Involvement of the MxtR/ErdR (CrbS/CrbR) Two-Component System in Acetate Metabolism in Pseudomonas putida KT2440
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Media
2.2. Generation of Mutants and Complemented Strains
2.3. Growth Experiments
2.4. Luciferase Assay
2.5. Gene Expression by qRT-PCR
2.6. Colony Morphology Assay
2.7. Susceptibility Assay
2.8. Growth Inhibition Assay
2.9. Protein Purification
2.10. EMSA
2.11. Statistical Analysis
3. Results
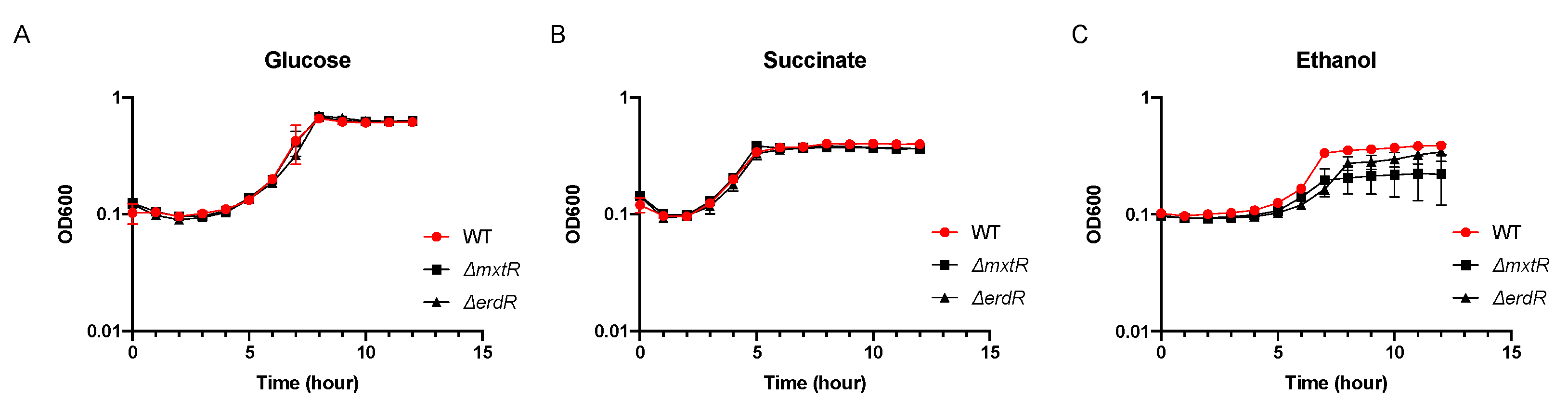
3.1. Contribution of MxtR/ErdR to the Growth of P. putida KT2440 in Different Carbon Sources
3.2. Contribution of MxtR/ErdR to the Regulation of Factors Related to Colonization in P. putida KT2440
3.3. Characterization of Genes Involved in Acetate Utilization and Regulated by MxtR/ErdR in P. putida KT2440
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bekker, M.; de Mattos, M.J.T.; Hellingwerf, K.J. The role of two-component regulation systems in the physiology of the bacterial cell. Sci. Prog. 2006, 89, 213–242. [Google Scholar] [CrossRef] [PubMed]
- Gumerov, V.M.; Ortega, D.R.; Adebali, O.; Ulrich, L.E.; Zhulin, I.B. MiST 3.0: An updated microbial signal transduction database with an emphasis on chemosensory systems. Nucleic Acids Res. 2019, 48, D459–D464. [Google Scholar] [CrossRef]
- Palleroni, N.J. Pseudomonas. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; p. 1. [Google Scholar] [CrossRef]
- Silby, M.W.; Winstanley, C.; Godfrey, S.A.C.; Levy, S.B.; Jackson, R.W. Pseudomonas genomes: Diverse and adaptable. FEMS Microbiol. Rev. 2011, 35, 652–680. [Google Scholar] [CrossRef]
- Rodrigue, A.; Quentin, Y.; Lazdunski, A.; Méjean, V.; Foglino, M. Two-component systems in Pseudomonas aeruginosa: Why so many? Trends Microbiol. 2000, 8, 498–504. [Google Scholar] [CrossRef]
- Nishijyo, T.; Haas, D.; Itoh, Y. The CbrA–CbrB two-component regulatory system controls the utilization of multiple carbon and nitrogen sources in Pseudomonas aeruginosa. Mol. Microbiol. 2001, 40, 917–931. [Google Scholar] [CrossRef]
- Valentini, M.; García-Mauriño, S.M.; Pérez-Martínez, I.; Santero, E.; Canosa, I.; Lapouge, K. Hierarchical management of carbon sources is regulated similarly by the CbrA/B systems in Pseudomonas aeruginosa and Pseudomonas putida. Microbiology 2014, 160, 2243–2252. [Google Scholar] [CrossRef]
- Zhang, X.-X.; Rainey, P.B. Dual involvement of CbrAB and NtrBC in the regulation of histidine utilization in Pseudomonas fluorescens SBW25. Genetics 2008, 178, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Monteagudo-Cascales, E.; García-Mauriño, S.M.; Santero, E.; Canosa, I. Unraveling the role of the CbrA histidine kinase in the signal transduction of the CbrAB two-component system in Pseudomonas putida. Sci. Rep. 2019, 9, 9110. [Google Scholar] [CrossRef] [PubMed]
- Wirtz, L.; Eder, M.; Schipper, K.; Rohrer, S.; Jung, H. Transport and kinase activities of CbrA of Pseudomonas putida KT2440. Sci. Rep. 2020, 10, 5400. [Google Scholar] [CrossRef] [PubMed]
- Hang, S.; Purdy, A.E.; Robins, W.P.; Wang, Z.; Mandal, M.; Chang, S.; Mekalanos, J.J.; Watnick, P.I. The acetate switch of an intestinal pathogen disrupts host insulin signaling and lipid metabolism. Cell Host Microbe 2014, 16, 592–604. [Google Scholar] [CrossRef]
- Jacob, K.; Rasmussen, A.; Tyler, P.; Servos, M.M.; Sylla, M.; Prado, C.; Daniele, E.; Sharp, J.S.; Purdy, A.E. Regulation of acetyl-CoA synthetase transcription by the CrbS/R two-component system is conserved in genetically diverse environmental pathogens. PLoS ONE 2017, 12, e0177825. [Google Scholar] [CrossRef]
- Muzhingi, I.; Prado, C.; Sylla, M.; Diehl, F.F.; Nguyen, D.K.; Servos, M.M.; Flores Ramos, S.; Purdy, A.E. Modulation of CrbS-dependent activation of the acetate switch in Vibrio cholerae. J. Bacteriol. 2018, 200, e00380-18. [Google Scholar] [CrossRef]
- Zaoui, C.; Overhage, J.; Löns, D.; Zimmermann, A.; Müsken, M.; Bielecki, P.; Pustelny, C.; Becker, T.; Nimtz, M.; Häussler, S. An orphan sensor kinase controls quinolone signal production via MexT in Pseudomonas aeruginosa. Mol. Microbiol. 2012, 83, 536–547. [Google Scholar] [CrossRef] [PubMed]
- Sepulveda, E.; Lupas, A.N. Characterization of the CrbS/R Two-Component System in Pseudomonas fluorescens reveals a new set of genes under its control and a DNA motif required for CrbR-mediated transcriptional activation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Kretzschmar, U.; Khodaverdi, V.; Adrian, L. Transcriptional regulation of the acetyl-CoA synthetase gene acsA in Pseudomonas aeruginosa. Arch. Microbiol. 2010, 192, 685–690. [Google Scholar] [CrossRef] [PubMed]
- Mern, D.S.; Ha, S.-W.; Khodaverdi, V.; Gliese, N.; Görisch, H. A complex regulatory network controls aerobic ethanol oxidation in Pseudomonas aeruginosa: Indication of four levels of sensor kinases and response regulators. Microbiology 2010, 156, 1505–1516. [Google Scholar] [CrossRef][Green Version]
- Bagdasarian, M.; Lurz, R.; Rückert, B.; Franklin, F.C.H.; Bagdasarian, M.M.; Frey, J.; Timmis, K.N. Specific-purpose plasmid cloning vectors II. Broad host range, high copy number, RSF 1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas. Gene 1981, 16, 237–247. [Google Scholar] [CrossRef]
- Miroux, B.; Walker, J.E. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol 1996, 260, 289–298. [Google Scholar] [CrossRef]
- Silva-Rocha, R.; Martinez-Garcia, E.; Calles, B.; Chavarria, M.; Arce-Rodriguez, A.; de Las Heras, A.; Paez-Espino, A.D.; Durante-Rodriguez, G.; Kim, J.; Nikel, P.I.; et al. The Standard European Vector Architecture (SEVA): A coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res. 2013, 41, D666–D675. [Google Scholar] [CrossRef] [PubMed]
- Gödeke, J.; Heun, M.; Bubendorfer, S.; Paul, K.; Thormann, K.M. Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Appl. Environ. Microbiol. 2011, 77, 5342–5351. [Google Scholar] [CrossRef]
- Henriquez, T.; Baldow, T.; Lo, Y.K.; Weydert, D.; Brachmann, A.; Jung, H. Involvement of MexS and MexEF-OprN in resistance to toxic ion chelators in Pseudomonas putida KT2440. Microorganisms 2020, 8, 1782. [Google Scholar] [CrossRef] [PubMed]
- Lassak, J.; Henche, A.-L.; Binnenkade, L.; Thormann, K.M. ArcS, the cognate sensor kinase in an atypical Arc system of Shewanella oneidensis MR-1. Appl. Environ. Microbiol. 2010, 76, 3263–3274. [Google Scholar] [CrossRef]
- Sakhtah, H.; Koyama, L.; Zhang, Y.; Morales, D.K.; Fields, B.L.; Price-Whelan, A.; Hogan, D.A.; Shepard, K.; Dietrich, L.E.P. The Pseudomonas aeruginosa efflux pump MexGHI-OpmD transports a natural phenazine that controls gene expression and biofilm development. Proc. Natl. Acad. Sci. USA 2016, 113, E3538–E3547. [Google Scholar] [CrossRef]
- Haber, M.; Ilan, M. Diversity and antibacterial activity of bacteria cultured from Mediterranean Axinella spp. sponges. J. Appl. Microbiol. 2014, 116, 519–532. [Google Scholar] [CrossRef]
- Arias-Barrau, E.; Olivera, E.R.; Sandoval, A.; Naharro, G.; Luengo, J.M. Acetyl-CoA synthetase from Pseudomonas putida U is the only acyl-CoA activating enzyme induced by acetate in this bacterium. FEMS Microbiol. Lett. 2006, 260, 36–46. [Google Scholar] [CrossRef][Green Version]
- Gimenez, R.; Nuñez, M.F.; Badia, J.; Aguilar, J.; Baldoma, L. The gene yjcG, cotranscribed with the gene acs, encodes an acetate permease in Escherichia coli. J. Bacteriol. 2003, 185, 6448–6455. [Google Scholar] [CrossRef]
- Jung, H. The sodium/substrate symporter family: Structural and functional features. FEBS Lett. 2002, 529, 73–77. [Google Scholar] [CrossRef]
- Henriquez, T.; Wirtz, L.; Su, D.; Jung, H. Prokaryotic solute/sodium symporters: Versatile functions and mechanisms of a transporter family. Int. J. Mol. Sci. 2021, 22, 1880. [Google Scholar] [CrossRef] [PubMed]
- Olivera, E.R.; Miñambres, B.; García, B.; Muñiz, C.; Moreno, M.A.; Ferrández, A.; Díaz, E.; García, J.L.; Luengo, J.M. Molecular characterization of the phenylacetic acid catabolic pathway in Pseudomonas putida U: The phenylacetyl-CoA catabolon. Proc. Natl. Acad. Sci. USA 1998, 95, 6419–6424. [Google Scholar] [CrossRef]
- Haller, T.; Buckel, T.; Rétey, J.; Gerlt, J.A. Discovering new enzymes and metabolic pathways: conversion of succinate to propionate by Escherichia coli. Biochemistry 2000, 39, 4622–4629. [Google Scholar] [CrossRef]
- Scott, J.W.; Hawley, S.A.; Green, K.A.; Anis, M.; Stewart, G.; Scullion, G.A.; Norman, D.G.; Hardie, D.G. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Investig. 2004, 113, 274–284. [Google Scholar] [CrossRef]
- Henry, P.M.; Gebben, S.J.; Tech, J.J.; Yip, J.L.; Leveau, J.H.J. Inhibition of Xanthomonas fragariae, causative agent of angular leaf spot of strawberry, through iron deprivation. Front. Microbiol. 2016, 7, 1589. [Google Scholar] [CrossRef] [PubMed]
- Bernal, P.; Allsopp, L.P.; Filloux, A.; Llamas, M.A. The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 2017, 11, 972–987. [Google Scholar] [CrossRef] [PubMed]






| Name (Strains) | Description | Source |
| Wild type (WT) | Pseudomonas putida KT2440 | [18] |
| ΔmxtR | Derived from WT strain by deletion of pp_1695 | This work |
| ΔerdR | Derived from WT strain by deletion of pp_1635 | This work |
| Pseudomonas syringae | Pseudomonas syringae van Hall 1902. Type strain. | DSM 10604 |
| Fusarium oxysporum | Fusarium oxysporum Schlechtendahl: Fries. Type strain | DSM 62297 |
| Escherichia coli C41 | BL21(DE3)-derived strain for protein overexpression | [19] |
| Name (plasmid) | Description | Source |
| pSEVA224 | KmR; pSEVA221 derivative with lacIq/Ptrc expression system | [20] |
| pSEVA224-mxtR | pSEVA224 derivative with pp_1695 cloned into the multicloning site | This work |
| pSEVA224-erdR | pSEVA224 derivative with pp_1635 cloned into the multicloning site | This work |
| pBBR1-MCS5-lux | pBBR1-based plasmid containing promoter-less luxCDABE, and the aacC1 gene (GenR) | [21] |
| pBBR1-MCS5-PmexE::lux | pBBR1-MCS5-lux derivative containing PmexE::luxCDABE | [22] |
| pBBR1-MCS5-Ppp_4243::lux | pBBR1-MCS5-lux derivative containing Ppp_4243::luxCDABE | This work |
| pET16b-erdR | pET16b derivative containing pp_1635 (erdR) | This work |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henriquez, T.; Jung, H. Involvement of the MxtR/ErdR (CrbS/CrbR) Two-Component System in Acetate Metabolism in Pseudomonas putida KT2440. Microorganisms 2021, 9, 1558. https://doi.org/10.3390/microorganisms9081558
Henriquez T, Jung H. Involvement of the MxtR/ErdR (CrbS/CrbR) Two-Component System in Acetate Metabolism in Pseudomonas putida KT2440. Microorganisms. 2021; 9(8):1558. https://doi.org/10.3390/microorganisms9081558
Chicago/Turabian StyleHenriquez, Tania, and Heinrich Jung. 2021. "Involvement of the MxtR/ErdR (CrbS/CrbR) Two-Component System in Acetate Metabolism in Pseudomonas putida KT2440" Microorganisms 9, no. 8: 1558. https://doi.org/10.3390/microorganisms9081558
APA StyleHenriquez, T., & Jung, H. (2021). Involvement of the MxtR/ErdR (CrbS/CrbR) Two-Component System in Acetate Metabolism in Pseudomonas putida KT2440. Microorganisms, 9(8), 1558. https://doi.org/10.3390/microorganisms9081558







