Dual RNA-Seq Enables Full-Genome Assembly of Measles Virus and Characterization of Host–Pathogen Interactions
Abstract
1. Introduction
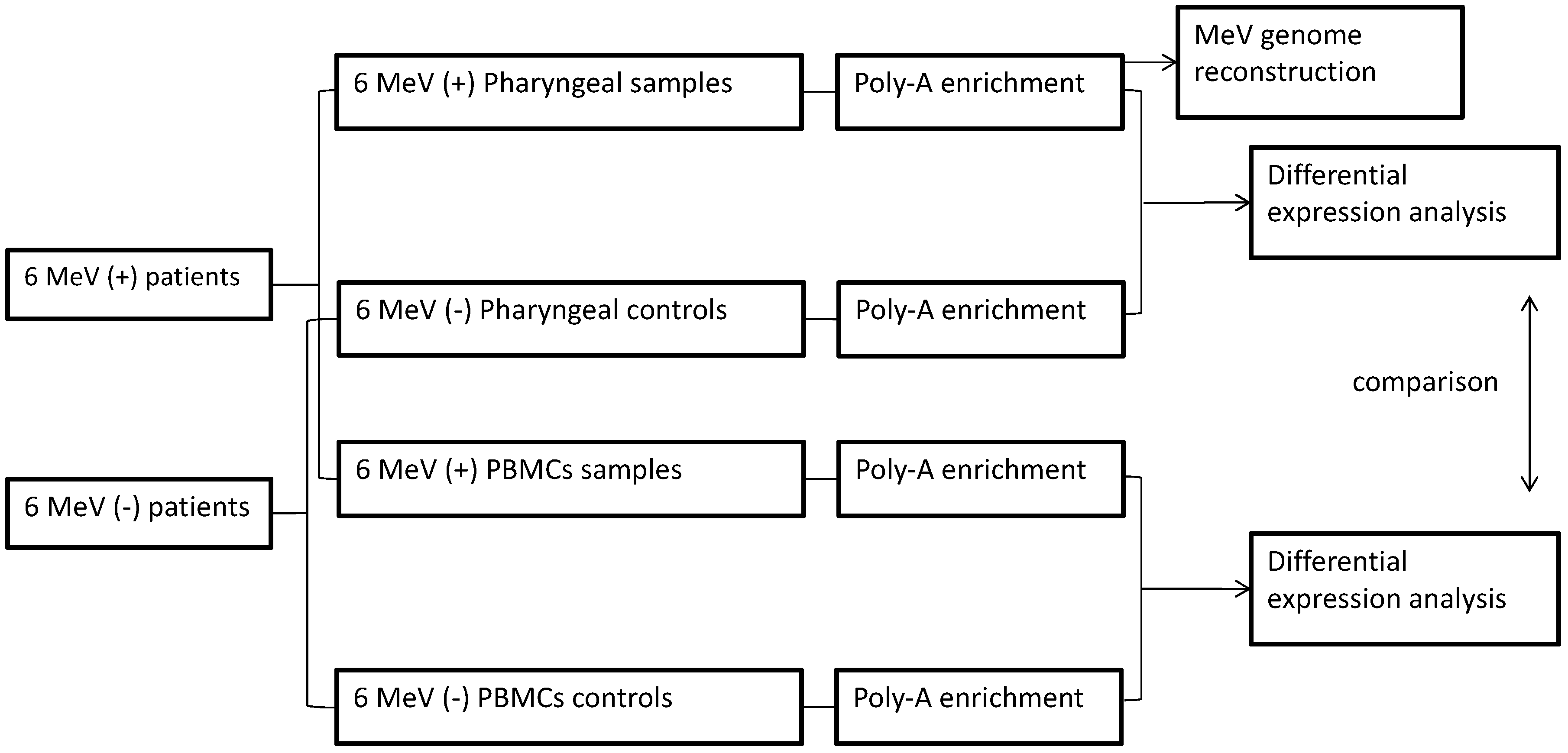
2. Materials and Methods
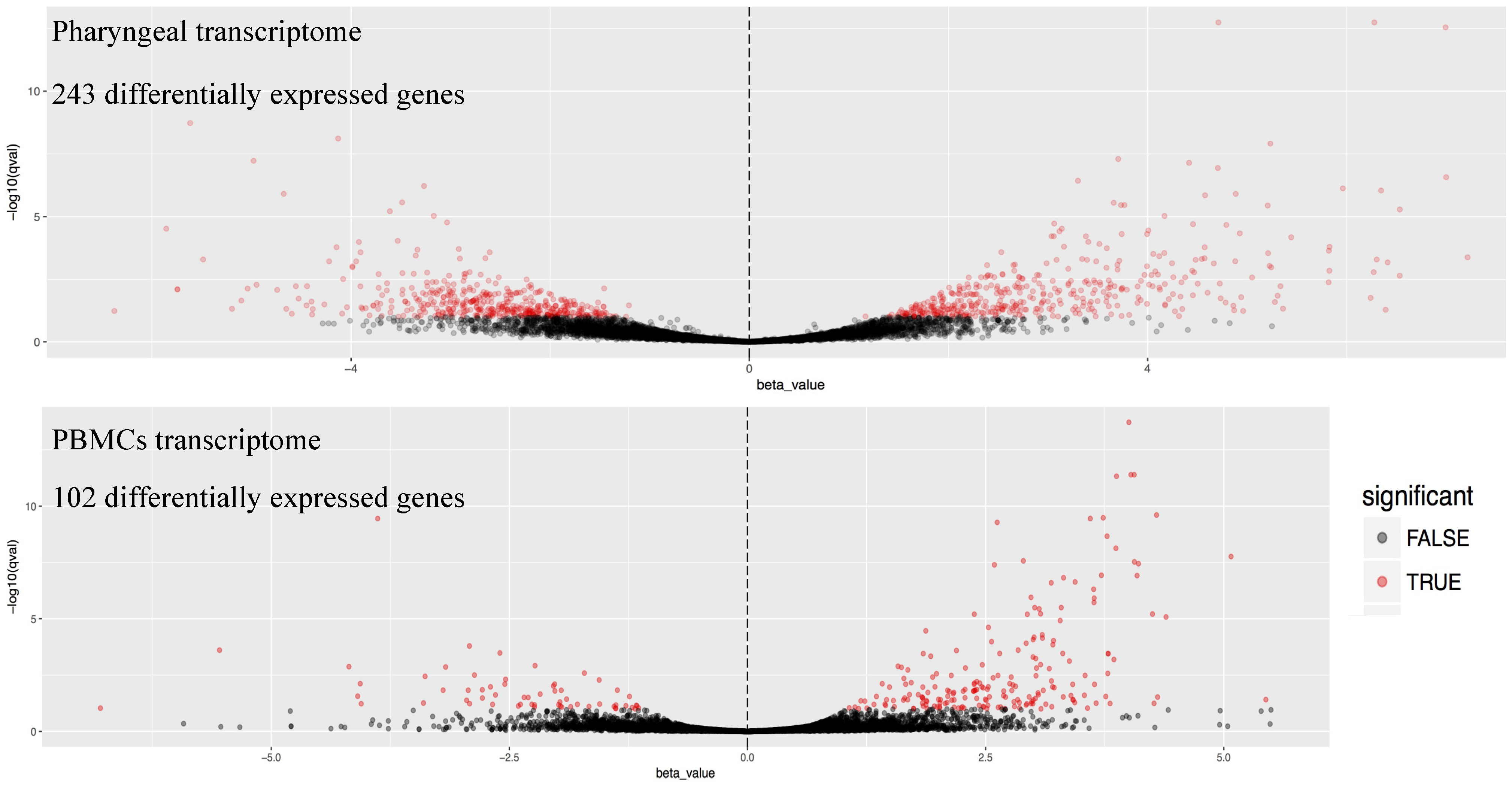
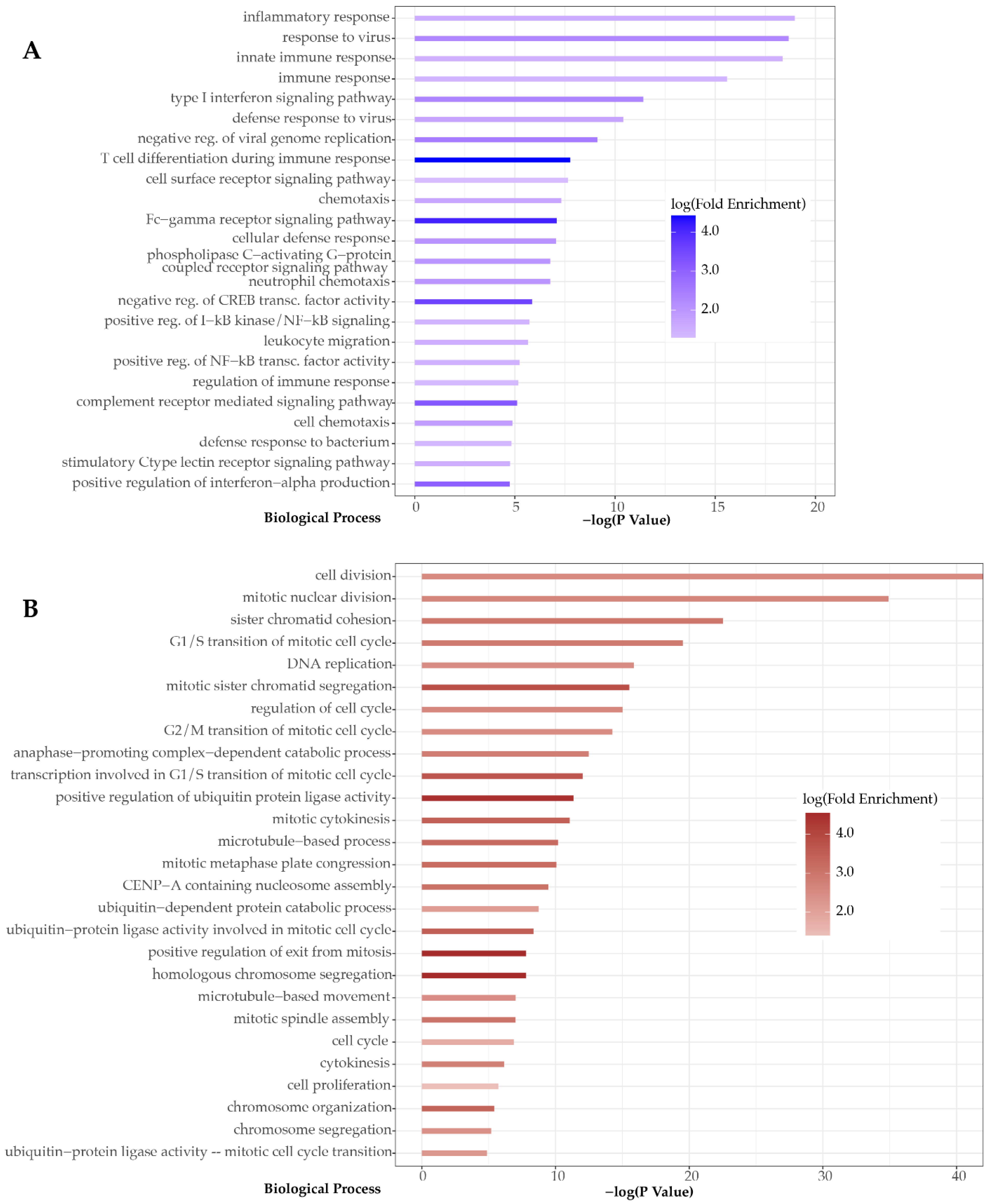
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rota, P.A.; Moss, W.J.; Takeda, M.; De Swart, R.L.; Thompson, K.M.; Goodson, J.L. Measles. Nat. Rev. Dis. Prim. 2016, 2. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Wassilak, S.; Emiroglu, N.; Uzicanin, A.; Deshesvoi, S.; Jankovic, D.; Goel, A.; Khetsuriani, N. What will it take to achieve measles elimination in the World Health Organization European Region: Progress from 2003–2009 and essential accelerated actions. J. Infect. Dis. 2011, 204, 325–334. [Google Scholar] [CrossRef]
- Muscat, M.; Bang, H.; Wohlfahrt, J.; Glismann, S.; Mølbak, K. Measles in Europe: An epidemiological assessment. Lancet 2009, 373, 383–389. [Google Scholar] [CrossRef]
- CfDCaP, C. Global Measles and Rubella Laboratory Network, January 2004–June 2005. Morb. Mortal. Wkly. Rep. 2005, 54, 1100–1104. [Google Scholar]
- WHO. Genetic diversity of wildtype measles viruses and the global measles nucleotide surveillance database (MeaNS) = La diversitι gιnιtique des virus rougeoleux de type sauvage et la base de donnιes MeaNS (Measles Nucleotide Surveillance). Wkly. Epidemiol. Rec. Relev. Ιpidιmiologique Hebd. 2015, 90, 373–380. [Google Scholar]
- Harvala, H.; Wiman, A.; Wallensten, A.; Zakikhany, K.; Englund, H.; Brytting, M. Role of sequencing the measles virus hemagglutinin gene and hypervariable region in the measles outbreak investigations in Sweden during 2013–2014. J. Infect. Dis. 2016, 213, 592–599. [Google Scholar] [CrossRef]
- Penedos, A.R.; Myers, R.; Hadef, B.; Aladin, F.; Brown, K.E. Assessment of the utility of whole genome sequencing of measles virus in the characterisation of outbreaks. PLoS ONE 2015, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Phan, M.V.T.; Schapendonk, C.M.E.; Oude Munnink, B.B.; Koopmans, M.P.G.; de Swart, R.L.; Cotten, M. Complete genome sequences of six measles virus strains. Genome Announc. 2018, 6, 1–2. [Google Scholar] [CrossRef]
- ROBBINS, F.C. Measles: Clinical Features. Am. J. Dis. Child. 1962, 103, 266. [Google Scholar] [CrossRef]
- Simani, O.E.; Adrian, P.V.; Violari, A.; Kuwanda, L.; Otwombe, K.; Nunes, M.C.; Cotton, M.F.; Madhi, S.A. Effect of in-utero HIV exposure and antiretroviral treatment strategies on measles susceptibility and immunogenicity of measles vaccine. Aids 2013, 27, 1583–1591. [Google Scholar] [CrossRef]
- Griffin, D.E. The immune response in measles: Virus control, clearance and protective immunity. Viruses 2016, 8, 282. [Google Scholar] [CrossRef]
- Mόhlebach, M.D.; Mateo, M.; Sinn, P.L.; Prόfer, S.; Katharina, M.; Leonard, V.H.J.; Navaratnarajah, C.K.; Frenzke, M.; Xiao, X.; Sawatsky, B.; et al. Adherens junction protein nectin-4 (PVRL4) is the epithelial receptor for measles virus. Nature 2012, 480, 530–533. [Google Scholar] [CrossRef]
- Moss, W.J. Seminar Measles. Lancet 2017, 6736, 12–19. [Google Scholar] [CrossRef]
- Plattet, P.; Alves, L.; Herren, M.; Aguilar, H.C. Measles virus fusion protein: Structure, function and inhibition. Viruses 2016, 8, 112. [Google Scholar] [CrossRef]
- Noyce, R.S.; Richardson, C.D. Nectin 4 is the epithelial cell receptor for measles virus. Trends Microbiol. 2012, 20, 429–439. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.D.; Mesman, A.W.; Geijtenbeek, T.B.H.; Duprex, W.P.; De Swart, R.L. The pathogenesis of measles. Curr. Opin. Virol. 2012, 2, 248–255. [Google Scholar] [CrossRef] [PubMed]
- Zilliox, M.J.; Parmigiani, G.; Griffin, D.E. Gene expression patterns in dendritic cells infected with measles virus compared with other pathogens. Proc. Natl. Acad. Sci. USA 2006, 103, 3363–3368. [Google Scholar] [CrossRef]
- Yu, X.; Cheng, Y.; Shi, B.; Qian, F.; Wang, F.; Liu, X.; Yang, H.; Xu, Q.; Qi, T.; Zha, L.; et al. Measles Virus Infection in Adults Induces Production of IL-10 and Is Associated with Increased CD4+ CD25+ Regulatory T Cells. J. Immunol. 2008, 181, 7356–7366. [Google Scholar] [CrossRef]
- Christ-Crain, M.; Jaccard-Stolz, D.; Bingisser, R.; Gencay, M.M.; Huber, P.R.; Tamm, M.; Mόller, B. Effect of procalcitonin-guided treatment on antibiotic use and outcome in lower respiratory tract infections: Cluster-randomised, single-blinded intervention trial. Lancet 2004, 363, 600–607. [Google Scholar] [CrossRef]
- Bergin, S.P.; Tsalik, E.L. Procalcitonin: The Right Answer but to Which Question? Clin. Infect. Dis. 2017, 65, 191–193. [Google Scholar] [CrossRef][Green Version]
- Holcomb, Z.E.; Tsalik, E.L.; Woods, C.W.; McChain, M.T. Host-Based Peripheral Blood Gene Expression Analysis for Diagnosis of Infectious Diseases. J. Clin. Microbiol. 2017, 55, 360–368. [Google Scholar] [CrossRef]
- Hu, X.; Yu, J.; Crosby, S.D.; Storch, G.A. Gene expression profiles in febrile children with defined viral and bacterial infection. Proc. Natl. Acad. Sci. USA 2013, 110, 12792–12797. [Google Scholar] [CrossRef] [PubMed]
- Tsalik, E.L.; Henao, R.; Nichols, M.; Burke, T.; Ko, E.R.; McClain, M.T.; Hudson, L.L.; Mazur, A.; Freeman, D.H.; Veldman, T.; et al. Host gene expression classifiers diagnose acute respiratory illness etiology. Sci. Transl. Med. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Suarez, N.M.; Bunsow, E.; Falsey, A.R.; Walsh, E.E.; Mejias, A.; Ramilo, O. Superiority of transcriptional profiling over procalcitonin for distinguishing bacterial from viral lower respiratory tract infections in hospitalized adults. J. Infect. Dis. 2015, 212, 213–222. [Google Scholar] [CrossRef]
- Westermann, A.J.; Gorski, S.A.; Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 2012, 10, 618–630. [Google Scholar] [CrossRef]
- Wesolowska-Andersen, A.; Everman, J.L.; Davidson, R.; Rios, C.; Herrin, R.; Eng, C.; Janssen, W.J.; Liu, A.H.; Oh, S.S.; Kumar, R.; et al. Dual RNA-seq reveals viral infections in asthmatic children without respiratory illness which are associated with changes in the airway transcriptome. Genome Biol. 2017, 18, 12. [Google Scholar] [CrossRef]
- Poole, A.; Urbanek, C.; Eng, C. Dissecting Childhood Asthma with Nasal Transcriptomics Distinguishes Subphenotypes of Disease. Bone 2008, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Bowtie2. Nat. Methods 2013, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Thorvaldsdσttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef]
- Church, D.M.; Schneider, V.A.; Steinberg, K.M.; Schatz, M.C.; Quinlan, A.R.; Chin, C.S.; Kitts, P.A.; Aken, B.; Marth, G.T.; Hoffman, M.M.; et al. Extending reference assembly models. Genome Biol. 2015, 16, 13. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Melchjorsen, J.; Kristiansen, H.; Christiansen, R.; Rintahaka, J.; Matikainen, S.; Paludan, S.R.; Hartmann, R. Differential regulation of the OASL and OAS1 genes in response to viral infections. J. Interf. Cytokine Res. 2009, 29, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Boucas, A.P.; Oliveira, F. dos S. de; Canani, L.H.; Crispim, D. The role of interferon induced with helicase C domain 1 (IFIH1) in the development of type 1 diabetes mellitus. Arq. Bras. Endocrinol. Metabol. 2013, 57, 667–676. [Google Scholar] [CrossRef]
- Durmus, S.; Cakir, T.; Ozgur, A.; Guthke, R. A review on computational systems biology of pathogen-host interactions. Front. Microbiol. 2015, 6, 1–19. [Google Scholar] [CrossRef]
- Sumegi, J.; Barnes, M.G.; Nestheide, S.V.; Molleran-Lee, S.; Villanueva, J.; Zhang, K.; Risma, K.A.; Grom, A.A.; Filipovich, A.H. Gene expression profiling of peripheral blood mononuclear cells from children with active hemophagocytic lymphohistiocytosis. Blood 2011, 117, 151–161. [Google Scholar] [CrossRef]
- Cilloniz, C.; Ebihara, H.; Ni, C.; Neumann, G.; Korth, M.J.; Kelly, S.M.; Kawaoka, Y.; Feldmann, H.; Katze, M.G. Functional Genomics Reveals the Induction of Inflammatory Response and Metalloproteinase Gene Expression during Lethal Ebola Virus Infection. J. Virol. 2011, 85, 9060–9068. [Google Scholar] [CrossRef]
- Chiffoleau, E. C-type lectin-like receptors as emerging orchestrators of sterile inflammation represent potential therapeutic targets. Front. Immunol. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Boro, M.; Balaji, K.N. CXCL1 and CXCL2 Regulate NLRP3 Inflammasome Activation via G-Protein–Coupled Receptor CXCR2. J. Immunol. 2017, 199, 1660–1671. [Google Scholar] [CrossRef] [PubMed]
- Komune, N.; Ichinohe, T.; Ito, M.; Yanagi, Y. Measles Virus V Protein Inhibits NLRP3 Inflammasome-Mediated Interleukin-1 Secretion. J. Virol. 2011, 85, 13019–13026. [Google Scholar] [CrossRef] [PubMed]
- Frigola, J.; He, J.; Kinkelin, K.; Pye, V.E.; Renault, L.; Douglas, M.E.; Remus, D.; Cherepanov, P.; Costa, A.; Diffley, J.F.X. Cdt1 stabilizes an open MCM ring for helicase loading. Nat. Commun. 2017, 8, 1–10. [Google Scholar] [CrossRef]
- Shen, T.; Huang, S. The Role of Cdc25A in the Regulation of Cell Proliferation and Apoptosis. Anticancer. Agents Med. Chem. 2012, 12, 631–639. [Google Scholar] [CrossRef]
- Hégarat, N.; Rata, S.; Hochegger, H. Bistability of mitotic entry and exit switches during open mitosis in mammalian cells. BioEssays 2016, 38, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Laksono, B.M.; de Vries, R.D.; Verburgh, R.J.; Visser, E.G.; de Jong, A.; Fraaij, P.L.A.; Ruijs, W.L.M.; Nieuwenhuijse, D.F.; van den Ham, H.J.; Koopmans, M.P.G.; et al. Studies into the mechanism of measles-associated immune suppression during a measles outbreak in the Netherlands. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Lemmens, B.; Hegarat, N.; Akopyan, K.; Sala-Gaston, J.; Bartek, J.; Hochegger, H.; Lindqvist, A. DNA Replication Determines Timing of Mitosis by Restricting CDK1 and PLK1 Activation. Mol. Cell 2018, 71, 117–128.e3. [Google Scholar] [CrossRef] [PubMed]
- Nakachi, I.; Helfrich, B.A.; Spillman, M.A.; Mickler, E.A.; Olson, C.J.; Rice, J.L.; Coldren, C.D.; Heasley, L.E.; Geraci, M.W.; Stearman, R.S. PTTG1 Levels Are Predictive of Saracatinib Sensitivity in Ovarian Cancer Cell Lines. Clin. Transl. Sci. 2016, 9, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, C.; Iankov, I.D.; Galanis, E. A key anti-viral protein, RSAD2/VIPERIN, restricts the release of Measles virus in infected cells. Physiol. Behav. 2016, 176, 100–106. [Google Scholar] [CrossRef]
- Landry, M.L.; Foxman, E.F. Antiviral Response in the Nasopharynx Identifies Patients with Respiratory Virus Infection. J. Infect. Dis. 2018, 217, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-Y.; Yaneva, R.; Cresswell, P. Viperin: A multifunctional, interferon-inducible protein that regulates virus replication. Bone 2012, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Jiang, D.; Weidner, J.M.; Qing, M.; Pan, X.-B.; Guo, H.; Xu, C.; Zhang, X.; Birk, A.; Chang, J.; Shi, P.-Y.; et al. Identification of Five Interferon-Induced Cellular Proteins That Inhibit West Nile Virus and Dengue Virus Infections. J. Virol. 2010, 84, 8332–8341. [Google Scholar] [CrossRef]
- John, W.; Schoggins, C.M.R. Interferon-stimulated genes and their antiviral effector functions. Curr. Opin. Virol. 2012, 1, 519–525. [Google Scholar] [CrossRef]
- Groom, J.R.; Luster, A.D. CXCR3 in T cell function. Bone 2012, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Oshiumi, H.; Miyashita, M.; Okamoto, M.; Morioka, Y.; Okabe, M.; Matsumoto, M.; Seya, T. DDX60 Is Involved in RIG-I-Dependent and Independent Antiviral Responses, and Its Function Is Attenuated by Virus-Induced EGFR Activation. Cell Rep. 2015, 11, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Childs, K.; Randall, R.; Goodbourn, S. Paramyxovirus V proteins interact with the RNA Helicase LGP2 to inhibit RIG-I-dependent interferon induction. J. Virol. 2012, 86, 3411–3421. [Google Scholar] [CrossRef] [PubMed]
- Ishizaki, Y.; Takemoto, M.; Kira, R.; Kusuhara, K.; Torisu, H.; Sakai, Y.; Sanefuji, M.; Yukaya, N.; Hara, T. Association of toll-like receptor 3 gene polymorphism with subacute sclerosing panencephalitis. J. Neurovirol. 2008, 14, 486–491. [Google Scholar] [CrossRef]
- Sato, H.; Honma, R.; Yoneda, M.; Miura, R.; Tsukiyama-Kohara, K.; Ikeda, F.; Seki, T.; Watanabe, S.; Kai, C. Measles virus induces cell-type specific changes in gene expression. Virology 2008, 375, 321–330. [Google Scholar] [CrossRef][Green Version]
- Donohue, R.C.; Pfaller, C.K.; Cattaneo, R. Cyclical adaptation of measles virus quasispecies to epithelial and lymphocytic cells: To V, or not to V. PLoS Pathog. 2019, 15, e1007605. [Google Scholar] [CrossRef]
- Marazzi, I.; Ho, J.S.Y.; Kim, J.; Manicassamy, B.; Dewell, S.; Albrecht, R.A.; Seibert, C.W.; Schaefer, U.; Jeffrey, K.L.; Prinjha, R.K.; et al. Suppression of the antiviral response by an influenza histone mimic. Nature 2012, 483, 428–433. [Google Scholar] [CrossRef]
- Law, G.L.; Korth, M.J.; Benecke, A.G.; Katze, M.G. Systems virology: Host-directed approaches to viral pathogenesis and drug targeting. Systems 2014, 11, 455–466. [Google Scholar] [CrossRef]
- Runge, S.; Sparrer, K.M.J.; Lδssig, C.; Hembach, K.; Baum, A.; Garcνa-Sastre, A.; Sφding, J.; Conzelmann, K.K.; Hopfner, K.P. In Vivo Ligands of MDA5 and RIG-I in Measles Virus-Infected Cells. PLoS Pathog. 2014, 10. [Google Scholar] [CrossRef]
- Guo, F.; Yuan, Y. Tumor necrosis factor alpha-induced proteins in malignant tumors: Progress and prospects. Onco. Targets. Ther. 2020, 13, 3303–3318. [Google Scholar] [CrossRef]
- Wenzl, K.; Hofer, S.; Troppan, K.; Lassnig, M.; Steinbauer, E.; Wiltgen, M.; Zulus, B.; Renner, W.; Beham-Schmid, C.; Neumeister, P.; et al. Higher incidence of the SNP Met 788 Ile in the coding region of A20 in diffuse large B cell lymphomas. Tumor Biol. 2016, 37, 4785–4789. [Google Scholar] [CrossRef] [PubMed]
- Mcelroy, R.; Ennis, M.; Schock, B.C. TNFAIP3 (Tumor Necrosis Factor, Alpha-Induced Protein 3). Encycl. Signal. Mol. 2017, 3. [Google Scholar] [CrossRef]
- Dyer, D.P.; Salanga, C.L.; Johns, S.C.; Valdambrini, E.; Fuster, M.M.; Milner, C.M.; Day, A.J.; Handel, T.M. The anti-inflammatory protein TSG-6 regulates chemokine function by inhibiting chemokine/glycosaminoglycan interactions. J. Biol. Chem. 2016, 291, 12627–12640. [Google Scholar] [CrossRef] [PubMed]
- Capp, E.; Milner, C.M.; Williams, J.; Hauck, L.; Jauckus, J.; Strowitzki, T.; Germeyer, A. Modulation of tumor necrosis factor-stimulated gene-6 (TSG-6) expression in human endometrium. Arch. Gynecol. Obstet. 2014, 289, 893–901. [Google Scholar] [CrossRef]
- Huysamen, C.; Brown, G.D. The fungal pattern recognition receptor, Dectin-1, and the associated cluster of C-type lectin-like receptors. FEMS Microbiol. Lett. 2009, 290, 121–128. [Google Scholar] [CrossRef]
- Patin, E.C.; Willcocks, S.; Orr, S.; Ward, T.H.; Lang, R.; Schaible, U.E. Mincle-mediated anti-inflammatory IL-10 response counter-regulates IL-12 in vitro. Innate Immun. 2016, 22, 181–185. [Google Scholar] [CrossRef]
- Méndez-Samperio, P. Expression and regulation of chemokines in mycobacterial infection. J. Infect. 2008, 57, 374–384. [Google Scholar] [CrossRef]
- Zilliox, M.J.; Moss, W.J.; Griffin, D.E. Gene expression changes in peripheral blood mononuclear cells during measles virus infection. Clin. Vaccine Immunol. 2007, 14, 918–923. [Google Scholar] [CrossRef]
- Tanaka, S.; Diffley, J.F.X. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2-7 during G1 phase. Nat. Cell Biol. 2002, 4, 198–207. [Google Scholar] [CrossRef]
- Zou, L.; Stillman, B. Assembly of a Complex Containing Cdc45p, Replication Protein A, and Mcm2p at Replication Origins Controlled by S-Phase Cyclin-Dependent Kinases and Cdc7p-Dbf4p Kinase. Mol. Cell. Biol. 2000, 20, 3086–3096. [Google Scholar] [CrossRef] [PubMed]
- Costa, A.; Ilves, I.; Tamberg, N.; Petojevic, T.; Nogales, E.; Botchan, M.R.; Berger, J.M. The structural basis for MCM2-7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 2011, 18, 471–479. [Google Scholar] [CrossRef] [PubMed]
- Broderick, R.; Ramadurai, S.; Tóth, K.; Togashi, D.M.; Ryder, A.G.; Langowski, J.; Nasheuer, H.P. Cell cycle-dependent mobility of Cdc45 determined in vivo by Fluorescence Correlation Spectroscopy. PLoS ONE 2012, 7. [Google Scholar] [CrossRef]
- Köhler, C.; Koalick, D.; Fabricius, A.; Parplys, A.C.; Borgmann, K.; Pospiech, H.; Grosse, F. Cdc45 is limiting for replication initiation in humans. Cell Cycle 2016, 15, 974–985. [Google Scholar] [CrossRef]
- Suryadinata, R.; Sadowski, M.; Sarcevic, B. Control of cell cycle progression by phosphorylation of cyclin-dependent kinase (CDK) substrates. Biosci. Rep. 2010, 30, 243–255. [Google Scholar] [CrossRef]
- Blais, A.; Dynlacht, B.D. E2F-associated chromatin modifiers and cell cycle control. Bone 2013, 23, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Rohe, A.; Erdmann, F.; Babler, C.; Wichapong, K.; Sippl, W.; Schmidt, M. In vitro and in silico studies on substrate recognition and acceptance of human PKMYT1, a Cdk1 inhibitory kinase. Bioorg. Med. Chem. Lett. 2012, 22, 1219–1223. [Google Scholar] [CrossRef]
- Toledo, C.M.; Ding, Y.; Hoellerbauer, P.; Davis, R.J.; Basom, R.; Girard, E.J.; Lee, E.; Corrin, P.; Hart, T.; Bolouri, H.; et al. Genome-wide CRISPR-Cas9 Screens Reveal Loss of Redundancy between PKMYT1 and WEE1 in Glioblastoma Stem-like Cells. Cell Rep. 2015, 13, 2425–2439. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, X.; Zhang, C.; Wang, W.; Li, F.; Liu, D.; Wu, K.; Zhu, D.; Liu, S.; Shen, C.; et al. Overexpressed PKMYT1 promotes tumor progression and associates with poor survival in esophageal squamous cell carcinoma. Cancer Manag. Res. 2019, 11, 7813–7824. [Google Scholar] [CrossRef] [PubMed]
- Petrova, V.N.; Sawatsky, B.; Han, A.X.; Laksono, B.M.; Walz, L.; Parker, E.; Pieper, K.; Anderson, C.A.; De Vries, R.D.; Lanzavecchia, A.; et al. Incomplete genetic reconstitution of B cell pools contributes to prolonged immunosuppression after measles. Sci. Immunol. 2019, 4. [Google Scholar] [CrossRef]
- Mina, M.J.; Kula, T.; Leng, Y.; Li, M.; Vries, R.D.; De Knip, M.; Siljander, H.; Rewers, M.; Choy, D.F.; Wilson, M.S.; et al. other pathogens. Science 2019, 606, 599–606. [Google Scholar] [CrossRef]
- Wesemann, D.R. Game of clones: How measles remodels the B cell landscape. Sci. Immunol. 2019, 4, 2–5. [Google Scholar] [CrossRef] [PubMed]
- De Vries, R.D.; McQuaid, S.; van Amerongen, G.; Yόksel, S.; Verburgh, R.J.; Osterhaus, A.D.M.E.; Duprex, W.P.; de Swart, R.L. Measles Immune Suppression: Lessons from the Macaque Model. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Moussallem, T.M.; Guedes, F.; Fernandes, E.R.; Pagliari, C.; Lancellotti, C.L.P.; de Andrade, H.F.; Duarte, M.I.S. Lung involvement in childhood measles: Severe immune dysfunction revealed by quantitative immunohistochemistry. Hum. Pathol. 2007, 38, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
| Sample | Enrichment Method | Total Reads | Total MeV Genome Reads | MeV Reads (%) | Average Depth (×) | MeV Genome Coverage (%) |
|---|---|---|---|---|---|---|
| Pharg_MeV1 | poly-A | 34,370,942 | 405,851 | 1.18 | 1953.95 | 100.00 |
| Pharg_MeV2 | poly-A | 34,817,617 | 282,092 | 0.81 | 1357.61 | 99.25 |
| Pharg_MeV3 | poly-A | 28,188,497 | 90,601 | 0.32 | 436.01 | 100.00 |
| Pharg_MeV4 | poly-A | 29,716,507 | 59,901 | 0.20 | 288.41 | 100.00 |
| Pharg_MeV5 | poly-A | 12,783,974 | 620,934 | 4.86 | 2990.41 | 100.00 |
| Pharg_MeV6 | poly-A | 3,126,258 | 198,309 | 6.34 | 955.13 | 100.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karamitros, T.; Pogka, V.; Papadopoulou, G.; Tsitsilonis, O.; Evangelidou, M.; Sympardi, S.; Mentis, A. Dual RNA-Seq Enables Full-Genome Assembly of Measles Virus and Characterization of Host–Pathogen Interactions. Microorganisms 2021, 9, 1538. https://doi.org/10.3390/microorganisms9071538
Karamitros T, Pogka V, Papadopoulou G, Tsitsilonis O, Evangelidou M, Sympardi S, Mentis A. Dual RNA-Seq Enables Full-Genome Assembly of Measles Virus and Characterization of Host–Pathogen Interactions. Microorganisms. 2021; 9(7):1538. https://doi.org/10.3390/microorganisms9071538
Chicago/Turabian StyleKaramitros, Timokratis, Vasiliki Pogka, Gethsimani Papadopoulou, Ourania Tsitsilonis, Maria Evangelidou, Styliani Sympardi, and Andreas Mentis. 2021. "Dual RNA-Seq Enables Full-Genome Assembly of Measles Virus and Characterization of Host–Pathogen Interactions" Microorganisms 9, no. 7: 1538. https://doi.org/10.3390/microorganisms9071538
APA StyleKaramitros, T., Pogka, V., Papadopoulou, G., Tsitsilonis, O., Evangelidou, M., Sympardi, S., & Mentis, A. (2021). Dual RNA-Seq Enables Full-Genome Assembly of Measles Virus and Characterization of Host–Pathogen Interactions. Microorganisms, 9(7), 1538. https://doi.org/10.3390/microorganisms9071538






