Sulfidogenic Microbial Communities of the Uzen High-Temperature Oil Field in Kazakhstan
Abstract
:1. Introduction
2. Materials and Methods
2.1. Characteristics of the Petroleum Reservoir and Sampling Procedures
2.2. DNA Isolation, Amplification, and Sequencing of the 16S rRNA Gene
2.3. Bioinformatics Analysis
2.4. Statistical Analysis and Functional Characterization
2.5. dsrA Illumina Sequencing, Bioinformatics Processing, and Data Analyses
2.6. Media Composition
2.7. Analytical Techniques and Biofilms Formation
2.8. Microscopic Techniques
2.9. Nucleotide Sequence Accession Numbers
3. Results
3.1. Environmental Characterization and Cultivable Microorganisms of the Injection and Production Water Samples
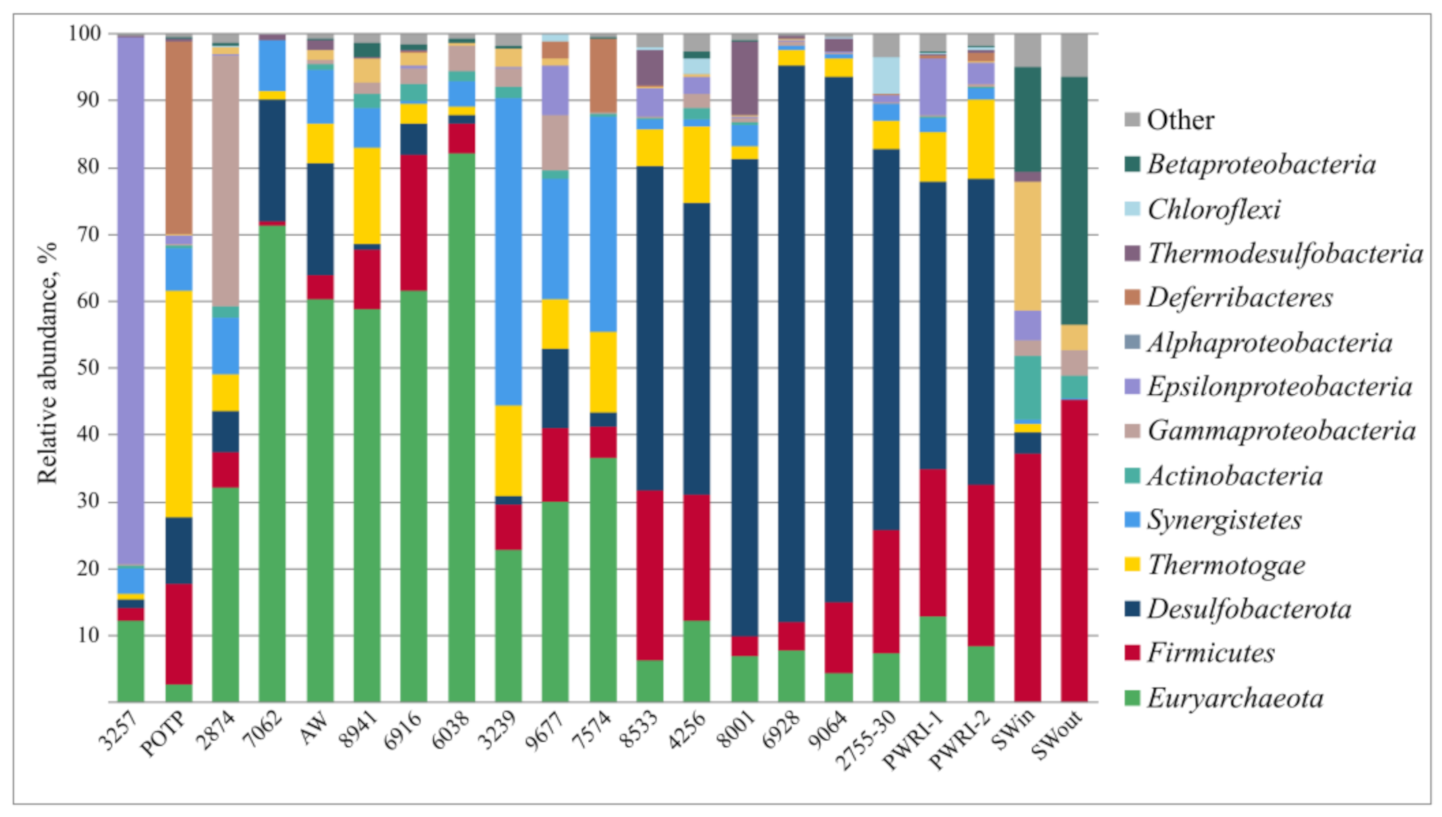
3.2. Microbial Diversity in Petroleum Reservoir Based on 16S rRNA and dsrA Gene Gequencing


3.3. Potential Functional Characteristics of Microbial Communities of Injection and Production Water Samples from the Uzen Oil Field

3.4. Effect of Nitrate Addition on Sulfide Production by SR and TSR Enrichments from the Oil Field
3.5. Molecular Analyses of Sulfate-, Thiosulfate-, and Nitrate-Reducing Enrichments
3.6. Effect of Biocides on Survival of Planktonic Cells and Biofilms of Desulfovibrio alaskensis Kaz19 and on Sulfide Production

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magot, M.; Ollivier, B.; Patel, B.K.C. Microbiology of petroleum reservoirs. Antonie Van Leeuwenhoek 2000, 77, 103–116. [Google Scholar] [CrossRef]
- Youssef, N.; Elshahed, M.S.; McInerney, M.J. Microbial processes in oil fields: Culprits, problems and opportunities. Adv. Appl. Microbiol. 2009, 66, 141–251. [Google Scholar] [CrossRef]
- Head, I.M.; Gray, N.D.; Larter, S.R. Life in the slow lane; biogeochemistry of biodegraded petroleum containing reservoirs and implications for energy recovery and carbon management. Front. Microbiol. 2014, 5, 566. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shestakova, N.M.; Ivoilov, V.S.; Tourova, T.P.; Belyaev, S.S.; Poltaraus, A.B.; Nazina, T.N. Application of clone libraries: Syntrophic acetate degradation to methane in a high-temperature petroleum reservoir: Culture-based and 16S rRNA genes characterization. In Applied Microbiology and Molecular Biology in Oil Field Systems; Whitby, C., Skovhus, T.L., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 45–53. [Google Scholar] [CrossRef]
- Gieg, L.M.; Davidova, I.A.; Duncan, K.E.; Suflita, J.M. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oil fields. Env. Microbiol. 2010, 12, 3074–3086. [Google Scholar] [CrossRef] [PubMed]
- Gieg, L.M.; Jack, T.R.; Foght, J.M. Biological souring and mitigation in oil reservoirs. Appl. Microbiol. Biotechnol. 2011, 92, 263–282. [Google Scholar] [CrossRef] [PubMed]
- Belyaev, S.S.; Laurinavichus, K.S.; Obraztsova, A.Y.; Gorlatov, S.N.; Ivanov, M.V. Microbiological processes in the near-bottom zone of injection wells of oil fields. Microbiology 1982, 51, 997–1001. [Google Scholar]
- Bonch-Osmolovskaya, E.A.; Miroshnichenko, M.L.; Lebedinsky, A.V.; Chernyh, N.A.; Nazina, T.N.; Ivoilov, V.S.; Belyaev, S.S.; Boulygina, E.S.; Lysov, Y.P.; Perov, A.N.; et al. Radioisotopic, culture-based, and oligonucleotide microchip analyses of thermophilic microbial communities in a continental high-temperature petroleum reservoir. Appl. Environ. Microbiol. 2003, 69, 6143–6151. [Google Scholar] [CrossRef] [Green Version]
- Nazina, T.N.; Shestakova, N.M.; Ivoilov, V.S.; Kostrukova, N.K.; Belyaev, S.S.; Ivanov, M.V. Radiotracer assay of microbial processes in petroleum reservoirs. Adv. Biotechnol. Microbiol. 2017, 2, 555591. [Google Scholar] [CrossRef]
- Nazina, T.N.; Shestakova, N.M.; Semenova, E.M.; Korshunova, A.V.; Kostrukova, N.K.; Tourova, T.P.; Min, L.; Feng, Q.; Poltaraus, A.B. Diversity of metabolically active Bacteria in water-flooded high-temperature heavy oil reservoir. Front. Microbiol. 2017, 8, 707. [Google Scholar] [CrossRef] [Green Version]
- Magot, M.; Ravot, G.; Campaignolle, X.; Ollivier, B.; Patel, B.K.C.; Fardeau, M.L.; Thomas, P.; Crolet, J.L.; Garcia, J.L. Dethiosulfovibrio peptidovorans gen. nov.; sp. nov.; a new anaerobic, slightly halophilic, thiosulfate-reducing bacterium from corroding offshore oil wells. Int. J. Syst. Bacteriol. 1997, 47, 818–824. [Google Scholar] [CrossRef] [Green Version]
- Liang, R.; Grizzle, R.S.; Duncan, K.E.; Mcinerney, M.J.; Suflita, J.M. Roles of thermophilic thiosulfate-reducing bacteria and methanogenic archaea in the biocorrosion of oil pipelines. Front. Microbiol. 2014, 5, 89. [Google Scholar] [CrossRef] [Green Version]
- Wikieł, A.J.; Datsenko, I.; Vera, M.; Sand, W. Impact of Desulfovibrio alaskensis biofilms on corrosion behaviour of carbon steel in marine environment. Bioelectrochemistry 2014, 97, 52–60. [Google Scholar] [CrossRef]
- Xue, Y.; Voordouw, G. Control of microbial sulfide production with biocides and nitrate in oil reservoir simulating bioreactors. Front. Microbiol. 2015, 6, 1387. [Google Scholar] [CrossRef] [PubMed]
- Bødtker, G.; Lysnes, K.; Torsvik, T.; Bjørnestad, E.Ø.; Sunde, E. Microbial analysis of backflowed injection water from a nitrate-treated North Sea oil reservoir. J. Ind. Microbiol. Biotechnol. 2009, 36, 439–450. [Google Scholar] [CrossRef] [PubMed]
- Gittel, A.; Sorensen, K.B.; Skovhus, T.L.; Ingvorsen, K.; Schramm, A. Prokaryotic community structure and activity of sulfate reducers in production water from high-temperature oil reservoirs with and without nitrate treatment. Appl. Environ. Microbiol. 2009, 75, 7086–7096. [Google Scholar] [CrossRef] [Green Version]
- Santillan, E.-F.U.; Choi, W.; Bennett, P.C.; Leyris, J.D. The effects of biocide use on the microbiology and geochemistry of produced water in the Eagle Ford formation, Texas, USA. J. Pet. Sci. Eng. 2015, 135, 1–9. [Google Scholar] [CrossRef]
- Nazina, T.N.; Ivanova, A.E.; Blagov, A.V. Microbiological characteristics of the Mangyshlak peninsula oil reservoirs. Microbiology 1992, 61, 316–322. (In Russian) [Google Scholar]
- Agleshov, R.M. Use of a new miscible agent as a perceived need for Uzen oil field development. Oil Gas Fields Dev. 2019, 17, 57–67. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C. UNOISE2: Improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv 2016, 081257. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.C.; Flyvbjerg, H. Error filtering, pair assembly and error correction for next-generation sequencing reads. Bioinformatics 2015, 31, 3476–3482. [Google Scholar] [CrossRef] [Green Version]
- Maidak, B.L.; Cole, J.R.; Lilburn, T.G.; Parker, C.T., Jr.; Saxman, P.R.; Stredwick, J.M.; Garrity, G.M.; Li, B.; Olsen, G.J.; Pramanik, S.; et al. The RDP (ribosomal database project) continues. Nucleic Acids Res. 2000, 28, 173–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.H.; Oksanen, M.J.; Suggests, M.A.S.S. The vegan package. Community Ecol. Package 2007, 10, 631–637. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Community Ecology Package. R Package. Version 2.5-6. 2019. Available online: https://cran.ism.ac.jp/web/packages/vegan/vegan.pdf (accessed on 1 September 2019).
- Nagpal, S.; Haque, M.M.; Singh, R.; Mande, S.S. iVikodak—A Platform and standard workflow for inferring, analyzing, comparing, and visualizing the functional potential of microbial communities. Front. Microbiol. 2019, 9, 3336. [Google Scholar] [CrossRef] [Green Version]
- Quillet, L.; Besaury, L.; Popova, M.; Paisse, S.; Deloffre, J.; Ouddane, B. Abundance, diversity and activity of sulfate-reducing prokaryotes in heavy metal-contaminated sediment from a salt marsh in the Medway Estuary (UK). Mar. Biotechnol. 2012, 14, 363–381. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Suzek, B.E.; Wang, Y.; Huang, H.; McGarvey, P.B.; Wu, C.H. UniProt Consortium, UniRef clusters: A comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 2015, 31, 926–932. [Google Scholar] [CrossRef] [Green Version]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, D.S.; Semenova, E.M.; Grouzdev, D.S.; Ershov, A.P.; Bidzhieva, S.K.; Ivanova, A.E.; Babich, T.L.; Sissenbayeva, M.R.; Bisenova, M.A.; Nazina, T.N. Microbial diversity and potential sulfide producers in the Karazhanbas oil field (Kazakhstan). Microbiology 2020, 89, 459–469. [Google Scholar] [CrossRef]
- Widdel, F.; Bak, F. Gram-negative mesophilic sulfate-reducing bacteria. In The Prokaryotes, 2nd ed.; Balows, A., Trüper, H.G., Dworkin, M., Harder, W., Schleifer, K.-H., Eds.; Springer: New York, NY, USA, 1992; Volume 4, pp. 3352–3378. [Google Scholar]
- Pfennig, N.; Lippert, K.D. Über das vitamin B12—Bedürfnis phototropher Schweferelbakterien. Arch. Microbiol. 1966, 55, 245–256. [Google Scholar]
- Postgate, J.R. The Sulfate-Reducing Bacteria, 2nd ed.; Cambridge University Press: Cambridge, UK, 1984; p. 208. [Google Scholar]
- Trüper, H.G.; Schlegel, H.G. Sulfur metabolism in Thiorhodaceae. I. Quantitative measurements on growing cells of Chromatium okenii. Antonie Van Leeuwenhoek 1964, 30, 321–323. [Google Scholar] [CrossRef] [PubMed]
- Semenova, E.M.; Ershov, A.P.; Sokolova, D.S.; Tourova, T.P.; Nazina, T.N. Diversity and biotechnological potential of nitrate-reducing bacteria from heavy-oil reservoirs (Russia). Microbiology 2020, 89, 685–696. [Google Scholar] [CrossRef]
- Wang, H.; Cheng, H.; Wang, F.; Wei, D.; Wang, X. An improved 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) reduction assay for evaluating the viability of Escherichia coli cells. J. Microbiol. Methods 2010, 82, 330–333. [Google Scholar] [CrossRef]
- Plakunov, V.K.; Mart’yanov, S.V.; Teteneva, N.A.; Zhurina, M.V. A universal method for quantitative characterization of growth and metabolic activity of microbial biofilms in static models. Microbiology 2016, 85, 509–513. [Google Scholar] [CrossRef]
- Li, X.X.; Mbadinga, S.M.; Liu, J.F.; Zhou, L.; Yang, S.Z.; Gu, J.D.; Mu, B.Z. Microbiota and their affiliation with physiochemical characteristics of different subsurface petroleum reservoirs. Int. Biodeterior. Biodegrad. 2017, 120, 170–185. [Google Scholar] [CrossRef]
- An, B.A.; Shen, Y.; Voordouw, G. Control of sulfide production in high salinity Bakken Shale oil reservoirs by halophilic bacteria reducing nitrate to nitrite. Front. Microbiol. 2017, 8, 1164. [Google Scholar] [CrossRef] [Green Version]
- Feio, M.J.; Zinkevich, V.; Beech, I.B.; Llobet-Brossa, E.; Eaton, P.; Schmitt, J.; Guezennec, J. Desulfovibrio alaskensis sp. nov.: A sulphate-reducing bacterium from a soured oil reservoir. Int. J. Syst. Evol. Microbiol. 2004, 54, 1747–1752. [Google Scholar] [CrossRef] [Green Version]
- Mayilraj, S.; Kaksonen, A.H.; Cord-Ruwisch, R.; Schumann, P.; Sproer, C.; Tindall, B.J.; Spring, S. Desulfonauticus autotrophicus sp. nov.; a novel thermophilic sulfate-reducing bacterium isolated from oil-production water and emended description of the genus Desulfonauticus. Extremophiles 2009, 13, 247–255. [Google Scholar] [CrossRef]
- Thevenieau, F.; Fardeau, M.L.; Ollivier, B.; Joulian, C.; Baena, S. Desulfomicrobium thermophilum sp. nov.; a novel thermophilic sulphate-reducing bacterium isolated from a terrestrial hot spring in Colombia. Extremophiles 2007, 11, 295–303. [Google Scholar] [CrossRef]
- Nilsen, R.K.; Torsvik, T. Methanococcus thermolithotrophicus isolated from North Sea oil field reservoir water. Appl. Environ. Microbiol. 1996, 62, 728–731. [Google Scholar] [CrossRef] [Green Version]
- Orphan, V.J.; Taylor, L.T.; Hafenbradl, D.; Delong, E.F. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 2000, 66, 700–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huber, H.; Thomm, M.; Konig, H.; Thies, G.; Stetter, K.O. Methanococcus thermolithotrophicus, a novel thermophilic lithotrophic methanogen. Arch. Microbiol. 1982, 132, 47–50. [Google Scholar] [CrossRef] [Green Version]
- Takai, K.; Inoue, A.; Horikoshi, K. Methanothermococcus okinawensis sp. nov.; A thermophilic methane-producing archaeon isolated from a Western Pacific deep-sea hydrothermal vent system. Int. J. Syst. Evol. Microbiol. 2002, 52, 1089–1095. [Google Scholar] [CrossRef] [PubMed]
- L’Haridon, S.; Reysenbach, A.-L.; Glenat, P.; Prieur, D.; Jeanthon, C. Hot subterranean biosphere in a continental oil reservoir. Nature 1995, 377, 223–224. [Google Scholar] [CrossRef]
- Slobodkin, A.I.; Jeanthon, C.; L’Haridon, S.; Nazina, T.; Miroshnichenko, M.; Bonch-Osmolovskaya, E. Dissimilatory reduction of Fe(III) by thermophilic bacteria and archaea in deep subsurface petroleum reservoirs of western Siberia. Curr. Microbiol. 1999, 39, 99–102. [Google Scholar] [CrossRef]
- Ravot, G.; Ollivier, B.; Patel, B.K.C.; Magot, M.; Garcia, J.L. Emended description of Thermosipho africanus as a carbohydrate-fermenting species using thiosulfate as an electron acceptor. Int. J. Syst. Bacteriol. 1996, 46, 321–323. [Google Scholar] [CrossRef]
- Dahle, H.; Birkeland, N.-K. Thermovirga lienii gen. nov.; sp. nov.; A novel moderately thermophilic, anaerobic, amino-acid-degrading bacterium isolated from a North Sea oil well. Int. J. Syst. Evol. Microbiol. 2006, 56, 1539–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben Hania, W.; Godbane, R.; Postec, A.; Hamdi, M.; Ollivier, B.; Fardeau, M.-L. Defluviitoga tunisiensis gen. nov.; sp. nov.; A thermophilic bacterium isolated from a mesothermic and anaerobic whey digester. Int. J. Syst. Evol. Microbiol. 2012, 62, 1377–1382. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Greenfield, P.; Rosewarne, C.P.; Midgley, J. Draft genome sequence of Thermoanaerobacter sp. strain A7A, reconstructed from a metagenome obtained from a high-temperature hydrocarbon reservoir in the Bass Strait, Australia. Genome Announc. 2013, 1, e00701–e00713. [Google Scholar] [CrossRef] [Green Version]
- Piceno, Y.M.; Reid, F.C.; Tom, L.M.; Conrad, M.E.; Bill, M.; Hubbard, C.G.; Fouke, B.W.; Graff, C.J.; Han, J.; Stringfellow, W.T.; et al. Temperature and injection water source influence microbial community structure in four Alaskan North Slope hydrocarbon reservoirs. Front. Microbiol. 2014, 5, 409. [Google Scholar] [CrossRef] [PubMed]
- Davidova, I.A.; Duncan, K.E.; Choi, O.K.; Suflita, J.M. Desulfoglaeba alkanexedens gen. nov., sp. nov., an n-alkane-degrading, sulfate-reducing bacterium. Int. J. Syst. Evol. Microbiol. 2006, 56, 2737–2742. [Google Scholar] [CrossRef]
- Mochimaru, H.; Tamaki, H.; Hanada, S.; Imachi, H.; Nakamura, K.; Sakata, S.; Kamagata, Y. Methanolobus profundi sp. nov., a methylotrophic methanogen isolated from deep subsurface sediments in a natural gas field. Int. J. Syst. Evol. Microbiol. 2009, 59, 714–718. [Google Scholar] [CrossRef] [Green Version]
- Fida, T.T.; Chen, C.; Okpala, G.; Voordouw, G. Implications of limited thermophilicity of nitrite reduction for control of sulfide production in oil reservoirs. Appl. Environ. Microbiol. 2016, 82, 4190–4199. [Google Scholar] [CrossRef] [Green Version]
- Gardner, L.; Stewart, P. Action of glutaraldehyde and nitrite against sulfate-reducing bacterial biofilms. J. Ind. Microbiol. Biotechnol. 2002, 29, 354–360. [Google Scholar] [CrossRef]
- Korte, H.L.; Fels, S.R.; Christensen, G.A.; Price, M.N.; Kuehl, J.V.; Zane, G.M.; Deutschbauer, A.M.; Arkin, A.P.; Wall, J.D. Genetic basis for nitrate resistance in Desulfovibrio strains. Front. Microbiol. 2014, 5, 153. [Google Scholar] [CrossRef] [PubMed]
- Plugge, C.M.; Zhang, W.; Scholten, J.C.; Stams, A.J.M. Metabolic flexibility of sulfate-reducing bacteria. Front. Microbiol. 2011, 2, 81. [Google Scholar] [CrossRef] [Green Version]
- Daniels, L.; Belay, N.; Rajagopal, B.S.; Weimer, P.J. Bacterial methanogenesis and growth from CO2 with elemental iron as the sole source of electrons. Science 1987, 237, 509–511. [Google Scholar] [CrossRef]
- Dinh, H.T.; Kuever, J.; Muszmann, M.; Hassel, A.W.; Stratmann, M.; Widdel, F. Iron corrosion by novel anaerobic microorganisms. Nature 2004, 427, 829–832. [Google Scholar] [CrossRef] [PubMed]
- Mand, J.; Park, H.S.; Okoro, C.; Lomans, B.P.; Smith, S.; Chiejina, L.; Voordouw, G. Microbial methane production associated with carbon steel corrosion in a Nigerian oil field. Front. Microbiol. 2016, 6, 1538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vigneron, A.; Alsop, E.B.; Chambers, B.; Lomans, B.P.; Head, I.M.; Tsesmetzis, N. Complementary microorganisms in highly corrosive biofilms from an offshore oil production facility. Appl. Environ. Microbiol. 2016, 82, 2545–2554. [Google Scholar] [CrossRef] [Green Version]
- Narasingarao, P.; Haggblom, M.M. Pelobacter seleniigenes sp. nov., a selenate-respiring bacterium. Int. J. Syst. Evol. Microbiol. 2007, 57, 1937–1942. [Google Scholar] [CrossRef]
- Duncan, K.E.; Gieg, L.M.; Parisi, V.A.; Tanner, R.S.; Tringe, S.G.; Bristow, J.; Suflita, J.M. Biocorrosive thermophilic microbial communities in Alaskan North Slope oil facilities. Environ. Sci. Technol. 2009, 43, 7977–7984. [Google Scholar] [CrossRef] [Green Version]
- Hubert, C.R.J.; Oldenburg, T.B.P.; Fustic, M.; Gray, N.D.; Larter, S.R.; Penn, K.; Rowan, A.K.; Seshadri, R.; Sherry, A.; Swainsbury, R.; et al. Massive dominance of Epsilonproteobacteria in formation waters from a Canadian oil sands reservoir containing severely biodegraded oil. Environ. Microbiol. 2012, 14, 387–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gorby, Y.A.; Yanina, S.; McLean, J.S.; Rosso, K.M.; Moyles, D.; Dohnalkova, A.; Beveridge, T.J.; Chang, I.S.; Kim, B.H.; Kim, K.S.; et al. Electrically conductive bacterial nanowires produced by Shewanella oneidensis strain MR-1 and other microorganisms. Proc. Natl. Acad. Sci. USA 2006, 103, 11358–11363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovley, D.; Malvankar, N.S. Seeing is believing: Novel imaging techniques help clarify microbial nanowire structure and function. Environ. Microbiol. 2015, 17, 2209–2215. [Google Scholar] [CrossRef]
- Mercuri, E.G.F.; Vitule, J.R.S.; Kumata, A.Y.J.; Amaral, E.B. Energy by microbial fuel cells: Scientometric global synthesis and challenges. Renew. Sustain. Energy Rev. 2016, 65, 832–840. [Google Scholar] [CrossRef]
- Krumholz, L.R.; Bradstock, P.; Sheik, C.S.; Diao, Y.; Gazioglu, O.; Gorby, Y.; McInerney, M.J. Syntrophic growth of Desulfovibrio alaskensis requires genes for H2 and formate metabolism as well as those for flagellum and biofilm formation. Appl. Environ. Microbiol. 2015, 81, 2339–2348. [Google Scholar] [CrossRef]
- Martins, M.; Mourato, C.; Morais-Silva, F.O.; Rodrigues-Pousada, C.; Voordouw, G.; Wall, J.D.; Pereira, I.A. Electron transfer pathways of formate-driven H2 production in Desulfovibrio. Appl. Microbiol. Biotechnol. 2016, 100, 8135–8146. [Google Scholar] [CrossRef] [PubMed]
| Well Number, Sample | pH | Total Salinity, mg·L−1 | Content 1, mg L−1 | H2S, mg L−1 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Ca2+ | Mg2+ | Na+ + K+ | Cl− | SO42− | HCO3− | ||||
| 6928 | 6.7 | 42,534.6 | 2004.0 | 1337.6 | 12,417.7 | 26,303.9 | 44.4 | 427.0 | 107.6 |
| 8001 | 6.7 | 47,127.6 | 2204.4 | 1337.6 | 14,043.8 | 29,310.0 | 0.0 | 231.8 | 91.0 |
| 9064 | 6.8 | 30,838.9 | 1803.6 | 1094.4 | 8369.7 | 18,788.5 | 233.7 | 549.0 | 161.0 |
| 8533 | 8.0 | 14,039.1 | 1202.4 | 729.6 | 2948.6 | 8266.9 | 86.4 | 805.2 | 106.3 |
| 2874 | 6.5 | 48,765.2 | 2404.8 | 1216.0 | 14,618.8 | 30,062.0 | 0.0 | 463.6 | 5.6 |
| 9677 | 7.0 | 24,210.4 | 1202.4 | 972.8 | 6587.2 | 14,655.0 | 0.0 | 793.0 | 12.0 |
| 6916 | 6.2 | 120,556.4 | 6412.8 | 3040.0 | 35,737.4 | 75,154.0 | 65.8 | 146.4 | 15.6 |
| 7062 | 6.3 | 43,039.0 | 2004.0 | 1337.6 | 12,627.0 | 26,680.0 | 0.0 | 390.4 | 18.3 |
| 4256 | 6.3 | 80,533.4 | 4809.6 | 2432.0 | 22,636.6 | 50,399.0 | 0.0 | 256.2 | 7.7 |
| 6038 | 6.4 | 49,192.2 | 3006.0 | 3040.0 | 11,362.0 | 31,564.6 | 0.0 | 219.6 | 2.4 |
| 3257 | 6.2 | 63,139.2 | 4008.0 | 1824.0 | 17,631.8 | 39,455.8 | 0.0 | 219.6 | 16.2 |
| 8941 | 6.3 | 62,736.8 | 4408.8 | 1702.4 | 17,204.0 | 39,080.0 | 0.0 | 341.6 | 6.4 |
| 7574 | 6.4 | 36,344.6 | 2404.8 | 1702.4 | 8965.4 | 22,546.2 | 433.0 | 292.8 | 1.6 |
| 3239 | 6.6 | 37,902.3 | 2404.8 | 1945.6 | 9128.7 | 22,921.9 | 1281.7 | 219.6 | 19.9 |
| 2755-30 m3 | 6.6 | 51,410.5 | 3807.6 | 1459.2 | 13,763.2 | 31,941.5 | 48.6 | 390.4 | 2.5 |
| PWRI-1 | 6.7 | 54,183.2 | 3607.2 | 1216.0 | 15,446.8 | 33,443.5 | 103.7 | 366.0 | 1.6 |
| PWRI-2 | 6.6 | 58,570.1 | 4008.0 | 1702.4 | 15,991.9 | 36,450.0 | 51.8 | 366.0 | 0.0 |
| POTP | 6.6 | 41,362.9 | 3006.0 | 1580.8 | 10,511.0 | 25,928.1 | 19.8 | 317.2 | 2.4 |
| SW inlet | 7.5 | 12,890.8 | 1202.4 | 729.6 | 2267.8 | 5260.7 | 3161.9 | 268.4 | 0.0 |
| SW outlet | 7.5 | 11,764.2 | 1402.8 | 851.2 | 1444.4 | 4885.0 | 2912.4 | 268.4 | 0.5 |
| AW | 8.5 | 9609.5 | 200.4 | 170.2 | 3064.0 | 3907.0 | 2181.5 | 73.2 | 0.0 |
| Standard error | ±0.1 | ±0.09 | ±0.09 | ±0.02 | ±0.004 | ±0.02 | ±0.001 | ±0.02 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokolova, D.S.; Semenova, E.M.; Grouzdev, D.S.; Bidzhieva, S.K.; Babich, T.L.; Loiko, N.G.; Ershov, A.P.; Kadnikov, V.V.; Beletsky, A.V.; Mardanov, A.V.; et al. Sulfidogenic Microbial Communities of the Uzen High-Temperature Oil Field in Kazakhstan. Microorganisms 2021, 9, 1818. https://doi.org/10.3390/microorganisms9091818
Sokolova DS, Semenova EM, Grouzdev DS, Bidzhieva SK, Babich TL, Loiko NG, Ershov AP, Kadnikov VV, Beletsky AV, Mardanov AV, et al. Sulfidogenic Microbial Communities of the Uzen High-Temperature Oil Field in Kazakhstan. Microorganisms. 2021; 9(9):1818. https://doi.org/10.3390/microorganisms9091818
Chicago/Turabian StyleSokolova, Diyana S., Ekaterina M. Semenova, Denis S. Grouzdev, Salimat K. Bidzhieva, Tamara L. Babich, Nataliya G. Loiko, Alexey P. Ershov, Vitaly V. Kadnikov, Alexey V. Beletsky, Andrey V. Mardanov, and et al. 2021. "Sulfidogenic Microbial Communities of the Uzen High-Temperature Oil Field in Kazakhstan" Microorganisms 9, no. 9: 1818. https://doi.org/10.3390/microorganisms9091818
APA StyleSokolova, D. S., Semenova, E. M., Grouzdev, D. S., Bidzhieva, S. K., Babich, T. L., Loiko, N. G., Ershov, A. P., Kadnikov, V. V., Beletsky, A. V., Mardanov, A. V., Zhaparov, N. S., & Nazina, T. N. (2021). Sulfidogenic Microbial Communities of the Uzen High-Temperature Oil Field in Kazakhstan. Microorganisms, 9(9), 1818. https://doi.org/10.3390/microorganisms9091818







