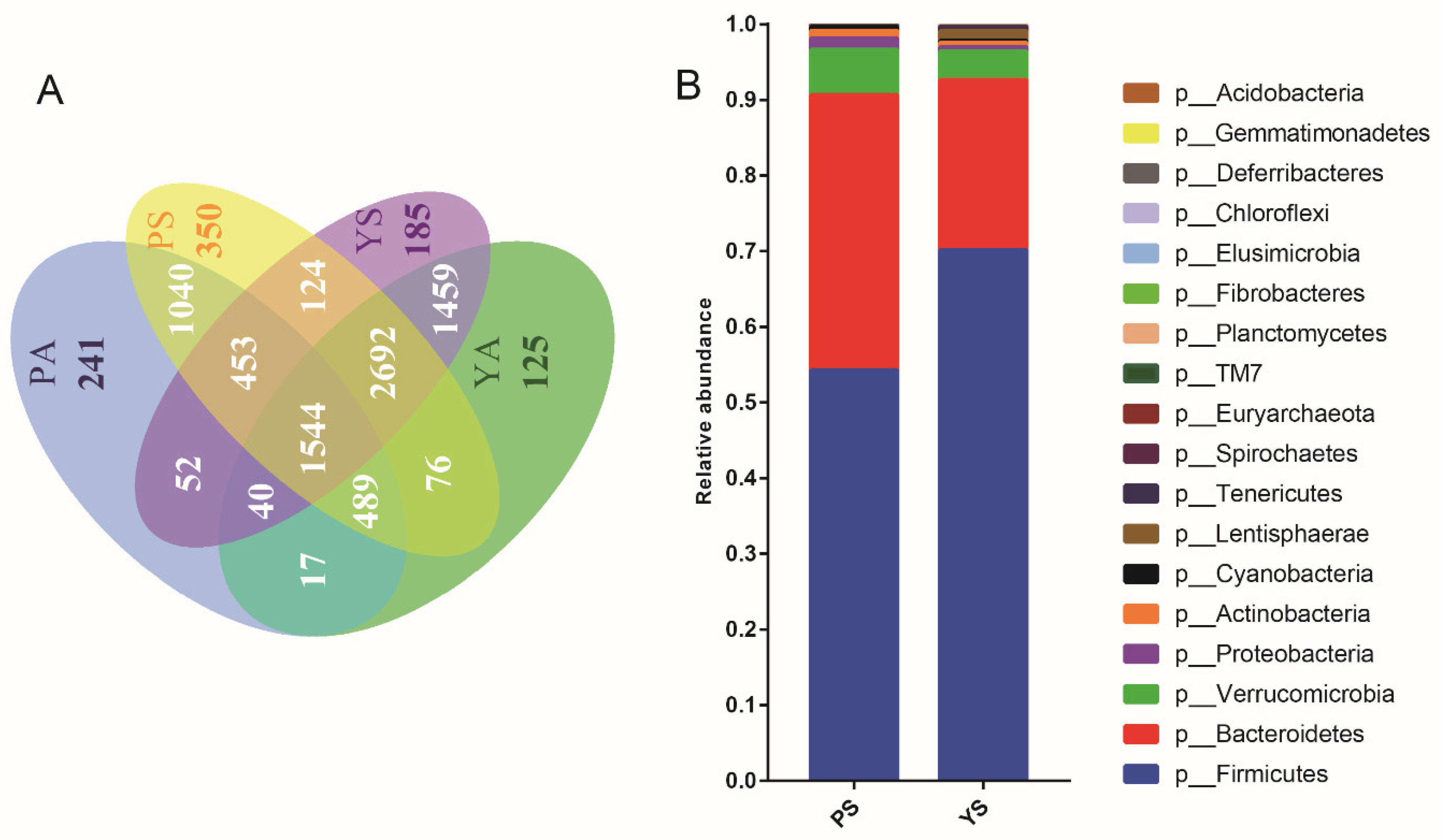
3.1. Differences in Gut Microbial Communities between Plateau Pikas and Yaks
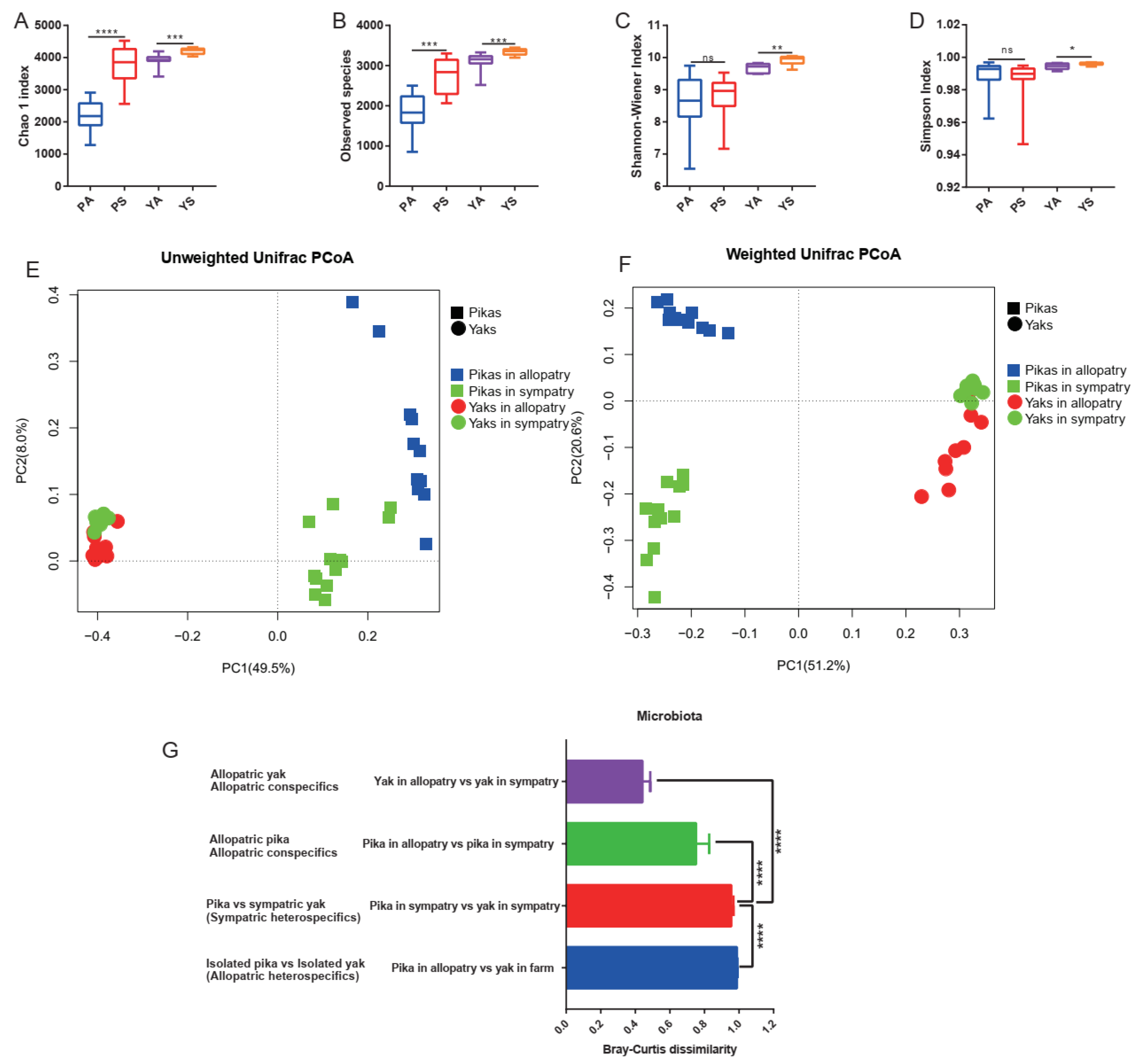
3.2. Elevated Microbial Diversity in Sympatry
3.3. Horizontally Transmitted OTU Clusters Shared between Plateau Pikas and Yaks
3.4. Functional Effects of Horizontally Transmitted Bacteria on Their New Hosts
3.5. Functional Differences of Gut Microbiota between the Allopatric and Sympatric Individuals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Begon, M.; Townsend, C.R.; Harper, J.L. Ecology: From Individuals to Ecosystems, 4th ed.; Blackwell Publishing: Hoboken, NJ, USA, 2005; p. 752. [Google Scholar]
- Hooper, D.U.; Chapin, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Harris, R.B.; Wenying, W.; Badinqiuying; Smith, A.T.; Bedunah, D.J. Herbivory and competition of Tibetan steppe vegetation in winter pasture: Effects of Livestock Exclosure and Plateau Pika Reduction. PLoS ONE 2015, 10, e0132897. [Google Scholar] [CrossRef] [Green Version]
- Cease, A.J.; Elser, J.J.; Ford, C.F.; Hao, S.; Kang, L.; Harrison, J.F. Heavy livestock grazing promotes locust outbreaks by lowering plant nitrogen content. Science 2012, 335, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Odadi, W.O.; Karachi, M.K.; Abdulrazak, S.A.; Young, T.P. African wild ungulates compete with or facilitate cattle depending on season. Science 2011, 333, 1753–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, F.; Nookaew, I.; Sommer, N.; Fogelstrand, P.; Backhed, F. Site-specific programming of the host epithelial transcriptome by the gut microbiota. Genome Biol. 2015, 16, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.E.; Demoor, T.; Rauch, M.; Faruqi, A.A.; Jang, S.; Johnson, C.C.; Boushey, H.A.; Zoratti, E.; Ownby, D.; Lukacs, N.W.; et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc. Natl. Acad. Sci. USA 2014, 111, 805–810. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, T.; Yao, M.; Li, J.; Zhang, S.; Wirth, S.; Cao, W.; Lin, Q.; Li, X. Pika gut may select for rare but diverse environmental bacteria. Front. Microbiol. 2016, 7, 1269. [Google Scholar] [CrossRef]
- Jaenike, J.; Unckless, R.; Cockburn, S.N.; Boelio, L.M.; Perlman, S.J. Adaptation via symbiosis: Recent spread of a drosophila defensive dymbiont. Science 2010, 329, 212–215. [Google Scholar] [CrossRef]
- Montllor, C.B.; Maxmen, A.; Purcell, A.H. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol. Entomol. 2002, 27, 189–195. [Google Scholar] [CrossRef]
- Wiener, G.; Han, J.; Long, R. The Yak; FAO Regional office for Asia and the Pacific: Bangkok, Thailand, 2003; pp. 57–58. [Google Scholar]
- Luo, Y.; Gao, W.; Gao, Y.; Tang, S.; Huang, Q.; Tan, X.; Chen, J.; Huang, T. Mitochondrial genome analysis of Ochotona curzoniae and implication of cytochrome c oxidase in hypoxic adaptation. Mitochondrion 2008, 8, 352–357. [Google Scholar] [CrossRef]
- Dahal, N.; Lissovsky, A.A.; Lin, Z.; Solari, K.; Hadly, E.A.; Zhan, X.; Ramakrishnan, U. Genetics, morphology and ecology reveal a cryptic pika lineage in the Sikkim Himalaya. Mol. Phylogenet. Evol. 2017, 106, 55–60. [Google Scholar] [CrossRef]
- Yu, N.; Zheng, C.L.; Zhang, Y.P.; Li, W.H. Molecular systematics of pikas (genus Ochotona) inferred from mitochondrial DNA sequences. Mol. Phylogenet. Evol. 2000, 16, 85–95. [Google Scholar] [CrossRef]
- Dong, Q.M.; Zhao, X.Q.; Wu, G.L.; Shi, J.J.; Ren, G.H. A review of formation mechanism and restoration measures of “black-soil-type” degraded grassland in the Qinghai-Tibetan Plateau. Environ. Earth Sci. 2013, 70, 2359–2370. [Google Scholar] [CrossRef]
- Qu, J.P.; Ji, W.H.; Russell, J.C.; Zhang, H.; Zhang, Y.M. The more the merrier? Multi-species grazing of small herbivores mediates plant community impacts. Biodivers. Conserv. 2016, 25, 2055–2069. [Google Scholar] [CrossRef]
- Qu, J.P.; Li, W.J.; Yang, M.; Li, K.X.; Zhang, Y.M. Methods for large scale assessment of small mammal abundance in open habitats: Plateau pika (Ochotona Curzoniae) in alpine grassland. Pol. J. Ecol. 2011, 59, 829–833. [Google Scholar]
- Wang, Y.; Wang, X.; Wang, Z.; Patrick, G.; Kenichi, T.; Graham, A. Primary Study on habitat choice of plateau pika (Ochotona curzoniae). J. Sichuan Univ. Nat. Sci. Ed. 2004, 41, 1041–1045. [Google Scholar]
- Dai, X.; Gu, X.; Shi, J.; Yuan, F.; Yin, B.; Wang, A.; Wei, W.; Yang Sheng, M. The seasonal changes of plant secondary metabolites and their influence on the food selection of plateau pika. Acta Theriol. Sin. 2012, 32, 306–317. [Google Scholar]
- Dai, X.; Zhang, B.; Wu, X.Y.; Jiang, L.Y.; Zou, Z.Z.; Wang, A.Q.; Wei, W.H.; Yang, S.M. Identification of tannin-degrading microorganisms in the gut of plateau pikas (Ochotona curzoniae) and root voles (Microtus oeconomus). Symbiosis 2014, 63, 1–9. [Google Scholar] [CrossRef]
- Fan, C.; Zhang, L.; Fu, H.; Liu, C.; Zhang, Y. Enterotypes of the gut microbial community and their response to plant secondary compounds in plateau pikas. Microorganisms 2020, 8, 1311. [Google Scholar] [CrossRef]
- Speakman, J.R.; Chi, Q.; Ołdakowski, Ł.; Fu, H.; Fletcher, Q.E.; Hambly, C.; Togo, J.; Liu, X.; Piertney, S.B.; Wang, X.; et al. Surviving winter on the Qinghai-Tibetan Plateau: Pikas suppress energy demands and exploit yak feces to survive winter. Proc. Natl. Acad. Sci. USA 2021, 118, e2100707118. [Google Scholar] [CrossRef] [PubMed]
- Moeller, A.H.; Peeters, M.; Ndjango, J.B.; Li, Y.; Hahn, B.H.; Ochman, H. Sympatric chimpanzees and gorillas harbor convergent gut microbial communities. Genome Res. 2013, 23, 1715–1720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Xing, S.; Ma, Y. Characteristics of Soil Moisture for Different Types of Vegetation in Gangcha County. Bull. Soil Water Conserv. 2012, 32, 14–18. [Google Scholar]
- Tan, C.; Yu, Y.; Jiang, Z.; Zhong, L.; Zhang, Y.; Qu, J. Differences in exploration and resting metabolic rates of plateau pikas (Ochotona curzoniae) at different altitudes. Acta Theriol. Sin. 2020, 40, 27–36. [Google Scholar]
- Wang, J.M.; Zhang, Y.M.; Wang, D.H. Seasonal thermogenesis and body mass regulation in plateau pikas (Ochotona curzoniae). Oecologia 2006, 149, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhang, L.; Fan, C.; Liu, C.; Li, W.; Cheng, Q.; Zhao, X.; Jia, S.; Zhang, Y. Environment and host species identity shape gut microbiota diversity in sympatric herbivorous mammals. Microb. Biotechnol. 2020, 14, 1300–1315. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Zhang, L.; Fan, C.; Liu, C.; Li, W.; Li, J.; Zhao, X.; Jia, S.; Zhang, Y. Domestication Shapes the Community Structure and Functional Metagenomic Content of the Yak Fecal Microbiota. Front. Microbiol. 2021, 12, 594075. [Google Scholar] [CrossRef]
- Claesson, M.J.; Wang, Q.; O’Sullivan, O.; Greene-Diniz, R.; Cole, J.R.; Ross, R.P.; O’Toole, P.W. Comparison of two next-generation sequencing technologies for resolving highly complex microbiota composition using tandem variable 16S rRNA gene regions. Nucleic Acids Res. 2010, 38, e200. [Google Scholar] [CrossRef]
- Vilo, C.; Dong, Q. Evaluation of the RDP Classifier Accuracy Using 16S rRNA Gene Variable Regions. Metagenomics 2012, 1, 104303. [Google Scholar] [CrossRef] [Green Version]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pena, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Shabat, S.K.; Sasson, G.; Doron-Faigenboim, A.; Durman, T.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Shterzer, N.; Mizrahi, I. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J. 2016, 10, 2958–2972. [Google Scholar] [CrossRef] [Green Version]
- Xue, Z.; Zhang, W.; Wang, L.; Hou, R.; Zhang, M.; Fei, L.; Zhang, X.; Huang, H.; Bridgewater, L.C.; Jiang, Y.; et al. The bamboo-eating giant panda harbors a carnivore-like gut microbiota, with excessive seasonal variations. MBio 2015, 6, e00022-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.D.; Martinez-Fernandez, G.; Padmanabha, J.; Long, R.J.; Denman, S.E.; McSweeney, C.S. Methanogen diversity in indigenous and introduced ruminant species on the Tibetan Plateau. Archaea 2016, 2016, 10. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. CB 2016, 26, 1873–1879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, W.; Penney, N.C.; Cronin, O.; Garcia-Perez, I.; Molloy, M.G.; Holmes, E.; Shanahan, F.; Cotter, P.D.; O’Sullivan, O. The microbiome of professional athletes differs from that of more sedentary subjects in composition and particularly at the functional metabolic level. Gut 2018, 67, 625–633. [Google Scholar] [CrossRef]
- Chevalier, C.; Stojanovic, O.; Colin, D.J.; Suarez-Zamorano, N.; Tarallo, V.; Veyrat-Durebex, C.; Rigo, D.; Fabbiano, S.; Stevanovic, A.; Hagemann, S.; et al. Gut microbiota orchestrates energy homeostasis during cold. Cell 2015, 163, 1360–1374. [Google Scholar] [CrossRef] [Green Version]
- Derrien, M.; van Passel, M.W.J.; van de Bovenkamp, J.H.B.; Schipper, R.G.; de Vos, W.M.; Dekker, J. Mucin-bacterial interactions in the human oral cavity and digestive tract. Gut Microbes 2010, 1, 254–268. [Google Scholar] [CrossRef] [Green Version]
- Song, S.J.; Lauber, C.; Costello, E.K.; Lozupone, C.A.; Humphrey, G.; Berg-Lyons, D.; Caporaso, J.G.; Knights, D.; Clemente, J.C.; Nakielny, S.; et al. Cohabiting family members share microbiota with one another and with their dogs. eLife 2013, 2, 22. [Google Scholar] [CrossRef]
- Li, H.; Qu, J.P.; Li, T.T.; Li, J.B.; Lin, Q.; Li, X.Z. Pika population density is associated with the composition and diversity of gut microbiota. Front. Microbiol. 2016, 7, 9. [Google Scholar] [CrossRef]
- Seedorf, H.; Griffin, N.W.; Ridaura, V.K.; Reyes, A.; Cheng, J.Y.; Rey, F.E.; Smith, M.I.; Simon, G.M.; Scheffrahn, R.H.; Woebken, D.; et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell 2014, 159, 253–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Q.; Wang, D. Coprophagy in herbivorous small mammals. Acta Theriol. Sin. 2004, 24, 333–338. [Google Scholar]
- Liu, Y.; Fan, J.; Shi, Z.; Yang, X.; Harris, W. Relationships between plateau pika (Ochotona curzoniae) densities and biomass and biodiversity indices of alpine meadow steppe on the Qinghai—Tibet Plateau China. Ecol. Eng. 2017, 102, 509–518. [Google Scholar] [CrossRef]
- Zhou, J.Z.; Kang, S.; Schadt, C.W.; Garten, C.T. Spatial scaling of functional gene diversity across various microbial taxa. Proc. Natl. Acad. Sci. USA 2008, 105, 7768–7773. [Google Scholar] [CrossRef] [Green Version]
- Huntly, N.J. Influence of refuging consumers (Pikas: Ochotona Princeps) on subalpine meadow vegetation. Ecology 1987, 68, 274–283. [Google Scholar] [CrossRef]
- Lai, C.H.; Smith, A.T. Keystone status of plateau pikas (Ochotona curzoniae): Effect of control on biodiversity of native birds. Biodivers. Conserv. 2003, 12, 1901–1912. [Google Scholar] [CrossRef]
- Long, R.J.; Apori, S.O.; Castro, F.B.; Orskov, E.R. Feed value of native forages of the Tibetan Plateau of China. Anim. Feed Sci. Technol. 1999, 80, 101–113. [Google Scholar] [CrossRef]
- Smith, A.T.; Badingqiuying; Wilson, M.C.; Hogan, B.W. Functional-trait ecology of the plateau pika Ochotona curzoniae in the Qinghai-Tibetan Plateau ecosystem. Integr. Zool. 2019, 14, 87–103. [Google Scholar] [CrossRef] [Green Version]
- Clarke, S.F.; Murphy, E.F.; O’Sullivan, O.; Lucey, A.J.; Humphreys, M.; Hogan, A.; Hayes, P.; O’Reilly, M.; Jeffery, I.B.; Wood-Martin, R.; et al. Exercise and associated dietary extremes impact on gut microbial diversity. Gut 2014, 63, 1913–1920. [Google Scholar] [CrossRef] [Green Version]
- Kohl, K.D.; Weiss, R.B.; Cox, J.; Dale, C.; Dearing, M.D. Gut microbes of mammalian herbivores facilitate intake of plant toxins. Ecol. Lett. 2014, 17, 1238–1246. [Google Scholar] [CrossRef]
- Li, G.L.; Li, J.; Kohl, K.D.; Yin, B.F.; Wei, W.H.; Wan, X.R.; Zhu, B.L.; Zhang, Z.B. Dietary shifts influenced by livestock grazing shape the gut microbiota composition and co-occurrence networks in a local rodent species. J. Anim. Ecol. 2019, 88, 302–314. [Google Scholar] [CrossRef]
- Yuan, F. Soil Ingestion Behavior of Yak Grazing on Alpine Grassland in Different Season. Master’s Thesis, Lanzhou University, Lanzhou, China, 2013. [Google Scholar]
- Zhang, X.; Sukhchuluun, G.; Bo, T.; Chi, Q.; Yang, J.; Chen, B.; Zhang, L.; Wang, D. Huddling remodels gut microbiota to reduce energy requirements in a small mammal species during cold exposure. Microbiome 2018, 6, 103. [Google Scholar] [CrossRef] [Green Version]
- Perofsky, A.C.; Lewis, R.J.; Meyers, L.A. Terrestriality and bacterial transfer: A comparative study of gut microbiomes in sympatric Malagasy mammals. ISME J. 2019, 1, 13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tallon, P.; Magajna, B.; Lofranco, C.; Leung, K.T. Microbial indicators of faecal contamination in water: A current perspective. Water Air Soil Pollut. 2005, 166, 139–166. [Google Scholar] [CrossRef]
- Yuan, M.L.; Dean, S.H.; Longo, A.V.; Rothermel, B.B.; Tuberville, T.D.; Zamudio, K.R. Kinship, inbreeding and fine-scale spatial structure influence gut microbiota in a hindgut-fermenting tortoise. Mol. Ecol. 2015, 24, 2521–2536. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.; Fang, Z.M.; Zhou, P.; Chang, F.; Hong, Y.Z.; Zhang, X.C.; Peng, H.; Xiao, Y.Z. Evidence for lignin oxidation by the giant panda fecal microbiome. PLoS ONE 2012, 7, 10. [Google Scholar] [CrossRef]
- Hehemann, J.H.; Correc, G.; Barbeyron, T.; Helbert, W.; Czjzek, M.; Michel, G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature 2010, 464, 908–912. [Google Scholar] [CrossRef] [PubMed]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashaolu, T.J. Immune boosting functional foods and their mechanisms: A critical evaluation of probiotics and prebiotics. Biomed. Pharmacother. 2020, 130, 110625. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Yu, Q.; Yang, S.; Xiao, N.; Ye, P.; Xue, J.; Tian, T.; Wu, W.; Zhou, X. Evaluation on the measures by a grid-based design for wildlife control in hyper-endemic areas for echinococcosis. Chin. J. Parasitol. Parasit. Dis. 2018, 36, 495–498. [Google Scholar]
- Haughn, G.W.; Davin, L.; Giblin, M.; Underhill, E.W. Biochemical genetics of plant secondary metabolites in Arabidopsis thaliana: The glucosinolates. Plant Physiol. 1991, 97, 217–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walker, C.H. Species differences in microsomal monooxygenase activity and their relationship to biological half-lives. Drug Metab. Rev. 1978, 7, 295–323. [Google Scholar] [CrossRef]
- Winter, H.; Seawright, A.A.; Noltie, H.J.; Mattocks, A.R.; Jukes, R.; Wangdi, K.; Gurung, J.B. Pyrrolizidine alkaloid poisoning of yaks: Identification of the plants involved. Vet. Rec. 1994, 134, 135–139. [Google Scholar] [CrossRef]
- Mondal, D.B.; Nandankar, U.A.; Mohanty, T.K.; Barari, S.K.; Pal, R.N.; Sarkar, M. Pyrrolizidine alkaloid poisoning in yak. Vet. Rec. 1999, 144, 508–509. [Google Scholar] [CrossRef]
- Bell, K.S.; Philp, J.C.; Aw, D.W.J.; Christofi, N. A review—The genus Rhodococcus. J. Appl. Microbiol. 1998, 85, 195–210. [Google Scholar] [CrossRef]
- Tsuchida, T.; Koga, R.; Fukatsu, T. Host plant specialization governed by facultative symbiont. Science 2004, 303, 1989. [Google Scholar] [CrossRef] [Green Version]
- Kohl, K.D.; Dearing, M.D. Experience matters: Prior exposure to plant toxins enhances diversity of gut microbes in herbivores. Ecol. Lett. 2012, 15, 1008–1015. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, H.; Zhang, L.; Fan, C.; Li, W.; Liu, C.; Zhang, H.; Cheng, Q.; Zhang, Y. Sympatric Yaks and Plateau Pikas Promote Microbial Diversity and Similarity by the Mutual Utilization of Gut Microbiota. Microorganisms 2021, 9, 1890. https://doi.org/10.3390/microorganisms9091890
Fu H, Zhang L, Fan C, Li W, Liu C, Zhang H, Cheng Q, Zhang Y. Sympatric Yaks and Plateau Pikas Promote Microbial Diversity and Similarity by the Mutual Utilization of Gut Microbiota. Microorganisms. 2021; 9(9):1890. https://doi.org/10.3390/microorganisms9091890
Chicago/Turabian StyleFu, Haibo, Liangzhi Zhang, Chao Fan, Wenjing Li, Chuanfa Liu, He Zhang, Qi Cheng, and Yanming Zhang. 2021. "Sympatric Yaks and Plateau Pikas Promote Microbial Diversity and Similarity by the Mutual Utilization of Gut Microbiota" Microorganisms 9, no. 9: 1890. https://doi.org/10.3390/microorganisms9091890
APA StyleFu, H., Zhang, L., Fan, C., Li, W., Liu, C., Zhang, H., Cheng, Q., & Zhang, Y. (2021). Sympatric Yaks and Plateau Pikas Promote Microbial Diversity and Similarity by the Mutual Utilization of Gut Microbiota. Microorganisms, 9(9), 1890. https://doi.org/10.3390/microorganisms9091890





