Deciphering the Dynamics of Signaling Cascades and Virulence Factors of B. cinerea during Tomato Cell Wall Degradation
Abstract
:1. Introduction
2. Material and Methods
2.1. Fungal Strains and Culture Conditions
2.2. Isolation of Proteins
2.3. Proteomes Data Integration
2.4. Bioinformatic Analysis
3. Results
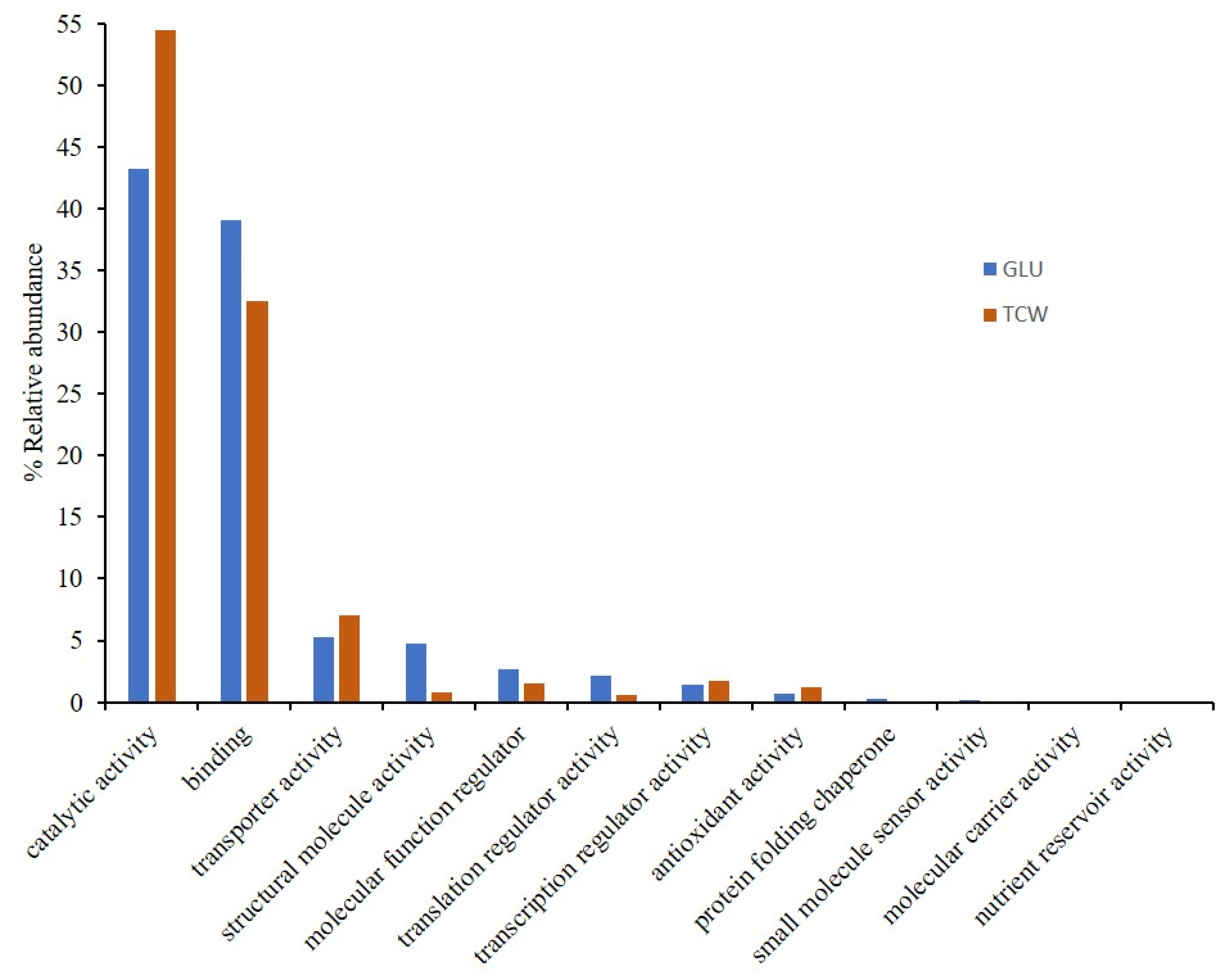
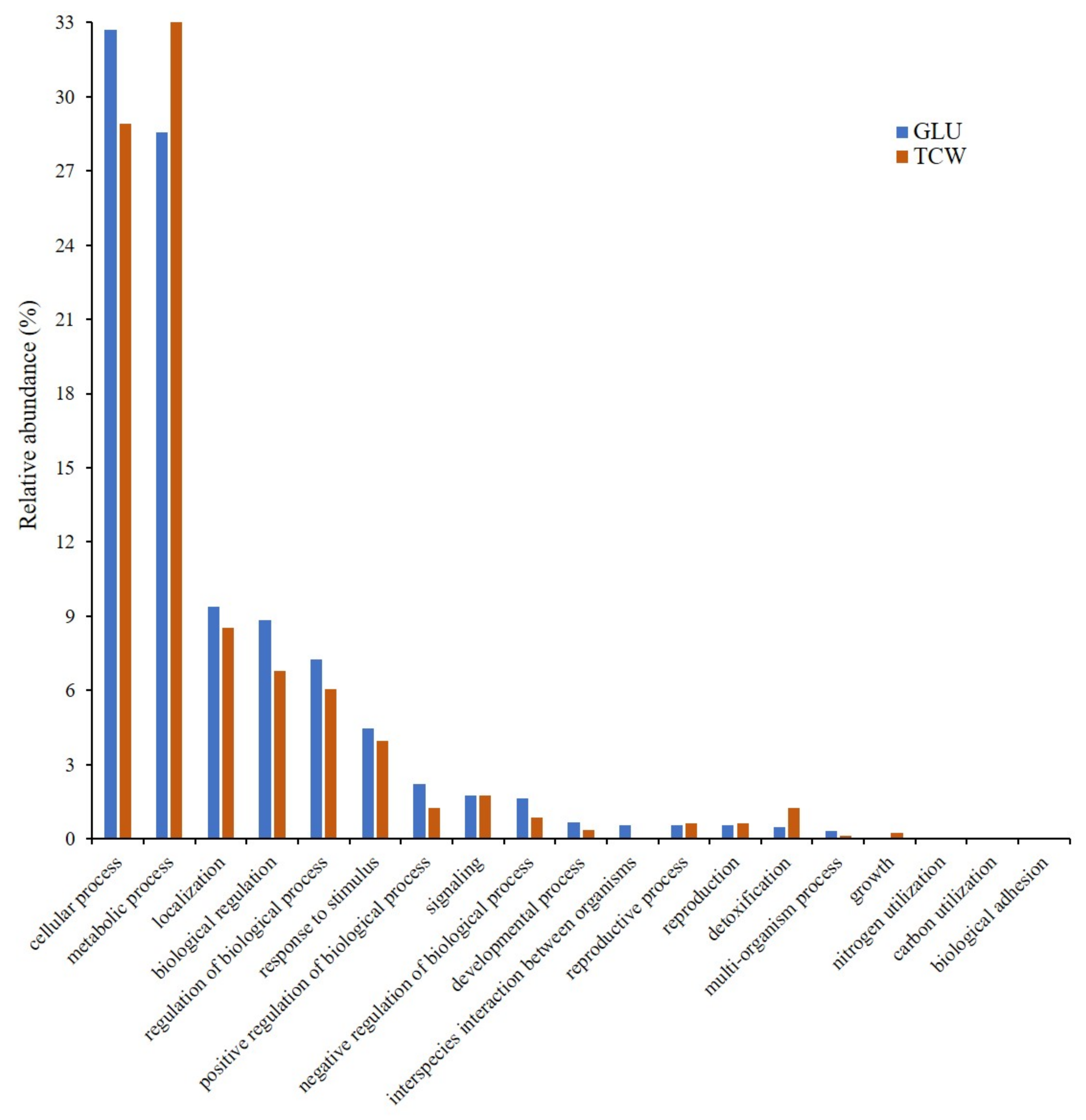
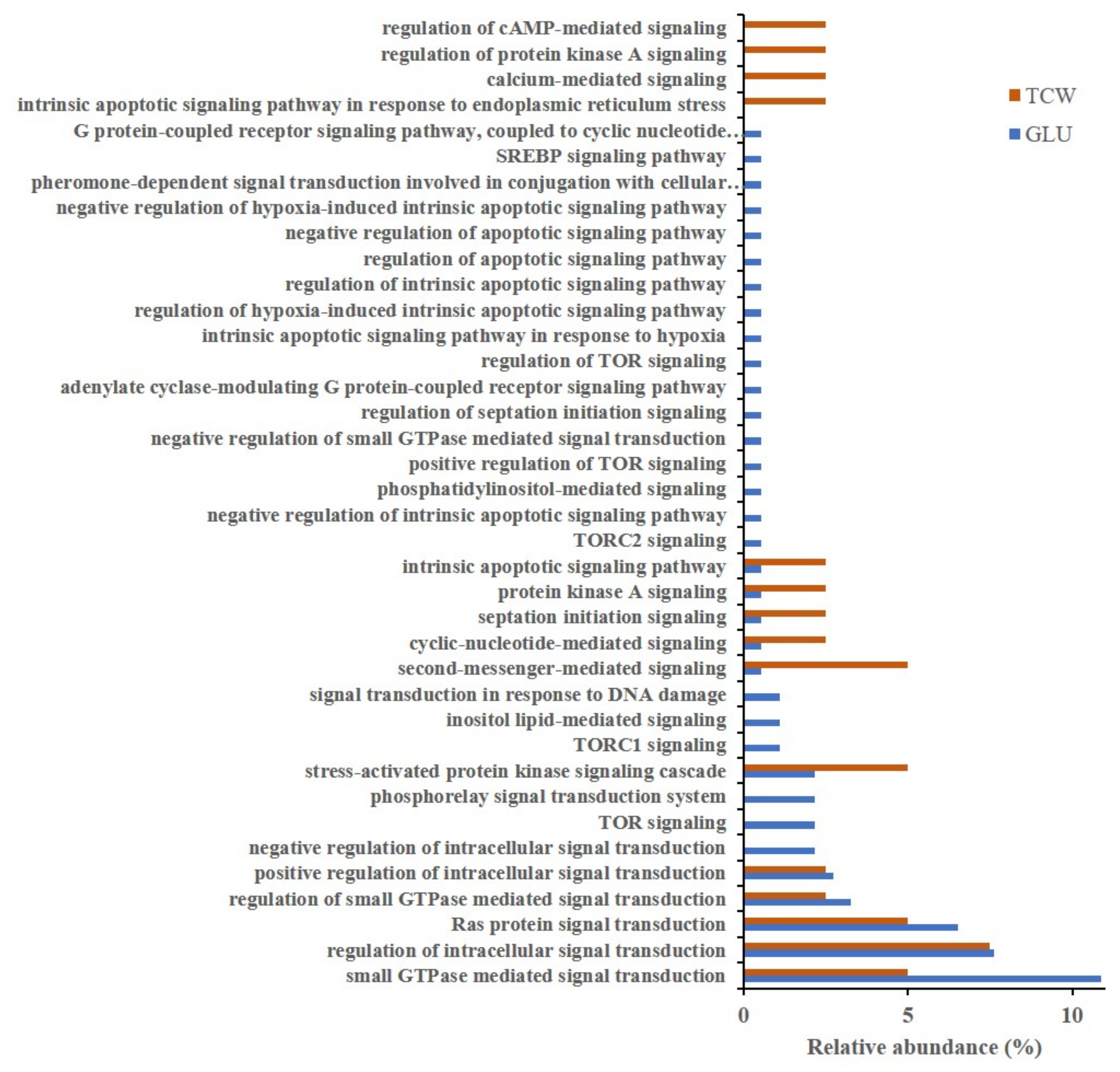
3.1. Gene Ontology Classification
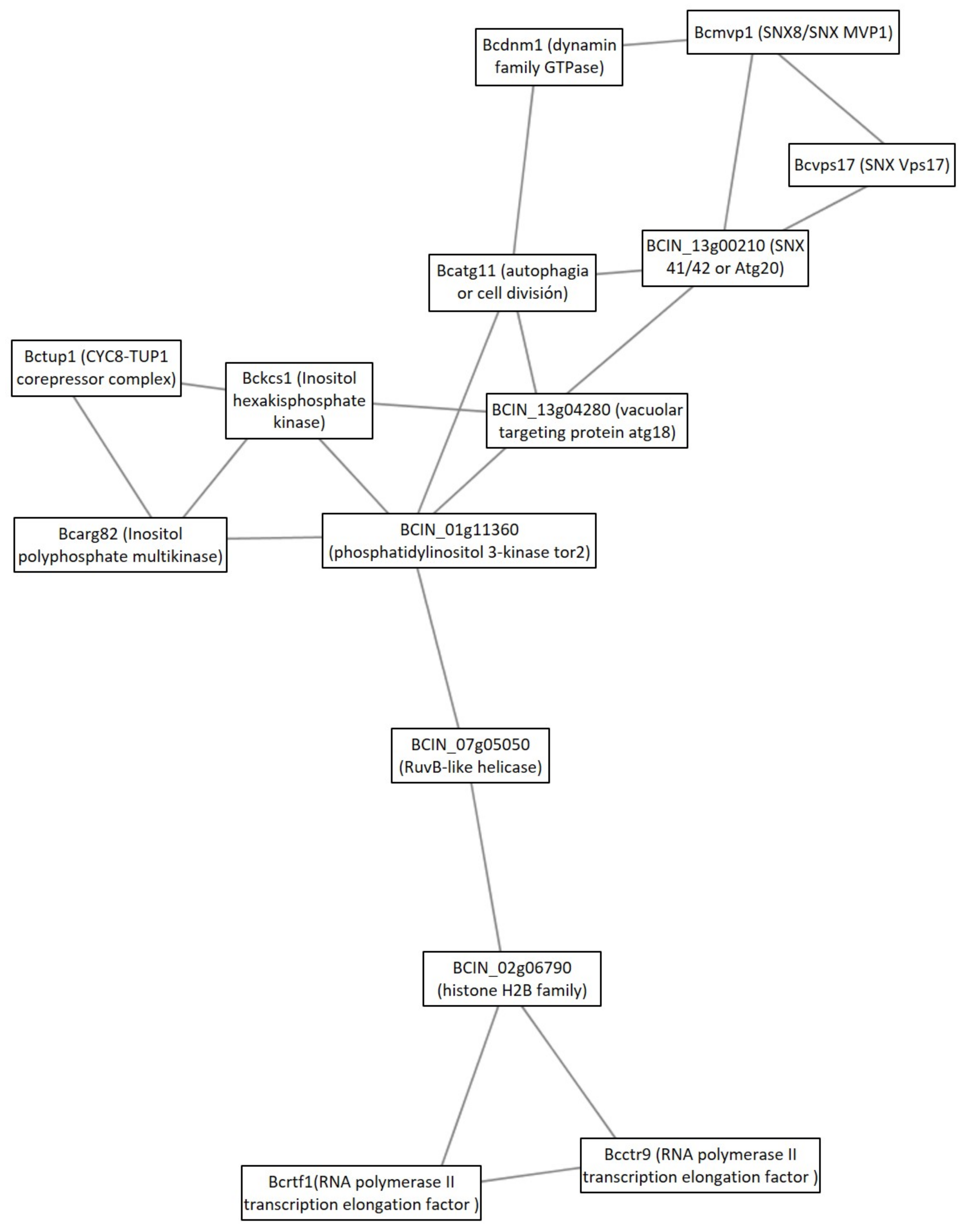
3.2. Protein Interaction Analysis
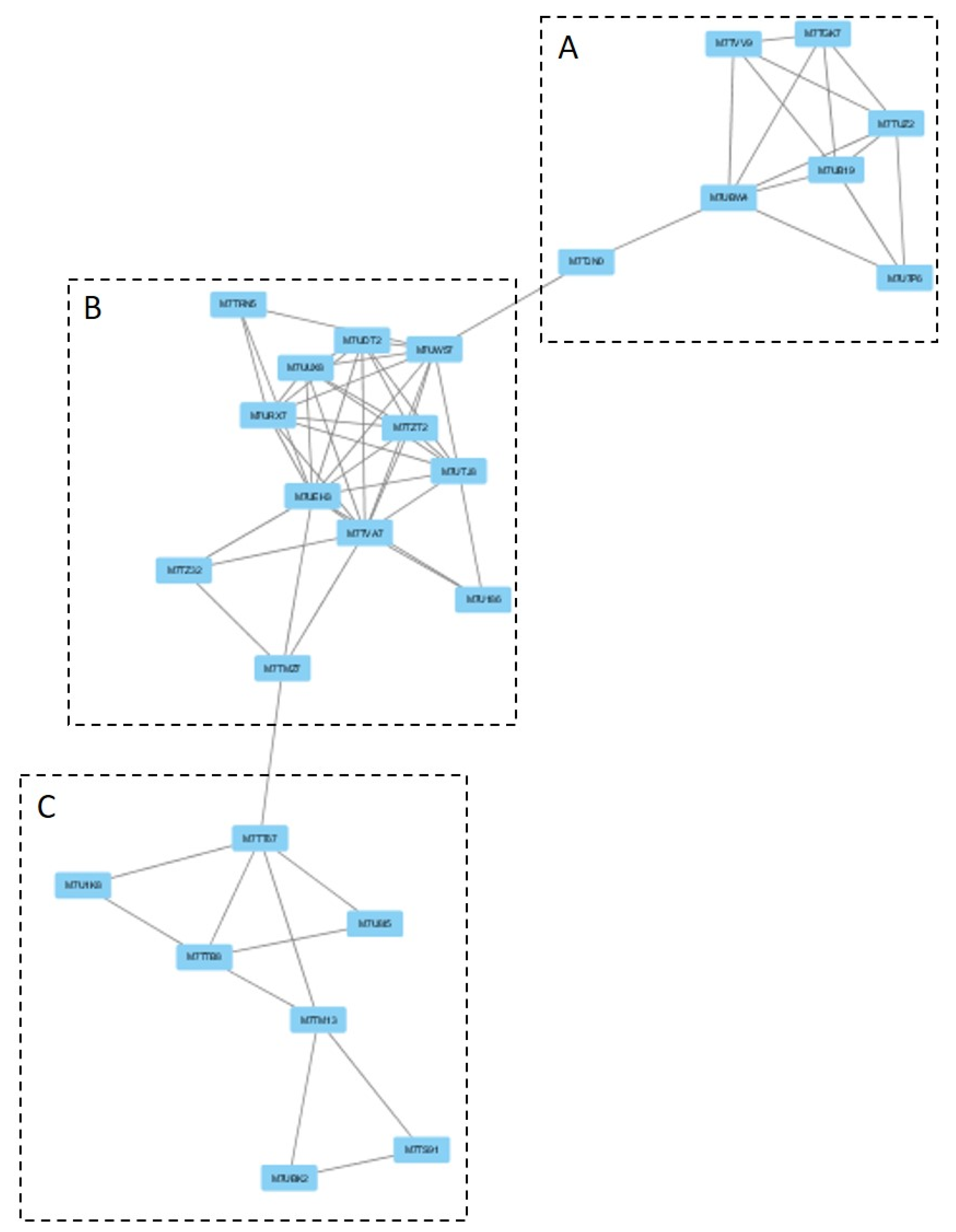
3.2.1. Cluster Analysis under the GLU Condition
3.2.2. Cluster Analysis under TCW Condition
4. Discussion
4.1. GO Analysis
4.2. Protein Interaction Analysis
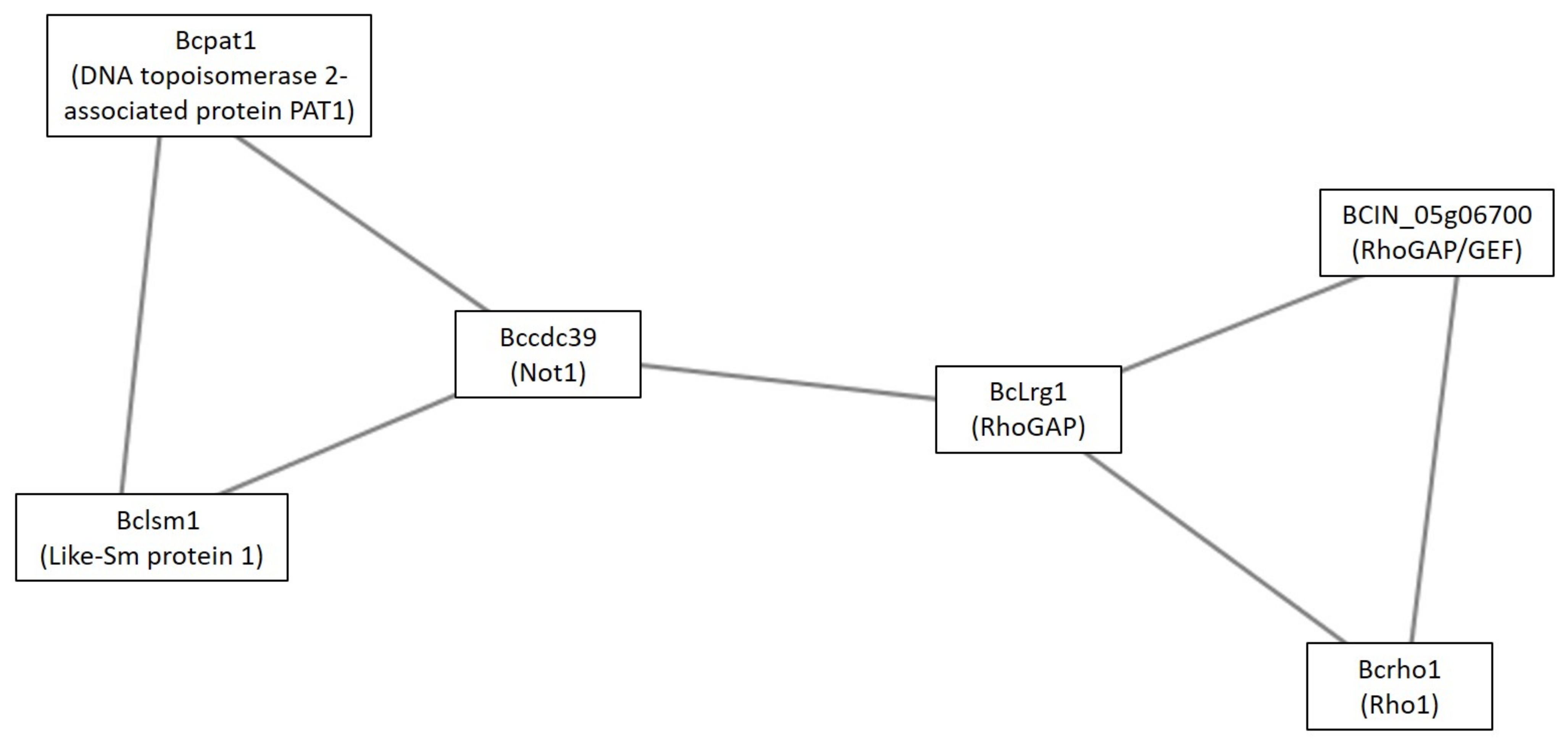
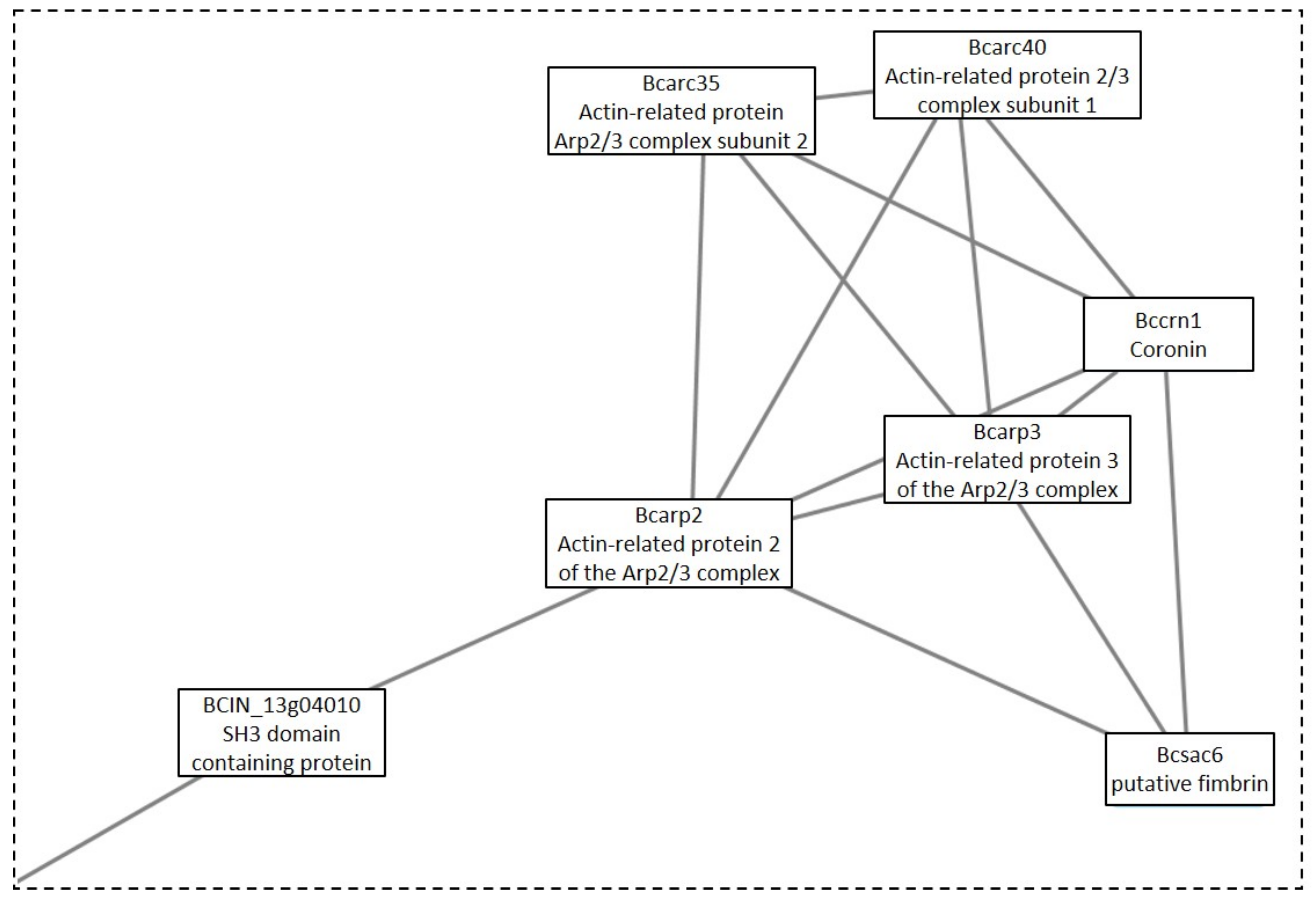
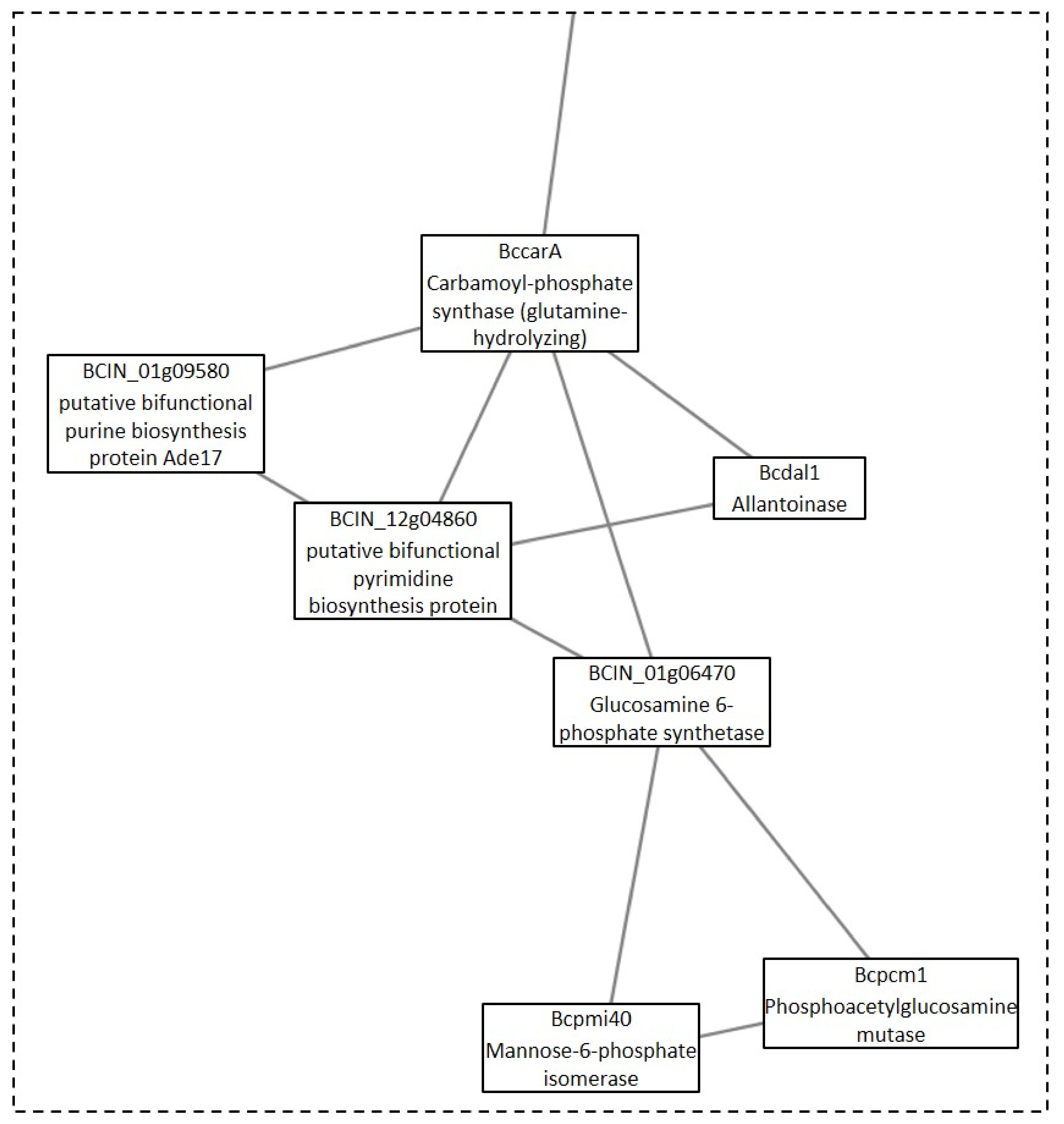
4.2.1. Clusters under GLU Condition
4.2.2. Clusters under the TCW Condition
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fillinger, S.; Elad, Y. Botrytis—The Fungus, The Pathogen and Its Management in Agricultural Systems; Springer International Publishing: New York, NY, USA, 2016. [Google Scholar]
- Staats, M.; van Kan, J.A. Genome Update of Botrytis cinerea Strains B05.10 and T4. Eukaryot. Cell 2012, 11, 1413–1414. [Google Scholar] [CrossRef] [Green Version]
- Atwell, S.; Corwin, J.; Soltis, N.; Subedy, A.; Denby, K.; Kliebenstein, D.J. Whole genome resequencing of Botrytis cinerea isolates identifies high levels of standing diversity. Front. Microbiol. 2015, 6, 996. [Google Scholar] [CrossRef] [Green Version]
- Urban, M.; Cuzick, A.; Rutherford, K.; Irvine, A.; Pedro, H.; Pant, R.; Sadanadan, V.; Khamari, L.; Billal, S.; Mohanty, S.; et al. PHI-base: A new interface and further additions for the multi-species pathogen-host interactions database. Nucleic Acids Res. 2017, 45, 604–610. [Google Scholar] [CrossRef]
- Acero, F.J.; Carbu, M.; El-Akhal, M.R.; Garrido, C.; Gonzalez-Rodríguez, V.E.; Cantoral, J.M. Development of Proteomics-Based Fungicides: New Strategies for Environmentally Friendly Control of Fungal Plant Diseases. Int. J. Mol. Sci. 2011, 12, 795–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández-Acero, F.J.; Colby, T.; Harzen, A.; Carbú, M.; Wieneke, U.; Cantoral, J.M.; Schmidt, J. 2-DE proteomic approach to the Botrytis cinerea secretome induced with different carbon sources and plant-based elicitors. Proteomics 2010, 10, 2270–2280. [Google Scholar] [CrossRef] [PubMed]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabidó, E.; Fernández-Acero, F.J. Phosphoproteome analysis of B. cinerea in response to different plant-based elicitors. J. Proteom. 2016, 139, 84–94. [Google Scholar] [CrossRef]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabido, E.; Fernandez-Acero, F.J. Modifications of fungal membrane proteins profile under pathogenicity induction: A proteomic analysis of Botrytis cinerea membranome. Proteomics 2016, 16, 2363–2376. [Google Scholar] [CrossRef]
- Escobar-Niño, A.E.; Liñeiro, E.; Amil, F.; Carrasco, R.; Chiva, C.; Fuentes, C.; Blanco-Ulate, B.; Fernández, J.M.C.; Sabidó, E.; Fernández-Acero, F.J. Proteomic study of the membrane components of signalling cascades of Botrytis cinerea controlled by phosphorylation. Sci. Rep. 2019, 9, 9860. [Google Scholar] [CrossRef]
- Liñeiro, E.; Macias-Sánchez, A.J.; Espinazo, M.; Cantoral, J.M.; Moraga, J.; Collado, I.G.; Fernández-Acero, F.J. Phenotypic Effects and Inhibition of Botrydial Biosynthesis Induced by Different Plant-Based Elicitors in Botrytis cinerea. Curr. Microbiol. 2018, 75, 431–440. [Google Scholar] [CrossRef] [Green Version]
- Röhrig, H.; Schmidt, J.; Colby, T.; Bräutigam, A.; Hufnagel, P.; Bartels, D. Desiccation of the resurrection plant Craterostigma plantagineum induces dynamic changes in protein phosphorylation. Plant Cell Environ. 2006, 29, 1606–1617. [Google Scholar] [CrossRef] [Green Version]
- Liñeiro, E.; Chiva, C.; Cantoral, J.M.; Sabido, E.; Fernández-Acero, F.J. Dataset of the Botrytis cinerea phosphoproteome induced by different plant-based elicitors. Data Brief 2016, 7, 1447–1450. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zhang, W.; Kho, Y.; Zhao, Y. Proteomic Analysis of Integral Plasma Membrane Proteins. Anal. Chem. 2004, 76, 1817–1823. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Kabbara, S.; Hérivaux, A.; Duge De Bernonville, T.D.; Courdavault, V.; Clastre, M.; Gastebois, A.; Osman, M.; Hamze, M.; Cock, J.M.; Schaap, P.; et al. Diversity and Evolution of Sensor Histidine Kinases in Eukaryotes. Genome Biol. Evol. 2019, 11, 86–108. [Google Scholar] [CrossRef]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; I Hurwitz, D.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González, A.; Hall, M.N. Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 2017, 36, 397–408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Moseley, J.B.; Goode, B.L. The Yeast Actin Cytoskeleton: From Cellular Function to Biochemical Mechanism. Microbiol. Mol. Biol. Rev. 2006, 70, 605–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pomraning, K.R.; Bredeweg, E.L.; Kerkhoven, E.J.; Barry, K.; Haridas, S.; Hundley, H.; LaButti, K.; Lipzen, A.; Yan, M.; Magnuson, J.K.; et al. Regulation of Yeast-to-Hyphae Transition in Yarrowia lipolytica. mSphere 2018, 3, e00541-18. [Google Scholar] [CrossRef] [Green Version]
- Veloso, J.; van Kan, J.A.L. Many Shades of Grey in Botrytis–Host Plant Interactions. Trends Plant Sci. 2018, 23, 613–622. [Google Scholar] [CrossRef]
- Xiong, F.; Liu, M.; Zhuo, F.; Yin, H.; Deng, K.; Feng, S.; Liu, Y.; Luo, X.; Feng, L.; Zhang, S.; et al. Host-induced gene silencing of BcTOR in Botrytis cinerea enhances plant resistance to grey mould. Mol. Plant Pathol. 2019, 20, 1722–1739. [Google Scholar] [CrossRef] [Green Version]
- Kamble, C.; Jain, S.; Murphy, E.; Kim, K. Requirements of Slm proteins for proper eisosome organization, endocytic trafficking and recycling in the yeast Saccharomyces cerevisiae. J. Biosci. 2011, 36, 79–96. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Gu, Q.; Yun, Y.; Yin, Y.; Xu, J.R.; Shim, W.B.; Ma, Z. The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014, 203, 219–232. [Google Scholar] [CrossRef]
- Liu, W.; Leroux, P.; Fillinger, S. The HOG1-like MAP kinase Sak1 of Botrytis cinerea is negatively regulated by the upstream histidine kinase Bos1 and is not involved in dicarboximide- and phenylpyrrole-resistance. Fungal Genet. Biol. 2008, 45, 1062–1074. [Google Scholar] [CrossRef]
- Jacob, S.; Foster, A.J.; Yemelin, A.; Thines, E. Histidine kinases mediate differentiation, stress response, and pathogenicity in Magnaporthe oryzae. Microbiologyopen 2014, 3, 668–687. [Google Scholar] [CrossRef]
- Chauhan, N. Two-component phosphorelays in fungal mitochondria and beyond. Mitochondrion 2015, 22, 60–65. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Cao, L. Starvation signals in yeast are integrated to coordinate metabolic reprogramming and stress response to ensure longevity. Curr. Genet. 2017, 63, 839–843. [Google Scholar] [CrossRef] [Green Version]
- Yin, Y.; Wu, S.; Chui, C.; Ma, T.; Jiang, H.; Hahn, M.; Ma, Z. The MAPK kinase BcMkk1 suppresses oxalic acid biosynthesis via impeding phosphorylation of BcRim15 by BcSch9 in Botrytis cinerea. PLoS Pathog. 2018, 14, e1007285. [Google Scholar] [CrossRef]
- Chen, L.; Zhang, L.; Mei, X.; Wang, C.; Guo, Z.; Li, L.; Li, B.; Liang, Y.; Zou, S.; Dong, H. The type II phosphoinositide 4-kinase FgLsb6 is important for the development and virulence of Fusarium graminearum. Fungal Genet. Biol. 2020, 144, 103443. [Google Scholar] [CrossRef]
- O’Meara, T.R.; Veri, A.O.; Polvi, E.J.; Li, X.; Valaei, S.F.; Diezmann, S.; Cowen, L.E. Mapping the Hsp90 Genetic Network Reveals Ergosterol Biosynthesis and Phosphatidylinositol-4-Kinase Signaling as Core Circuitry Governing Cellular Stress. PLoS Genet. 2016, 12, e1006142. [Google Scholar] [CrossRef] [Green Version]
- Barman, A.; Gohain, D.; Bora, U.; Tamuli, R. Phospholipases play multiple cellular roles including growth, stress tolerance, sexual development, and virulence in fungi. Microbiol. Res. 2018, 209, 55–69. [Google Scholar] [CrossRef]
- D’Ambrosio, J.M.; Gonorazky, G.; Sueldo, D.J.; Moraga, J.; Di Palma, A.A.; LaMattina, L.; Collado, I.G.; Laxalt, A.M. The sesquiterpene botrydial from Botrytis cinerea induces phosphatidic acid production in tomato cell suspensions. Planta 2018, 247, 1001–1009. [Google Scholar] [CrossRef]
- Ding, M.; Zhu, Q.; Liang, Y.; Li, J.; Fan, X.; Yu, X.; He, F.; Xu, H.; Liang, Y.; Yu, J. Differential roles of three FgPLD genes in regulating development and pathogenicity in Fusarium graminearum. Fungal Genet. Biol. 2017, 109, 46–52. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Dickman, M.B.; Wang, Z. Cytoprotective Co-chaperone BcBAG1 Is a Component for Fungal Development, Virulence, and Unfolded Protein Response (UPR) of Botrytis cinerea. Front. Microbiol. 2019, 10, 685. [Google Scholar] [CrossRef]
- Van Dalfsen, K.M.; Hodapp, S.; Keskin, A.; Otto, G.M.; Berdan, C.A.; Higdon, A.; Cheunkarndee, T.; Nomura, D.K.; Jovanovic, M.; Brar, G.A. Global Proteome Remodeling during ER Stress Involves Hac1-Driven Expression of Long Undecoded Transcript Isoforms. Dev. Cell 2018, 46, 219–235.e8. [Google Scholar] [CrossRef] [Green Version]
- Tabas, I.; Ron, D. Integrating the mechanisms of apoptosis induced by endoplasmic reticulum stress. Nat. Cell Biol. 2011, 13, 184–190. [Google Scholar] [CrossRef]
- Heimel, K.; Freitag, J.; Hampel, M.; Ast, J.; Bölker, M.; Kämper, J. Crosstalk between the Unfolded Protein Response and Pathways That Regulate Pathogenic Development in Ustilago maydis. Plant Cell 2013, 25, 4262–4277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lo Presti, L.; López Díaz, C.; Turrá, D.; Di Pietro, A.; Hampel, M.; Heimel, K.; Kahmann, R. A conserved co-chaperone is required for virulence in fungal plant pathogens. New Phytol. 2016, 209, 1135–1148. [Google Scholar] [CrossRef] [Green Version]
- Schumacher, J.; Kokkelink, L.; Huesmann, C.; Jimenez-Teja, D.; Collado, I.G.; Barakat, R.; Tudzynski, P.; Tudzynski, B. The cAMP-Dependent Signaling Pathway and Its Role in Conidial Germination, Growth, and Virulence of the Gray Mold Botrytis cinerea. Mol. Plant-Microbe Interact. 2008, 21, 1443–1459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, D.; Bloom, A.L.; Panepinto, J.C. Opposing PKA and Hog1 signals control the post-transcriptional response to glucose availability in Cryptococcus neoformans. Mol. Microbiol. 2016, 102, 306–320. [Google Scholar] [CrossRef] [Green Version]
- Caza, M.; Kronstad, J.W. The cAMP/Protein Kinase A Pathway Regulates Virulence and Adaptation to Host Conditions in Cryptococcus neoformans. Front. Cell. Infect. Microbiol. 2019, 9, 212. [Google Scholar] [CrossRef]
- Schumacher, J. Signal Transduction Cascades Regulating Differentiation and Virulence in Botrytis cinerea. In Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Fillinger, S., Elad, Y., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 247–267. [Google Scholar]
- Minz-Dub, A.; Sharon, A. The Botrytis cinerea PAK kinase BcCla4 mediates morphogenesis, growth and cell cycle regulating processes downstream of BcRac. Mol. Microbiol. 2017, 104, 487–498. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Ju, Z.; Wang, J.; Wang, J.; Wang, H.; Wu, J.; Yin, Y.; Zhao, Y.; Ma, Z.; Chen, Y. The RasGEF FgCdc25 regulates fungal development and virulence in Fusarium graminearum via cAMP and MAPK signalling pathways. Environ. Microbiol. 2020, 22, 5109–5124. [Google Scholar] [CrossRef]
- Goshima, G.; Iwasaki, O.; Obuse, C.; Yanagida, M. The role of Ppe1/PP6 phosphatase for equal chromosome segregation in fission yeast kinetochore. EMBO J. 2003, 22, 2752–2763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Wang, G.; Jiang, C.; Xu, J.-R.; Wang, C. Fgk3 glycogen synthase kinase is important for development, pathogenesis and stress responses in Fusarium graminearum. Sci. Rep. 2015, 5, 8504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariño, J.; Velazquez, D.; Casamayor, A. Ser/Thr protein phosphatases in fungi: Structure, regulation and function. Microb. Cell 2019, 6, 217–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adám, C.; Erdei, E.; Casado, C.; Kovacs, L.; Gonzalez, A.; Majoros, L.; Petrényi, K.; Bagossi, P.; Farkas, I.; Molnar, M.; et al. Protein phosphatase CaPpz1 is involved in cation homeostasis, cell wall integrity and virulence of Candida albicans. Microbiology 2012, 158 Pt 5, 1258–1267. [Google Scholar] [CrossRef] [Green Version]
- Carlton, J.G.; Cullen, P.J. Coincidence detection in phosphoinositide signaling. Trends Cell Biol. 2005, 15, 540–547. [Google Scholar] [CrossRef] [Green Version]
- Ekena, K.; Stevens, T.H. The Saccharomyces cerevisiae MVP1 gene interacts with VPS1 and is required for vacuolar protein sorting. Mol. Cell. Biol. 1995, 15, 1671–1678. [Google Scholar] [CrossRef] [Green Version]
- Zientara-Rytter, K.; Subramani, S. Mechanistic Insights into the Role of Atg11 in Selective Autophagy. J. Mol. Biol. 2020, 432, 104–122. [Google Scholar] [CrossRef]
- Rieter, E.; Vinke, F.; Bakula, D.; Cebollero, E.; Ungermann, C.; Proikas-Cezanne, T.; Reggiori, F. Atg18 function in autophagy is regulated by specific sites within its beta-propeller. J. Cell Sci. 2013, 126, 593–604. [Google Scholar] [CrossRef] [Green Version]
- Takahara, T.; Maeda, T. Evolutionarily conserved regulation of TOR signalling. J. Biochem. 2013, 154, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Zheng, M.; Chu, X.; Chen, Y.; Xu, H.; Wang, J.; Zhou, H.; Long, J. Paf1 and Ctr9 subcomplex formation is essential for Paf1 complex assembly and functional regulation. Nat. Commun. 2018, 9, 3795. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lev, S.; Saiardi, A.; Desmarini, D.; Sorrell, T.C.; Djordjevic, J.T. Inositol Polyphosphate Kinases, Fungal Virulence and Drug Discovery. J. Fungi 2016, 2, 24. [Google Scholar] [CrossRef] [Green Version]
- Tharun, S.; Muhlrad, D.; Chowdhury, A.; Parker, R. Mutations in the Saccharomyces cerevisiae LSM1 Gene That Affect mRNA Decapping and 3′ End Protection. Genetics 2005, 170, 33–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonnerot, C.; Boeck, R.; Lapeyre, B. The Two Proteins Pat1p (Mrt1p) and Spb8p Interact In Vivo, Are Required for mRNA Decay, and Are Functionally Linked to Pab1p. Mol. Cell. Biol. 2000, 20, 5939–5946. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.Y.; Badarinarayana, V.; Audino, D.C.; Rappsilber, J.; Mann, M.; Denis, C.L. The NOT proteins are part of the CCR4 transcriptional complex and affect gene expression both positively and negatively. EMBO J. 1998, 17, 1096–1106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roumanie, O.; Weinachter, C.; Larrieu, I.; Crouzet, M.; Doignon, F. Functional characterization of the Bag7, Lrg1 and Rgd2 RhoGAP proteins from Saccharomyces cerevisiae. FEBS Lett. 2001, 506, 149–156. [Google Scholar] [CrossRef]
- Li, Y.; Yue, X.; Que, Y.; Yan, X.; Ma, Z.; Talbot, N.J.; Wang, Z. Characterisation of Four LIM Protein-Encoding Genes Involved in Infection-Related Development and Pathogenicity by the Rice Blast Fungus Magnaporthe oryzae. PLoS ONE 2014, 9, e88246. [Google Scholar] [CrossRef]
- Vogt, N.; Seiler, S. The RHO1-specific GTPase-activating Protein LRG1 Regulates Polar Tip Growth in Parallel to Ndr Kinase Signaling inNeurospora. Mol. Biol. Cell 2008, 19, 4554–4569. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, K.; Tanaka, K.; Imamura, H.; Hihara, T.; Kameyama, T.; Nonaka, H.; Hirano, H.; Matsuura, Y.; Takai, Y. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 1996, 15, 2196–2207. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Schmelzle, T.; Hall, M.N. The RHO1-GAPs SAC7, BEM2 and BAG7 control distinct RHO1 functions in Saccharomyces cerevisiae. Mol. Microbiol. 2002, 45, 1433–1441. [Google Scholar] [CrossRef] [PubMed]
- Pérez, P.; Rincón, S.A. Rho GTPases: Regulation of cell polarity and growth in yeasts. Biochem. J. 2010, 426, 243–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Jia, X.; Tian, S.; Zhang, C.; Lu, Z.; Chen, Y.; Chen, F.; Li, Z.; Su, X.; Han, X.; et al. Role of the small GTPase Rho1 in cell wall integrity, stress response, and pathogenesis of Aspergillus fumigatus. Fungal Genet. Biol. 2018, 120, 30–41. [Google Scholar] [CrossRef]
- Martínez-Rocha, A.L.; Roncero, M.I.; López-Ramirez, A.; Mariné, M.; Guarro, J.; Martínez-Cadena, G.; Di Pietro, A. Rho1 has distinct functions in morphogenesis, cell wall biosynthesis and virulence of Fusarium oxysporum. Cell. Microbiol. 2008, 10, 1339–1351. [Google Scholar] [CrossRef]
- Panepinto, J.C.; Heinz, E.; Traven, A. The cellular roles of Ccr4-NOT in model and pathogenic fungi—Implications for fungal virulence. Front. Genet. 2013, 4, 302. [Google Scholar] [CrossRef] [Green Version]
- Ito, W.; Li, X.; Irie, K.; Mizuno, T.; Irie, K. RNA-Binding Protein Khd1 and Ccr4 Deadenylase Play Overlapping Roles in the Cell Wall Integrity Pathway in Saccharomyces cerevisiae. Eukaryot. Cell 2011, 10, 1340–1347. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Z.; Gao, T.; Zhang, Y.; Hou, Y.; Wang, J.; Zhou, M. FgFim, a key protein regulating resistance to the fungicide JS399-19, asexual and sexual development, stress responses and virulence in Fusarium graminearum. Mol. Plant Pathol. 2014, 15, 488–499. [Google Scholar] [CrossRef]
- Echauri-Espinosa, R.O.; Callejas-Negrete, O.A.; Roberson, R.W.; Bartnicki-García, S.; Mouriño-Pérez, R.R. Coronin Is a Component of the Endocytic Collar of Hyphae of Neurospora crassa and Is Necessary for Normal Growth and Morphogenesis. PLoS ONE 2012, 7, e38237. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Yu, Q.; Wang, Y.; Xiao, C.; Li, J.; Huo, D.; Zhang, D.; Jia, C.; Li, M. The Candida albicans fimbrin Sac6 regulates oxidative stress response (OSR) and morphogenesis at the transcriptional level. Biochim. Biophys. Acta 2016, 1863, 2255–2266. [Google Scholar] [CrossRef]
- Dewar, H.; Warren, D.T.; Gardiner, F.C.; Gourlay, C.G.; Satish, N.; Richardson, M.R.; Andrews, P.; Ayscough, K.R. Novel Proteins Linking the Actin Cytoskeleton to the Endocytic Machinery inSaccharomyces cerevisiae. Mol. Biol. Cell 2002, 13, 3646–3661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Likus, W.; Siemianowicz, K.; Bieńk, K.; Pakuła, M.; Pathak, H.; Dutta, C.; Wang, Q.; Shojaei, S.; Assaraf, Y.G.; Ghavami, S.; et al. Could drugs inhibiting the mevalonate pathway also target cancer stem cells? Drug Resist. Updat. 2016, 25, 13–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giesbert, S.; Siegmund, U.; Schumacher, J.; Kokkelink, L.; Tudzynski, P. Functional Analysis of BcBem1 and Its Interaction Partners in Botrytis cinerea: Impact on Differentiation and Virulence. PLoS ONE 2014, 9, e95172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riquelme, M.; Aguirre, J.; Bartnicki-García, S.; Braus, G.H.; Feldbrügge, M.; Fleig, U.; Hansberg, W.; Herrera-Estrella, A.; Kämper, J.; Kück, U.; et al. Fungal Morphogenesis, from the Polarized Growth of Hyphae to Complex Reproduction and Infection Structures. Microbiol. Mol. Biol. Rev. 2018, 82, e00068-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gulli, M.-P.; Jaquenoud, M.; Shimada, Y.; Niederhäuser, G.; Wiget, P.; Peter, M. Phosphorylation of the Cdc42 Exchange Factor Cdc24 by the PAK-like Kinase Cla4 May Regulate Polarized Growth in Yeast. Mol. Cell 2000, 6, 1155–1167. [Google Scholar] [CrossRef]
- Salazar, M.A.; Kwiatkowski, A.V.; Pellegrini, L.; Cestra, G.; Butler, M.H.; Rossman, K.L.; Serna, D.M.; Sondek, J.; Gertler, F.B.; De Camilli, P. Tuba, a Novel Protein Containing Bin/Amphiphysin/Rvs and Dbl Homology Domains, Links Dynamin to Regulation of the Actin Cytoskeleton. J. Biol. Chem. 2003, 278, 49031–49043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cestra, G.; Kwiatkowski, A.; Salazar, M.; Gertler, F.; De Camilli, P. Tuba, A GEF for CDC42, Links Dynamin to Actin Regulatory Proteins. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2005; pp. 537–545. [Google Scholar]
- Fort, P.; Blangy, A. The Evolutionary Landscape of Dbl-Like RhoGEF Families: Adapting Eukaryotic Cells to Environmental Signals. Genome Biol. Evol. 2017, 9, 1471–1486. [Google Scholar] [CrossRef] [Green Version]
- Weerasinghe, H.; Bugeja, H.E.; Andrianopoulos, A. The novel Dbl homology/BAR domain protein, MsgA, of Talaromyces marneffei regulates yeast morphogenesis during growth inside host cells. Sci. Rep. 2021, 11, 2334. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar-Niño, A.; Morano Bermejo, I.M.; Carrasco Reinado, R.; Fernandez-Acero, F.J. Deciphering the Dynamics of Signaling Cascades and Virulence Factors of B. cinerea during Tomato Cell Wall Degradation. Microorganisms 2021, 9, 1837. https://doi.org/10.3390/microorganisms9091837
Escobar-Niño A, Morano Bermejo IM, Carrasco Reinado R, Fernandez-Acero FJ. Deciphering the Dynamics of Signaling Cascades and Virulence Factors of B. cinerea during Tomato Cell Wall Degradation. Microorganisms. 2021; 9(9):1837. https://doi.org/10.3390/microorganisms9091837
Chicago/Turabian StyleEscobar-Niño, Almudena, Inés M. Morano Bermejo, Rafael Carrasco Reinado, and Francisco Javier Fernandez-Acero. 2021. "Deciphering the Dynamics of Signaling Cascades and Virulence Factors of B. cinerea during Tomato Cell Wall Degradation" Microorganisms 9, no. 9: 1837. https://doi.org/10.3390/microorganisms9091837
APA StyleEscobar-Niño, A., Morano Bermejo, I. M., Carrasco Reinado, R., & Fernandez-Acero, F. J. (2021). Deciphering the Dynamics of Signaling Cascades and Virulence Factors of B. cinerea during Tomato Cell Wall Degradation. Microorganisms, 9(9), 1837. https://doi.org/10.3390/microorganisms9091837






