Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide
Abstract
1. Introduction
2. Material and Methods
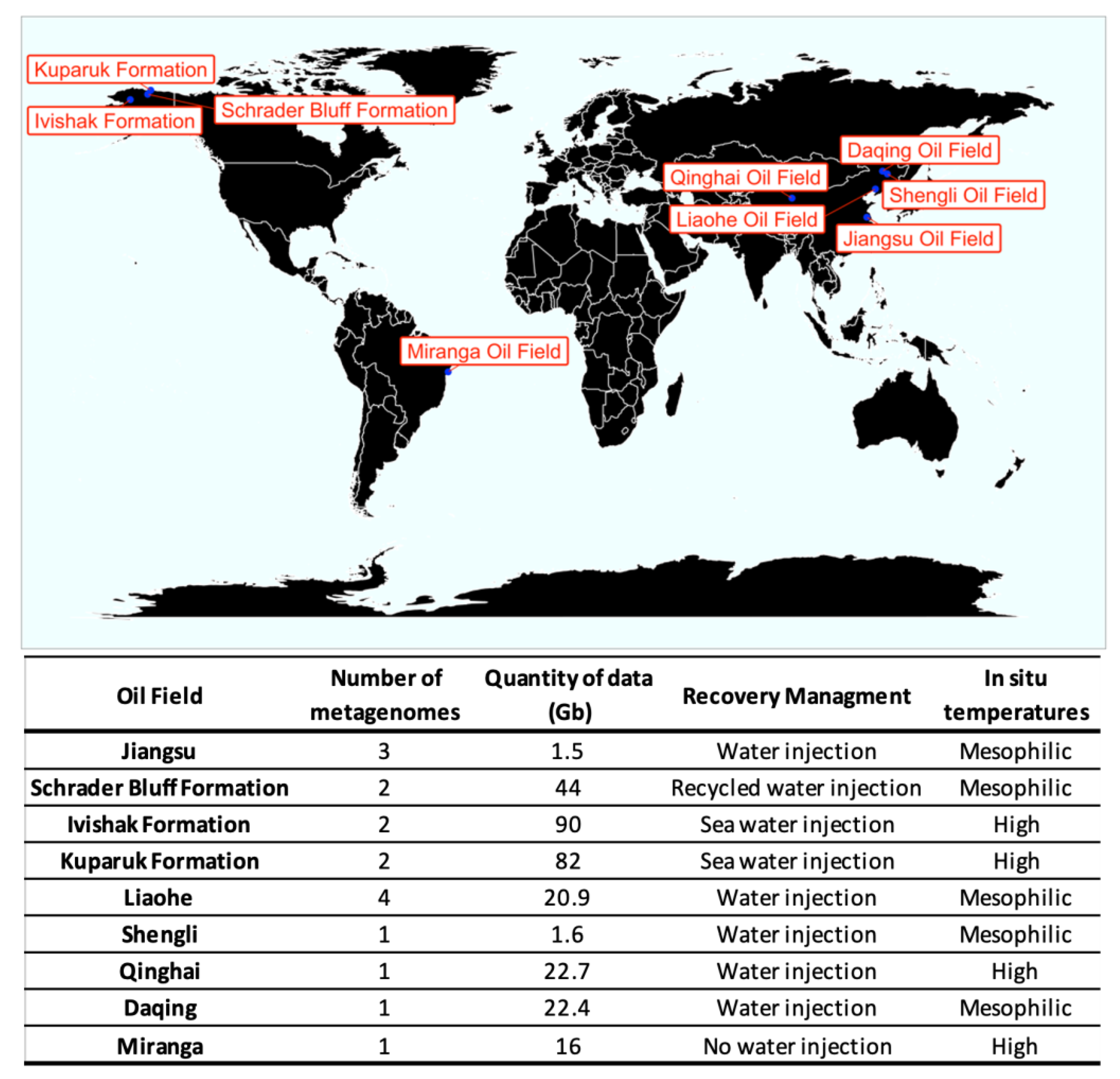
2.1. Data Acquisition
Description of Datasets
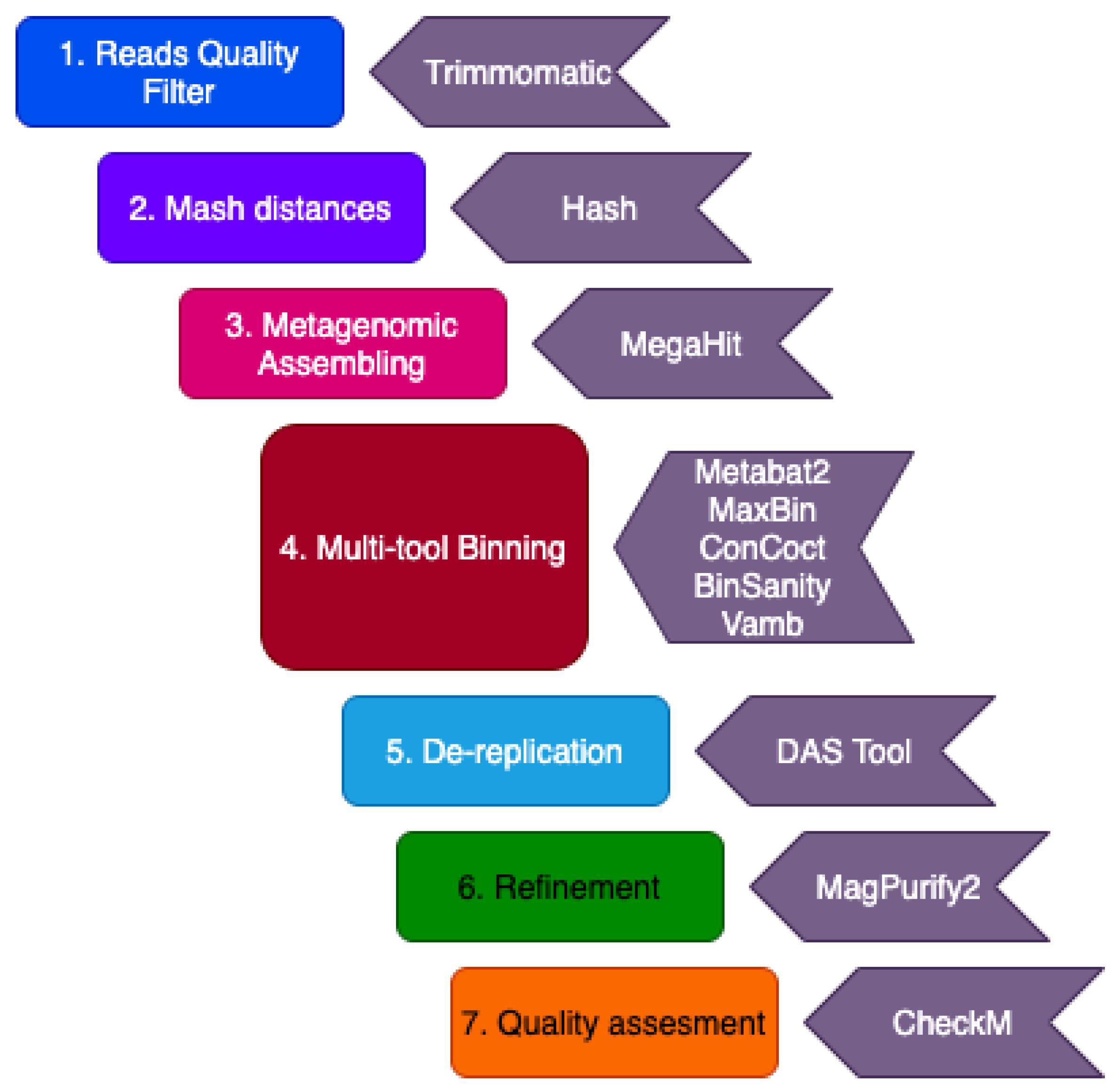
2.2. Computational Analysis
2.2.1. Quality Control and Filtering
2.2.2. Metagenome Assembly and Binning
2.3. Taxonomic Assignment and Phylogeny
2.4. Functional Assignment
2.5. Environment-Specific Orthogroups
2.6. Functional Core Analysis
3. Results & Discussion
3.1. Co-Assembling Statistics
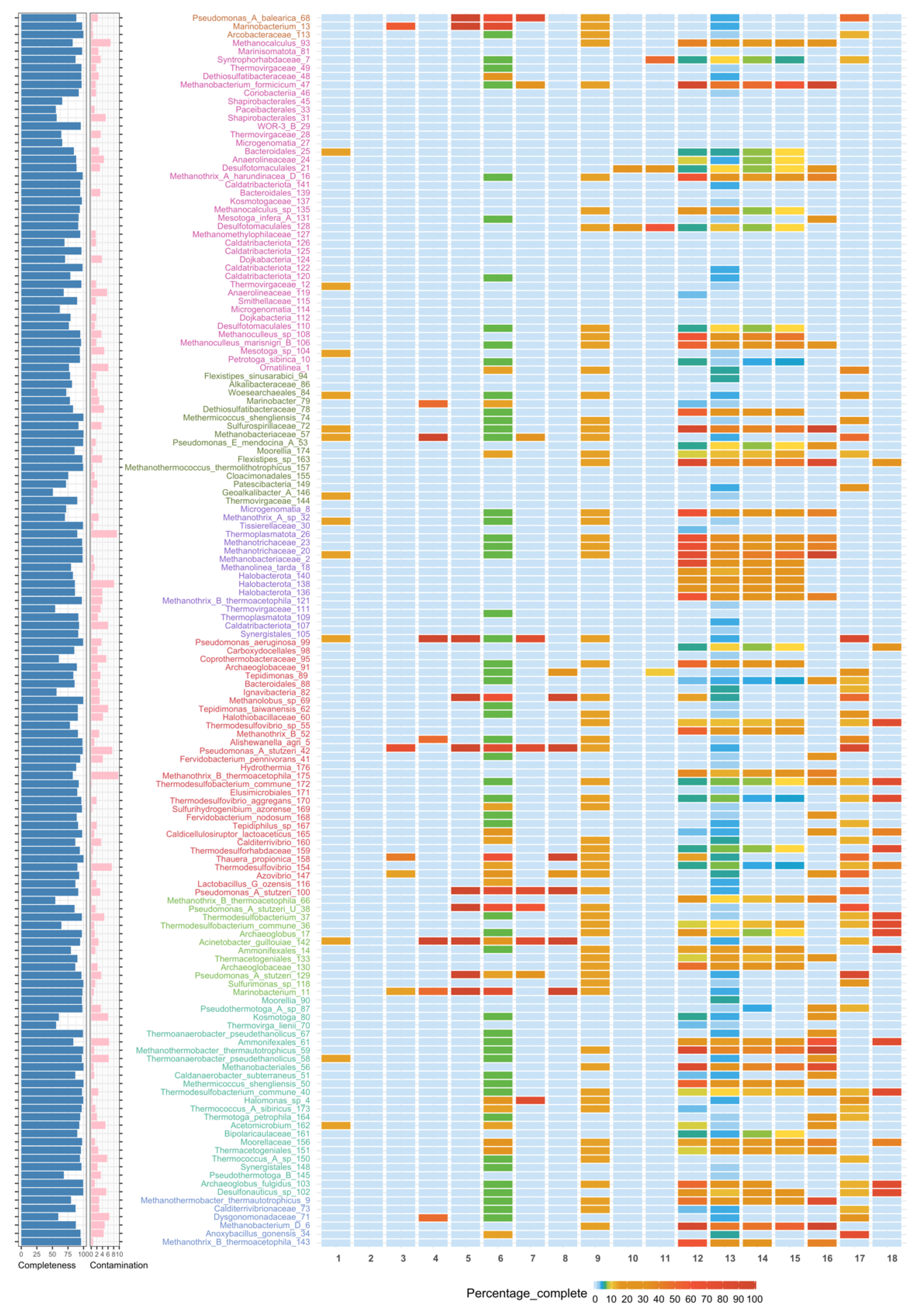
3.2. Binning
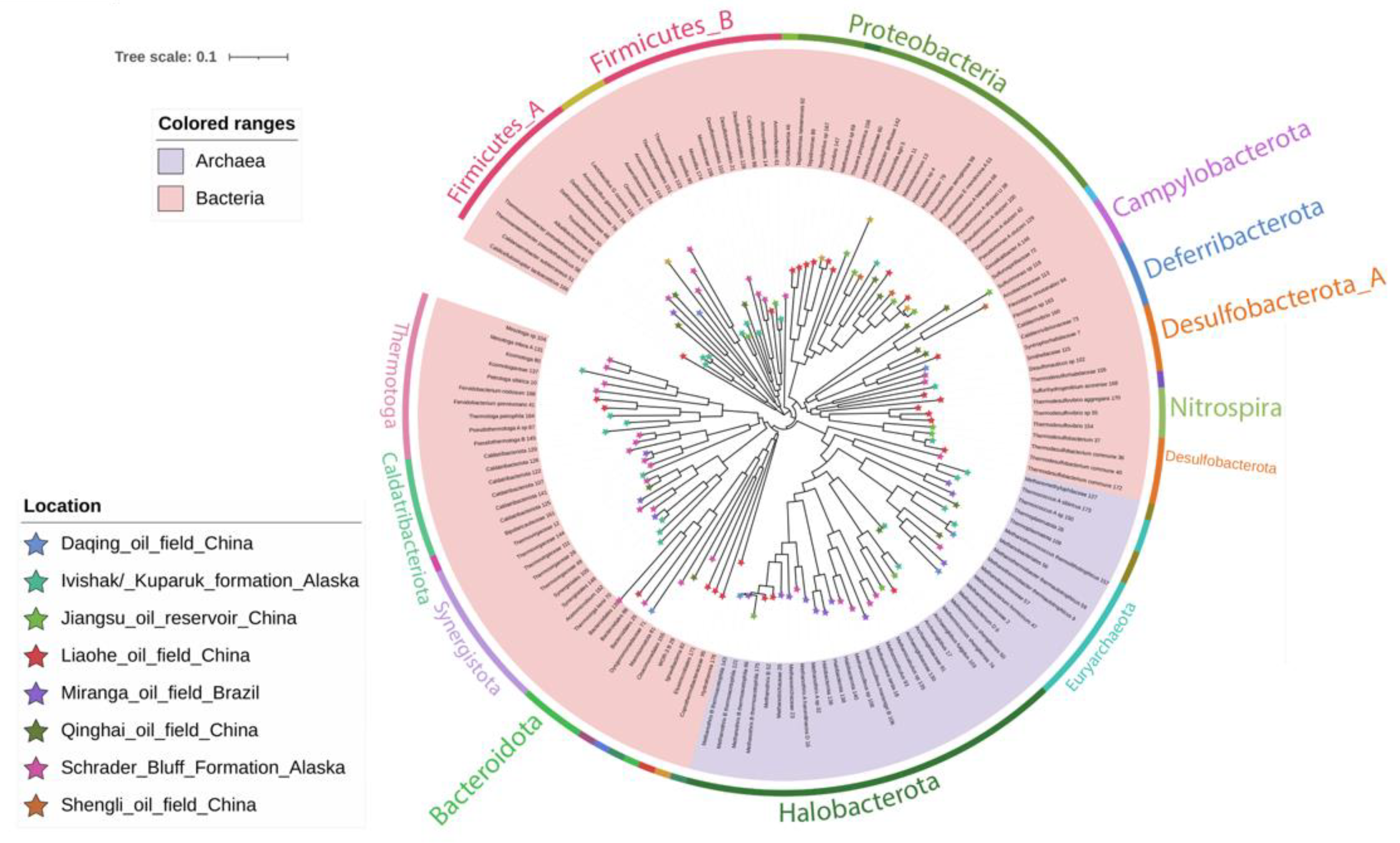
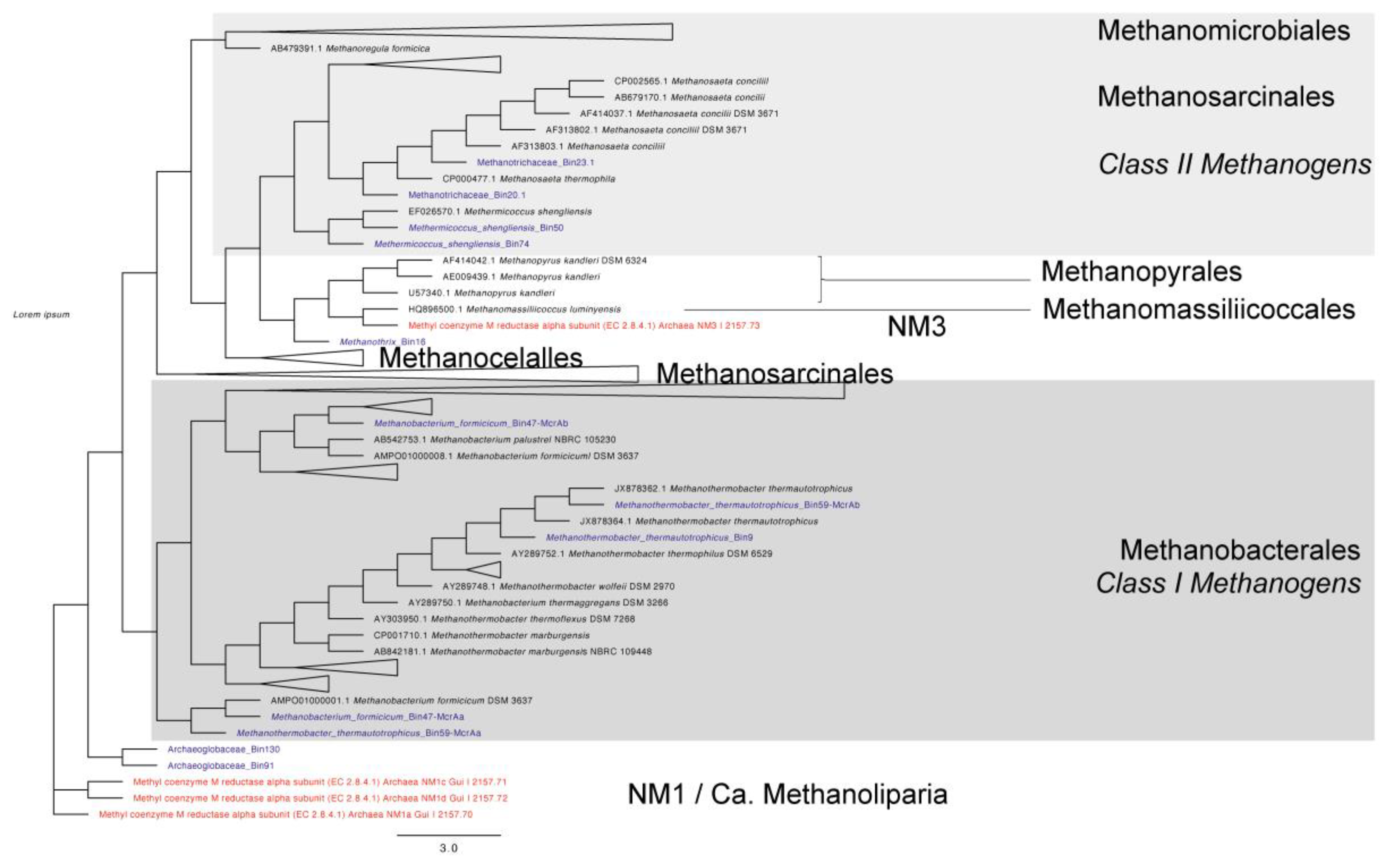
3.3. Taxonomic Affiliation and Phylogeny
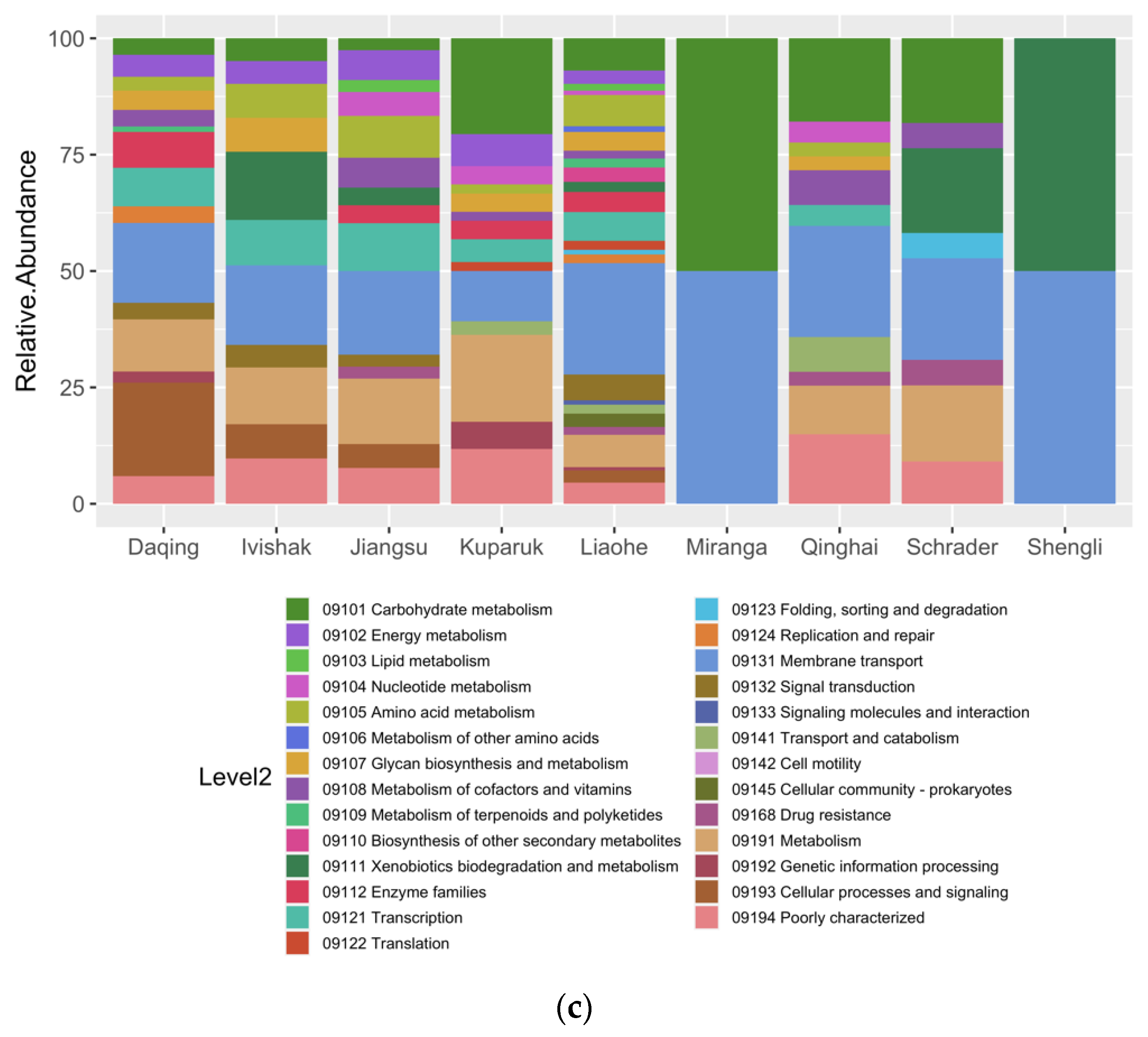
3.4. Metabolic Potential
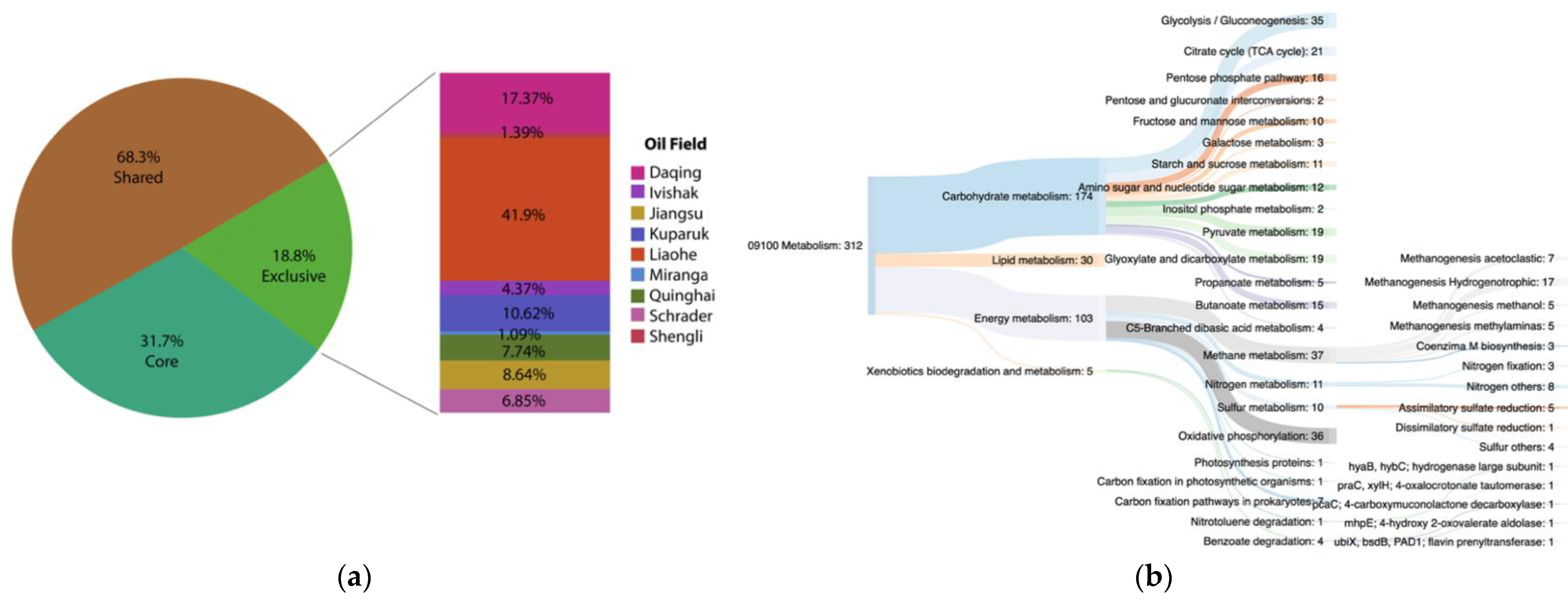
3.5. Functional Core Analysis
Environment-Specific Orthogroups
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Wang, X.; Li, X.; Yu, L.; Huang, L.; Xiu, J.; Lin, W.; Zhang, Y. Characterizing the microbiome in petroleum reservoir flooded by different water sources. Sci. Total Environ. 2018, 653, 872–885. [Google Scholar] [CrossRef]
- Nie, Y.; Zhao, J.-Y.; Tang, Y.-Q.; Guo, P.; Yang, Y.; Wu, X.-L.; Zhao, F. Species divergence vs. functional convergence characterizes crude oil microbial community assembly. Front. Microbiol. 2016, 7, 1254. [Google Scholar] [CrossRef]
- Sierra-Garcia, I.N.; Belgini, D.R.B.; Torres-Ballesteros, A.; Paez-Espino, D.; Capilla, R.; Santos Neto, E.V.; Gray, N.; de Oliveira, V.M. In depth metagenomic analysis in contrasting oil wells reveals syntrophic bacterial and archaeal associations for oil biodegradation in petroleum reservoirs. Sci. Total Environ. 2020, 715, 136646. [Google Scholar] [CrossRef]
- Sierra-Garcia, I.N.; Dellagnezze, B.M.; Santos, V.P.; Capilla, R.; Santos Neto, E.V.S.; Gray, N.; Oliveira, V.M. Microbial diversity in degraded and non-degraded petroleum samples and comparison across oil reservoirs at local and global scales. Extremophiles 2017, 21, 211–229. [Google Scholar] [CrossRef]
- An, D.; Caffrey, S.M.; Soh, J.; Agrawal, A.; Brown, D.; Budwill, K.; Dong, X.; Dunfield, P.F.; Foght, J.; Gieg, L.M.; et al. Metagenomics of hydrocarbon resource environments indicates aerobic taxa and genes to be unexpectedly common. Environ. Sci. Technol. 2013, 47, 10708–10717. [Google Scholar] [CrossRef]
- Hu, P.; Tom, L.; Singh, A.; Thomas, B.C.; Baker, B.J.; Piceno, Y.M.; Andersen, G.L.; Banfield, J.F. Genome-resolved metagenomic analysis reveals roles for candidate phyla and other microbial community members in biogeochemical transformations in oil reservoirs. mBio 2016, 7, e01669-15. [Google Scholar] [CrossRef]
- Kotlar, H.K.; Lewin, A.; Johansen, J.; Throne-Holst, M.; Haverkamp, T.; Markussen, S.; Winnberg, A.; Ringrose, P.; Aakvik, T.; Ryeng, E. High coverage sequencing of DNA from microorganisms living in an oil reservoir 2.5 kilometres subsurface. Environ. Microbiol. Rep. 2011, 3, 674–681. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A new view of the tree of life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.-A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Tyson, G.W.; Chapman, J.; Hugenholtz, P.; Allen, E.E.; Ram, R.J.; Richardson, P.M.; Solovyev, V.V.; Rubin, E.M.; Rokhsar, D.S.; Banfield, J.F. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004, 428, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.-F.; Galzerani, D.D.; Mbadinga, S.M.; Zaramela, L.S.; Gu, J.-D.; Mu, B.-Z.; Zengler, K. Metabolic capability and in situ activity of microorganisms in an oil reservoir. Microbiome 2018, 6, 5. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Chen, S.; Zhao, F.; Zhu, W. Whole metagenome of injected and produced fluids reveal the heterogenetic characteristics of the microbial community in a water-flooded oil reservoir. J. Pet. Sci. Eng. 2019, 176, 1198–1207. [Google Scholar] [CrossRef]
- Vigneron, A.; Alsop, E.B.; Lomans, B.P.; Kyrpides, N.C.; Head, I.M.; Tsesmetzis, N. Succession in the petroleum reservoir microbiome through an oil field production lifecycle. ISME J. 2017, 11, 2141–2154. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2010. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 7 September 2020).
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Ondov, B.D.; Treangen, T.J.; Melsted, P.; Mallonee, A.B.; Bergman, N.H.; Koren, S.; Phillippy, A.M. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016, 17, 132. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Liu, C.-M.; Luo, R.; Sadakane, K.; Lam, T.-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 2015, 31, 1674–1676. [Google Scholar] [CrossRef]
- Mikheenko, A.; Saveliev, V.; Gurevich, A. MetaQUAST: Evaluation of metagenome assemblies. Bioinformatics 2016, 32, 1088–1090. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-W.; Simmons, B.A.; Singer, S.W. MaxBin 2.0: An automated binning algorithm to recover genomes from multiple metagenomic datasets. Bioinformatics 2015, 32, 605–607. [Google Scholar] [CrossRef]
- Alneberg, J.; Bjarnason, B.S.; de Bruijn, I.; Schirmer, M.; Quick, J.; Ijaz, U.Z.; Lahti, L.; Loman, N.J.; Andersson, A.F.; Quince, C. Binning metagenomic contigs by coverage and composition. Nat. Methods 2014, 11, 1144–1146. [Google Scholar] [CrossRef]
- Graham, E.D.; Heidelberg, J.F.; Tully, B.J. BinSanity: Unsupervised clustering of environmental microbial assemblies using coverage and affinity propagation. PeerJ 2017, 5, e3035. [Google Scholar] [CrossRef]
- Nissen, J.N.; Johansen, J.; Allesøe, R.L.; Sønderby, C.K.; Armenteros, J.J.A.; Grønbech, C.H.; Jensen, L.J.; Nielsen, H.B.; Petersen, T.N.; Winther, O.; et al. Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 2021, 39, 555–560. [Google Scholar] [CrossRef]
- Sieber, C.M.K.; Probst, A.J.; Sharrar, A.; Thomas, B.C.; Hess, M.; Tringe, S.G.; Banfield, J.F. Recovery of genomes from metagenomes via a dereplication, aggregation and scoring strategy. Nat. Microbiol. 2018, 3, 836–843. [Google Scholar] [CrossRef]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Wheeler, T.J.; Eddy, S.R. nhmmer: DNA homology search with profile HMMs. Bioinformatics 2013, 29, 2487–2489. [Google Scholar] [CrossRef]
- Bowers, R.M.; Kyrpides, N.C.; Stepanauskas, R.; Harmon-Smith, M.; Doud, D.; Reddy, T.B.K.; Schulz, F.; Jarett, J.; Rivers, A.R.; Eloe-Fadrosh, E.A.; et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 2017, 35, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Chaumeil, P.-A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the genome taxonomy database. Bioinformatics 2019, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Chaumeil, P.-A.; Rinke, C.; Mussig, A.J.; Hugenholtz, P. A complete domain-to-species taxonomy for Bacteria and Archaea. Nat. Biotechnol. 2020, 38, 1079–1086. [Google Scholar] [CrossRef]
- Parks, D.H.; Chuvochina, M.; Waite, D.W.; Rinke, C.; Skarshewski, A.; Chaumeil, P.-A.; Hugenholtz, P. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat. Biotechnol. 2018, 36, 996–1004. [Google Scholar] [CrossRef]
- Matsen, F.A.; Kodner, R.B.; Armbrust, E.V. Pplacer: Linear time maximum-likelihood and Bayesian phylogenetic placement of sequences onto a fixed reference tree. BMC Bioinform. 2010, 11, 538. [Google Scholar] [CrossRef]
- Segata, N.; Börnigen, D.; Morgan, X.C.; Huttenhower, C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat. Commun. 2013, 4, 1–11. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2014, 12, 59–60. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef]
- Camargo, A.P.; Vasconcelos, A.A.; Fiamenghi, M.B.; Pereira, G.A.; Carazzolle, M.F. Tspex: A Tissue-Specificity Calculator for Gene Expression Data. 2020. Available online: https://assets.researchsquare.com/files/rs-51998/v1/ec31ccc0-329a-46e0-bfef-c9e409b1edcb.pdf?c=1596577796 (accessed on 5 May 2021). [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Jensen, L.J.; Julien, P.; Kuhn, M.; von Mering, C.; Muller, J.; Doerks, T.; Bork, P. EggNOG: Automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2007, 36, D250–D254. [Google Scholar] [CrossRef]
- Brown, C.T.; Hug, L.A.; Thomas, B.C.; Sharon, I.; Castelle, C.J.; Singh, A.; Wilkins, M.J.; Wrighton, K.C.; Williams, K.H.; Banfield, J.F. Unusual biology across a group comprising more than 15% of domain bacteria. Nature 2015, 523, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef]
- Herrmann, M.; Wegner, C.-E.; Taubert, M.; Geesink, P.; Lehmann, K.; Yan, L.; Lehmann, R.; Totsche, K.U.; Küsel, K. Predominance of cand. patescibacteria in groundwater is caused by their preferential mobilization from soils and flourishing under oligotrophic conditions. Front. Microbiol. 2019, 10, 1407. [Google Scholar] [CrossRef]
- Hubalek, V.; Wu, X.; Eiler, A.; Buck, M.; Heim, C.; Dopson, M.; Bertilsson, S.; Ionescu, D. Connectivity to the surface determines diversity patterns in subsurface aquifers of the Fennoscandian shield. ISME J. 2016, 10, 2447–2458. [Google Scholar] [CrossRef]
- León-Zayas, R.; Peoples, L.; Biddle, J.F.; Podell, S.; Novotny, M.; Cameron, J.; Lasken, R.S.; Bartlett, D.H. The metabolic potential of the single cell genomes obtained from the Challenger Deep, Mariana Trench within the candidate superphylum P arcubacteria (OD 1). Environ. Microbiol. 2017, 19, 2769–2784. [Google Scholar] [CrossRef] [PubMed]
- Luef, B.; Frischkorn, K.R.; Wrighton, K.C.; Holman, H.-Y.N.; Birarda, G.; Thomas, B.C.; Singh, A.; Williams, K.H.; Siegerist, C.E.; Tringe, S.G.; et al. Diverse uncultivated ultra-small bacterial cells in groundwater. Nat. Commun. 2015, 6, 6372. [Google Scholar] [CrossRef]
- Antunes, T.C.; Marconatto, L.; Borges, L.G.d.A.; Giongo, A.; Sand, S.T.V.D. Analysis of microbial community biodiversity in activated sludge from a petrochemical plant. Rev. Ambiente Água 2021, 16. [Google Scholar] [CrossRef]
- Eze, M.O.; Lütgert, S.A.; Neubauer, H.; Balouri, A.; Kraft, A.A.; Sieven, A.; Daniel, R.; Wemheuer, B. Metagenome assembly and metagenome-assembled genome sequences from a historical oil field located in Wietze, Germany. Microbiol. Resour. Announc. 2020, 9. [Google Scholar] [CrossRef]
- Tian, R.; Ning, D.; He, Z.; Zhang, P.; Spencer, S.J.; Gao, S.; Shi, W.; Wu, L.; Zhang, Y.; Yang, Y.; et al. Small and mighty: Adaptation of superphylum Patescibacteria to groundwater environment drives their genome simplicity. Microbiome 2020, 8, 51. [Google Scholar] [CrossRef]
- Castelle, C.J.; Brown, C.T.; Anantharaman, K.; Probst, A.J.; Huang, R.H.; Banfield, J.F. Biosynthetic capacity, metabolic variety and unusual biology in the CPR and DPANN radiations. Nat. Rev. Microbiol. 2018, 16, 629–645. [Google Scholar] [CrossRef]
- Shelton, J.L.; Andrews, R.S.; Akob, D.M.; DeVera, C.A.; Mumford, A.; McCray, J.E.; McIntosh, J.C. Microbial community composition of a hydrocarbon reservoir 40 years after a CO2 enhanced oil recovery flood. FEMS Microbiol. Ecol. 2018, 94. [Google Scholar] [CrossRef]
- Alsouleman, K.; Linke, B.; Klang, J.; Klocke, M.; Krakat, N.; Theuerl, S. Reorganisation of a mesophilic biogas microbiome as response to a stepwise increase of ammonium nitrogen induced by poultry manure supply. Bioresour. Technol. 2016, 208, 200–204. [Google Scholar] [CrossRef]
- Gao, P.; Tian, H.; Wang, Y.; Li, Y.; Li, Y.; Xie, J.; Zeng, B.; Zhou, J.; Li, G.; Ma, T. Spatial isolation and environmental factors drive distinct bacterial and archaeal communities in different types of petroleum reservoirs in China. Sci. Rep. 2016, 6, 20174. [Google Scholar] [CrossRef]
- Zhao, J.-Y.; Hu, B.; Dolfing, J.; Li, Y.; Tang, Y.-Q.; Jiang, Y.; Chi, C.-Q.; Xing, J.; Nie, Y.; Wu, X.-L. Thermodynamically favorable reactions shape the archaeal community affecting bacterial community assembly in oil reservoirs. Sci. Total Environ. 2021, 781, 146506. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Z.; Gou, M.; Yi, Y.; Xia, Z.-Y.; Tang, Y.-Q. Identification of novel potential acetate-oxidizing bacteria in an acetate-fed methanogenic chemostat based on DNA stable isotope probing. J. Gen. Appl. Microbiol. 2018, 64, 221–231. [Google Scholar] [CrossRef]
- Sengupta, K.; Pal, S. A review on microbial diversity and genetic markers involved in methanogenic degradation of hydrocarbons: Futuristic prospects of biofuel recovery from contaminated regions. Environ. Sci. Pollut. Res. 2021, 28, 40288–40307. [Google Scholar] [CrossRef]
- Oren, A. The family Methanotrichaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Gemany, 2014; pp. 298–306. [Google Scholar]
- Akinyemi, T.S.; Shao, N.; Whitman, W.B. Methanothrix. In Bergey’s Manual of Systematics of Archaea and Bacteria; Springer: Berlin/Heidelberg, Germany, 2014; pp. 297–306. [Google Scholar] [CrossRef]
- Holmes, D.E.; Shrestha, P.M.; Walker, D.J.F.; Dang, Y.; Nevin, K.P.; Woodard, T.L.; Lovley, D.R. Metatranscriptomic evidence for direct interspecies electron transfer between geobacter and methanothrix species in methanogenic rice paddy soils. Appl. Environ. Microbiol. 2017, 83, e00223-17. [Google Scholar] [CrossRef]
- John, E.S.; Flores, G.E.; Meneghin, J.; Reysenbach, A.-L. Deep-sea hydrothermal vent metagenome–Assembled genomes provide insight into the phylum Nanoarchaeota. Environ. Microbiol. Rep. 2019, 11, 262–270. [Google Scholar] [CrossRef]
- Munson-McGee, J.H.; Field, E.K.; Bateson, M.; Rooney, C.; Stepanauskas, R.; Young, M.J.; Wommack, K.E. Nanoarchaeota, their sulfolobales host, and nanoarchaeota virus distribution across Yellowstone national park hot springs. Appl. Environ. Microbiol. 2015, 81, 7860–7868. [Google Scholar] [CrossRef] [PubMed]
- Carrier, V.; Svenning, M.M.; Gründger, F.; Niemann, H.; Dessandier, P.-A.; Panieri, G.; Kalenitchenko, D. The impact of methane on microbial communities at marine arctic gas hydrate bearing sediment. Front. Microbiol. 2020, 11, 1932. [Google Scholar] [CrossRef]
- Casanueva, A.; Galada, N.; Baker, G.C.; Grant, W.D.; Heaphy, S.; Jones, B.; Yanhe, M.; Ventosa, A.; Blamey, J.; Cowan, D.A. Nanoarchaeal 16S rRNA gene sequences are widely dispersed in hyperthermophilic and mesophilic halophilic environments. Extremophiles 2008, 12, 651–656. [Google Scholar] [CrossRef]
- Chernyh, N.A.; Mardanov, A.V.; Gumerov, V.M.; Miroshnichenko, M.L.; Lebedinsky, A.V.; Merkel, A.Y.; Crowe, D.; Pimenov, N.V.; Rusanov, I.I.; Ravin, N.V.; et al. Microbial life in Bourlyashchy, the hottest thermal pool of Uzon Caldera, Kamchatka. Extremophiles 2015, 19, 1157–1171. [Google Scholar] [CrossRef]
- Clingenpeel, S.; Kan, J.; Macur, R.; Woyke, T.; Lovalvo, D.; Varley, J.; Inskeep, W.P.; Nealson, K.; McDermott, T. Yellowstone lake nanoarchaeota. Front. Microbiol. 2013, 4, 274. [Google Scholar] [CrossRef]
- Flores, G.; Shakya, M.; Meneghin, J.; Yang, Z.; Seewald, J.; Geoff Wheat, C.; Podar, M.; Reysenbach, A.L. Inter-field variability in the microbial communities of hydrothermal vent deposits from a back-arc basin. Geobiology 2012, 10, 333–346. [Google Scholar] [CrossRef]
- Flores, G.E.; Campbell, J.H.; Kirshtein, J.D.; Meneghin, J.; Podar, M.; Steinberg, J.I.; Seewald, J.S.; Tivey, M.K.; Voytek, M.A.; Yang, Z.K.; et al. Microbial community structure of hydrothermal deposits from geochemically different vent fields along the Mid-Atlantic Ridge. Environ. Microbiol. 2011, 13, 2158–2171. [Google Scholar] [CrossRef]
- Hohn, M.J.; Hedlund, B.P.; Huber, H. Detection of 16S rDNA sequences representing the novel phylum “Nanoarchaeota”: Indication for a wide distribution in high temperature biotopes. Syst. Appl. Microbiol. 2002, 25, 551–554. [Google Scholar] [CrossRef] [PubMed][Green Version]
- McCliment, E.A.; Voglesonger, K.M.; O’Day, P.A.; Dunn, E.E.; Holloway, J.R.; Cary, S.C. Colonization of nascent, deep-sea hydrothermal vents by a novel Archaeal and Nanoarchaeal assemblage. Environ. Microbiol. 2006, 8, 114–125. [Google Scholar] [CrossRef] [PubMed]
- Zemskaya, T.I.; Cabello-Yeves, P.J.; Pavlova, O.N.; Rodriguez-Valera, F. Microorganisms of lake Baikal—The deepest and most ancient lake on Earth. Appl. Microbiol. Biotechnol. 2020, 104, 6079–6090. [Google Scholar] [CrossRef]
- Mayumi, D.; Mochimaru, H.; Tamaki, H.; Yamamoto, K.; Yoshioka, H.; Suzuki, Y.; Kamagata, Y.; Sakata, S. Methane production from coal by a single methanogen. Science 2016, 354, 222–225. [Google Scholar] [CrossRef]
- Cheng, L.; Ding, C.; Li, Q.; He, Q.; Dai, L.-R.; Zhang, H. DNA-SIP reveals that syntrophaceae play an important role in methanogenic hexadecane degradation. PLoS ONE 2013, 8, e66784. [Google Scholar] [CrossRef]
- Gao, P.; Tian, H.; Li, G.; Sun, H.; Ma, T. Microbial diversity and abundance in the Xinjiang Luliang long-term water-flooding petroleum reservoir. Microbiol. Open 2015, 4, 332–342. [Google Scholar] [CrossRef]
- Walker, D.J.F.; Adhikari, R.Y.; Holmes, D.E.; Ward, J.E.; Woodard, T.L.; Nevin, K.P.; Lovley, D.R. Electrically conductive pili from pilin genes of phylogenetically diverse microorganisms. ISME J. 2018, 12, 48–58. [Google Scholar] [CrossRef]
- Greene, A.C.; Patel, B.K.C.; Yacob, S. Geoalkalibacter subterraneus sp. nov., an anaerobic Fe(III)- and Mn(IV)-reducing bacterium from a petroleum reservoir, and emended descriptions of the family Desulfuromonadaceae and the genus Geoalkalibacter. Int. J. Syst. Evol. Microbiol. 2009, 59, 781–785. [Google Scholar] [CrossRef]
- Piceno, Y.M.; Reid, F.C.; Tom, L.M.; Conrad, M.E.; Bill, M.; Hubbard, C.G.; Fouke, B.W.; Graff, C.J.; Han, J.; Stringfellow, W.T.; et al. Temperature and injection water source influence microbial community structure in four Alaskan North Slope hydrocarbon reservoirs. Front. Microbiol. 2014, 5, 409. [Google Scholar] [CrossRef]
- Mayilraj, S.; Kaksonen, A.H.; Cord-Ruwisch, R.; Schumann, P.; Spröer, C.; Tindall, B.J.; Spring, S. Desulfonauticus autotrophicus sp. nov., a novel thermophilic sulfate-reducing bacterium isolated from oil-production water and emended description of the genus Desulfonauticus. Extremophiles 2009, 13, 247–255. [Google Scholar] [CrossRef]
- Orphan, V.; Taylor, L.; Hafenbradl, D.; Delong, E. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 2000, 66, 700–711. [Google Scholar] [CrossRef]
- Shestakova, N.; Ivoilov, V.; Tourova, T.; Belyaev, S.; Poltaraus, A.; Nazina, T. Application of clone libraries: Syntrophic acetate degradation to methane in a high-temperature petroleum reservoir: Culture-based and 16S rRNA genes characterization. In Applied Microbiology and Molecular Biology in Oil Field Systems; Springer: Cham, Switzerland, 2011; pp. 45–53. [Google Scholar]
- Mnif, S.; Chamkha, M.; Sayadi, S. Isolation and characterization of Halomonas sp. strain C2SS100, a hydrocarbon-degrading bacterium under hypersaline conditions. J. Appl. Microbiol. 2009, 107, 785–794. [Google Scholar] [CrossRef]
- Cheng, L.; He, Q.; Ding, C.; Dai, L.-R.; Li, Q.; Zhang, H. Novel bacterial groups dominate in a thermophilic methanogenic hexadecane-degrading consortium. FEMS Microbiol. Ecol. 2013, 85, 568–577. [Google Scholar] [CrossRef]
- Mbadinga, S.M.; Li, K.-P.; Zhou, L.; Wang, L.-Y.; Yang, S.-Z.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z. Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl. Microbiol. Biotechnol. 2012, 96, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, K.-P.; Mbadinga, S.M.; Yang, S.-Z.; Gu, J.-D.; Mu, B.-Z. Analyses of n-alkanes degrading community dynamics of a high-temperature methanogenic consortium enriched from production water of a petroleum reservoir by a combination of molecular techniques. Ecotoxicology 2012, 21, 1680–1691. [Google Scholar] [CrossRef]
- Pournia, M.; Bahador, N.; Tabatabaei, M.; Azarbayjani, R.; Salekdeh, G.H. Microbial diversity of non-flooded high temperature petroleum reservoir in South of Iran. Biol. J. Microorg. 2020, 8, 15–23. [Google Scholar]
- Dahle, H.; Garshol, F.; Madsen, M.; Birkeland, N.-K. Microbial community structure analysis of produced water from a high-temperature North Sea oil-field. Antonie Van Leeuwenhoek 2008, 93, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Magot, M.; Ollivier, B.; Patel, B.K.C. Microbiology of petroleum reservoirs. Antonie Van Leeuwenhoek 2000, 77, 103–116. [Google Scholar] [CrossRef]
- Henry, E.A.; Devereux, R.; Maki, J.S.; Gilmour, C.C.; Woese, C.R.; Mandelco, L.; Schauder, R.; Remsen, C.C.; Mitchell, R. Characterization of a new thermophilic sulfate-reducing bacterium. Arch. Microbiol. 1994, 161, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Meslé, M.; Dromart, G.; Oger, P. Microbial methanogenesis in subsurface oil and coal. Res. Microbiol. 2013, 164, 959–972. [Google Scholar] [CrossRef] [PubMed]
- Nazina, T.N.; Shestakova, N.M.; Semenova, E.M.; Korshunova, A.V.; Kostrukova, N.K.; Tourova, T.P.; Min, L.; Feng, Q.; Poltaraus, A.B. Diversity of metabolically active bacteria in water-flooded high-temperature heavy oil reservoir. Front. Microbiol. 2017, 8, 707. [Google Scholar] [CrossRef]
- Silva, T.R.; Verde, L.C.L.; Santos Neto, E.V.; Oliveira, V.M. Diversity analyses of microbial communities in petroleum samples from Brazilian oil fields. Int. Biodeterior. Biodegrad. 2013, 81, 14. [Google Scholar] [CrossRef]
- Liu, J.-F.; Wu, W.-L.; Yao, F.; Wang, B.; Zhang, B.-L.; Mbadinga, S.M.; Gu, J.-D.; Mu, B.-Z. A thermophilic nitrate-reducing bacterium isolated from production water of a high temperature oil reservoir and its inhibition on sulfate-reducing bacteria. Appl. Environ. Biotechnol. 2016, 2, 35–42. [Google Scholar] [CrossRef]
- Tüccar, T.; Ilhan-Sungur, E.; Abbas, B.; Muyzer, G. Coexistence of sulfate reducers with the other oil bacterial groups in Diyarbakır oil fields. Anaerobe 2019, 59, 19–31. [Google Scholar] [CrossRef]
- Nazaries, L.; Murrell, J.C.; Millard, P.; Baggs, L.; Singh, B.K. Methane, microbes and models: Fundamental understanding of the soil methane cycle for future predictions. Environ. Microbiol. 2013, 15, 2395–2417. [Google Scholar] [CrossRef] [PubMed]
- Penger, J.; Conrad, R.; Blaser, M. Stable carbon isotope fractionation by methylotrophic methanogenic archaea. Appl. Environ. Microbiol. 2012, 78, 7596–7602. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.C.D.; Lopes, D.R.G.; Da Silva, J.D.; De Oliveira, M.D.; Dias, R.S.; Lima, H.S.; De Sousa, M.P.; De Paula, S.O.; Silva, C.C.D. Diversity of sulfate-reducing prokaryotes in petroleum production water and oil samples. Int. Biodeterior. Biodegrad. 2020, 151, 104966. [Google Scholar] [CrossRef]
- Summers, Z.M.; Belahbib, H.; Pradel, N.; Bartoli, M.; Mishra, P.; Tamburini, C.; Dolla, A.; Ollivier, B.; Armougom, F. A novel Thermotoga strain TFO isolated from a Californian petroleum reservoir phylogenetically related to Thermotoga petrophila and T. naphthophila, two thermophilic anaerobic isolates from a Japanese reservoir: Taxonomic and genomic considerations. Syst. Appl. Microbiol. 2020, 43, 126132. [Google Scholar] [CrossRef] [PubMed]
- Gieg, L. Microbial communities in oil shales, biodegraded and heavy oil reservoirs, and bitumen deposits. In Microbial Communities Utilizing Hydrocarbons and Lipids: Members, Metagenomics and Ecophysiology; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Qiu, Y.-L.; Hanada, S.; Ohashi, A.; Harada, H.; Kamagata, Y.; Sekiguchi, Y. Syntrophorhabdus aromaticivorans gen. nov., sp. nov., the first cultured anaerobe capable of degrading phenol to acetate in obligate syntrophic associations with a hydrogenotrophic methanogen. Appl. Environ. Microbiol. 2008, 74, 2051–2058. [Google Scholar] [CrossRef]
- Kuever, J. The family Syntrophorhabdaceae. In The Prokaryotes: Deltaproteobacteria and Epsilonproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 301–303. [Google Scholar]
- Dong, X.; Greening, C.; Rattray, J.E.; Chakraborty, A.; Chuvochina, M.; Mayumi, D.; Dolfing, J.; Li, C.; Brooks, J.M.; Bernard, B.B.; et al. Metabolic potential of uncultured bacteria and archaea associated with petroleum seepage in deep-sea sediments. Nat. Commun. 2019, 10, 1816. [Google Scholar] [CrossRef]
- Aitken, C.M.; Jones, D.M.; Maguire, M.J.; Gray, N.D.; Sherry, A.; Bowler, B.F.J.; Ditchfield, A.K.; Larter, S.R.; Head, I.M. Evidence that crude oil alkane activation proceeds by different mechanisms under sulfate-reducing and methanogenic conditions. Geochim. Cosmochim. Acta 2013, 109, 162–174. [Google Scholar] [CrossRef]
- Davis, J.J.; Wattam, A.R.; Aziz, R.K.; Brettin, T.; Butler, R.; Butler, R.M.; Chlenski, P.; Conrad, N.; Dickerman, A.; Dietrich, E.M.; et al. The PATRIC bioinformatics resource center: Expanding data and analysis capabilities. Nucleic Acids Res. 2020, 48, D606–D612. [Google Scholar] [CrossRef]
- Wang, S.C. Studies of Bacterial Catabolic Enzymes: Implications for the Evolution of Enzymes and Metabolic Pathways. Ph.D. Thesis, The University of Texas, Austin, TX, USA, 2003. [Google Scholar]
- Davidson, R.; Baas, B.-J.; Akiva, E.; Holliday, G.L.; Polacco, B.J.; LeVieux, J.A.; Pullara, C.R.; Zhang, Y.J.; Whitman, C.P.; Babbitt, P.C. A global view of structure–function relationships in the tautomerase superfamily. J. Biol. Chem. 2018, 293, 2342–2357. [Google Scholar] [CrossRef]
- Michał, B.; Gagat, P.; Jabłoński, S.; Chilimoniuk, J.; Gaworski, M.; Mackiewicz, P.; Marcin, Ł.; Burdukiewicz, M.; Łukaszewicz, M. PhyMet2: A database and toolkit for phylogenetic and metabolic analyses of methanogens. Environ. Microbiol. Rep. 2018, 10, 378–382. [Google Scholar] [CrossRef]
- Borrel, G.; Adam, P.S.; McKay, L.J.; Chen, L.-X.; Sierra-García, I.N.; Sieber, C.M.K.; Letourneur, Q.; Ghozlane, A.; Andersen, G.L.; Li, W.-J.; et al. Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat. Microbiol. 2019, 4, 603–613. [Google Scholar] [CrossRef]
- Laso-Pérez, R.; Hahn, C.; Vliet, D.M.V.; Tegetmeyer, H.E.; Schubotz, F.; Smit, N.T.; Pape, T.; Sahling, H.; Bohrmann, G.; Boetius, A.; et al. Anaerobic degradation of non-methane alkanes by candidatus methanoliparia in hydrocarbon seeps of the gulf of Mexico. mBio 2019, 10, e01814–e01819. [Google Scholar] [CrossRef]
- Lahme, S.; Callbeck, C.M.; Eland, L.E.; Wipat, A.; Enning, D.; Head, I.M.; Hubert, C.R.J. Comparison of sulfide-oxidizing Sulfurimonas strains reveals a new mode of thiosulfate formation in subsurface environments. Environ. Microbiol. 2020, 22, 1784–1800. [Google Scholar] [CrossRef] [PubMed]
- Dahle, H.; Roalkvam, I.; Thorseth, I.H.; Pedersen, R.B.; Steen, I.H. The versatile in situ gene expression of an E psilonproteobacteria-dominated biofilm from a hydrothermal chimney. Environ. Microbiol. Rep. 2013, 5, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Hensen, D.; Sperling, D.; Trüper, H.G.; Brune, D.C.; Dahl, C. Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol. Microbiol. 2006, 62, 794–810. [Google Scholar] [CrossRef] [PubMed]
- Zumft, W.; Kroneck, P. Respiratory transformation of nitrous oxide (N2O) to dinitrogen by bacteria and archaea. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2007; Volume 52. [Google Scholar]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hidalgo, K.J.; Sierra-Garcia, I.N.; Zafra, G.; de Oliveira, V.M. Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide. Microorganisms 2021, 9, 1812. https://doi.org/10.3390/microorganisms9091812
Hidalgo KJ, Sierra-Garcia IN, Zafra G, de Oliveira VM. Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide. Microorganisms. 2021; 9(9):1812. https://doi.org/10.3390/microorganisms9091812
Chicago/Turabian StyleHidalgo, Kelly J., Isabel N. Sierra-Garcia, German Zafra, and Valéria M. de Oliveira. 2021. "Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide" Microorganisms 9, no. 9: 1812. https://doi.org/10.3390/microorganisms9091812
APA StyleHidalgo, K. J., Sierra-Garcia, I. N., Zafra, G., & de Oliveira, V. M. (2021). Genome-Resolved Meta-Analysis of the Microbiome in Oil Reservoirs Worldwide. Microorganisms, 9(9), 1812. https://doi.org/10.3390/microorganisms9091812





