Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review
Abstract
1. Introduction
2. Physiochemical Profiling
3. Microbiological Analysis
3.1. Cultivation-Dependent Studies
3.2. Cultivation-Independent Studies
4. Correlation between the Geophysiology and Microbiology of Hot Springs in the Lesser Caucasus
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Hreggvidsson, G.O.; Petursdottir, S.K.; Bjornsdottir, S.H.; Fridjonsson, O.H. Microbial speciation in the geothermal ecosystem. In Adaption of Microbial Life to Environmental Extremes: Novel Research Results and Application; Stan, H., Fendrihan, L.S., Eds.; Springer: Vienna, Austria, 2012; pp. 37–68. [Google Scholar]
- Deepika, M.; Satyanarayana, T. Diversity of Hot Environments and Thermophilic Microbes. In Thermophilic Microbes in Environmental and Industrial Biotechnology Biotechnology of Thermophiles; Satyanarayana, T., Littlechild, J., Kawarabayasi, Y., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 3–60. [Google Scholar]
- DeCastro, M.E.; Rodríguez-Belmonte, E.; González-Siso, M.I. Metagenomics of Thermophiles with a Focus on Discovery of Novel Thermozymes. Front. Microbiol. 2016, 7, 1521. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.; Singhal, P.; Singh, H.; Damle, D.; Sharma, A.K. Insight into thermophiles and their wide-spectrum applications. 3 Biotech 2016, 6, 81. [Google Scholar] [CrossRef] [PubMed]
- Stan-Lotter, H.; Fendrihan, S. Adaption of Microbial Life to Environmental Extremes; Springer: Vienna, Austria, 2012. [Google Scholar]
- Das Sarma, S.; Das Sarma, P.; Laye, V.J.; Schwieterman, E.W. Extremophilic models for astrobiology: Haloarchaeal survival strategies and pigments for remote sensing. Extremophiles 2020, 24, 31–41. [Google Scholar] [CrossRef]
- López-López, O.; Cerdán, M.E.; González-Siso, M.I. Hot spring metagenomics. Life 2013, 3, 308–320. [Google Scholar] [CrossRef]
- Sahm, K.; John, P.; Nacke, H.; Wemheuer, B.; Grote, R.; Daniel, R.; Antranikian, G. High abundance of heterotrophic prokaryotes in hydrothermal springs of the Azores as revealed by a network of 16S rRNA gene-based methods. Extremophiles 2013, 17, 649–662. [Google Scholar] [CrossRef] [PubMed]
- Krebs, J.E.; Vaishampayan, P.; Probst, A.J.; Tom, L.M.; Marteinsson, V.T.; Andersen, G.L.; Venkateswaran, K. Microbial community structures of novel Icelandic hot spring systems revealed by PhyloChip G3 analysis. Astrobiology 2014, 14, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Stefanova, K.; Tomova, I.; Tomova, A.; Radchenkova, N.; Atanassov, I.; Kambourova, M. Archaeal and bacterial diversity in two hot springs from geothermal regions in Bulgaria as demonstrated by 16S rRNA and GH-57 genes. Int. Microbiol. 2015, 18, 217–223. [Google Scholar] [PubMed]
- López-López, O.; Knapik, K.; Cerdán, M.E.; González-Siso, M.I. Metagenomics of an alkaline hot spring in Galicia (Spain): Microbial diversity analysis and screening for novel lipolytic enzymes. Front. Microbiol. 2015, 6, 1291. [Google Scholar] [CrossRef]
- Hou, W.; Wang, S.; Dong, H.; Jiang, H.; Briggs, B.R.; Peacock, J.P.; Huang, Q.; Huang, L.; Wu, G.; Zhi, X.; et al. A Comprehensive census of microbial diversity in hot springs of Tengchong, Yunnan province China Using 16S rRNA Gene Pyrosequencing. PLoS ONE 2013, 8, e53350. [Google Scholar]
- Chan, C.S.; Chan, K.G.; Tay, Y.L.; Chua, Y.H.; Goh, K.M. Diversity of thermophiles in a Malaysian hot spring determined using 16S rRNA and shotgun metagenome sequencing. Front. Microbiol. 2015, 6, 177. [Google Scholar] [CrossRef]
- Merkel, A.Y.; Pimenov, N.V.; Rusanov, I.I.; Slobodkin, A.I.; Slobodkina, G.B.; Tarnovetckii, I.Y.; Frolov, E.; Dubin, A.V.; Perevalova, A.A.; Bonch-Osmolovskaya, E.A. Microbial diversity and autotrophic activity in Kamchatka hot springs. Extremophiles 2017, 21, 307–317. [Google Scholar] [CrossRef]
- Amin, A.; Ahmed, I.; Salam, N.; Kim, B.Y.; Singh, D.; Zhi, X.Y.; Xiao, M.; Li, W.J. Diversity and distribution of thermophilic bacteria in hot springs of Pakistan. Microb. Ecol. 2017, 74, 116–127. [Google Scholar] [CrossRef]
- Saxena, R.; Dhakan, D.B.; Mittal, P.; Waiker, P.; Chowdhury, A.; Ghatak, A.; Sharma, V.K. Metagenomic analysis of hot springs in Central India reveals hydrocarbon degrading thermophiles and pathways essential for survival in extreme environments. Front. Microbiol. 2017, 7, 21–23. [Google Scholar] [CrossRef] [PubMed]
- Hussein, E.I.; Jacob, J.H.; Shakhatreh, M.A.K.; Al-razaq, M.A.A.; Juhmani, A.F.; Cornelison, C.T. Exploring the microbial diversity in Jordanian hot springs by comparative metagenomic analysis. Microbiol. Open 2017, 6, e521. [Google Scholar] [CrossRef] [PubMed]
- Guven, K.; Bekler, F.M.; Guven, R.G. Thermophilic and halophilic microorganisms isolated from extreme environments of Turkey, with potential biotechnological applications. In Extremophiles in Eurasian Ecosystems: Ecology, Diversity, and Applications; Egamberdieva, D., Birkeland, N.K., Panosyan, H., Li, W.J., Eds.; Springer: Singapore, 2018; pp. 219–264. [Google Scholar]
- De León, B.K.; Gerlach, R.; Peyton, B.M.; Fields, M.W. Archaeal and bacterial communities in three alkaline hot springs in Heart Lake Geyser Basin, Yellowstone National Park. Front. Microbiol. 2013, 4, 330. [Google Scholar] [CrossRef]
- Urbieta, M.S.; Gonzalez-Toril, E.; Bazan, A.A.; Giaveno, M.A.; Donati, E. Comparison of the microbial communities of hot springs waters and the microbial biofilms in the acidic geothermal area of Copahue (Neuquen, Argentina). Extremophiles 2015, 19, 437–450. [Google Scholar] [CrossRef]
- Sayeh, R.; Birrien, J.L.; Alain, K.; Barbier, G.; Hamdi, M.; Prieur, D. Microbial diversity in Tunisian geothermal springs as detected by molecular and culture-based approaches. Extremophiles 2010, 14, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Magnabosco, C.; Tekere, M.; Lau, M.C.Y.; Linage, B.; Kuloyo, O.; Erasmus, M.; Cason, E.; van Heerden, E.; Borgonie, G.; Kieft, T.L.; et al. Comparisons of the composition and biogeographic distribution of the bacterial communities occupying South African thermal springs with those inhabiting deep subsurface fracture water. Front. Microbiol. 2014, 5, 679. [Google Scholar] [CrossRef] [PubMed]
- Ogg, C.D.; Spanevello, M.D.; Patel, B.K.C. Exploring the ecology of thermophiles from Australia’s Great Artesian Basin during the genomic era. In Thermophilic Microbes in Environmental and Industrial Biotechnology; Satyanarayana, T., Littlechild, J., Kawarabayasi, Y., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 61–97. [Google Scholar]
- Power, J.F.; Carere, C.R.; Lee, C.K.; Wakerley, G.L.J.; Evans, D.W.; Button, M.; White, D.; Climo, M.D.; Hinze, A.M.; Morgan, X.C.; et al. Microbial biogeography of 925 geothermal springs in New Zealand. Nat. Commun. 2018, 9, 2876. [Google Scholar] [CrossRef]
- Flores, P.A.; Amenábar, M.J.; Blamey, J.M. Hot environments from Antarctica: Source of thermophiles and hyperthermophiles, with potential biotechnological applications. In Thermophilic Microbes in Environmental and Industrial Biotechnology; Satyanarayana, T., Littlechild, J., Kawarabayasi, Y., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 99–118. [Google Scholar]
- Elleuche, S.; Antranikian, G. Starch-hydrolyzing enzymes from thermophiles. In Thermophilic Microbes in Environmental and Industrial Biotechnology; Satyanarayana, T., Littlechild, J., Kawarabayasi, Y., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 509–533. [Google Scholar]
- Sharma, R.; Thakur, V.; Sharma, M.; Birkeland, N.K. Biocatalysis through thermostable lipases: Adding flavor to chemistry. In Thermophilic Microbes in Environmental and Industrial Biotechnology; Satyanarayana, T., Littlechild, J., Kawarabayasi, Y., Eds.; Springer: Dordrecht, The Netherlands, 2013; pp. 905–927. [Google Scholar]
- Henneberger, R.; Cooksley, D.; Hallberg, J. Geothermal resources of Armenia. In Proceedings of the World Geothermal Congress, Kyushu-Tohoku, Japan, 28 May–10 June 2000; Volume 28, pp. 1217–1222. [Google Scholar]
- Badalyan, M. Geothermal features of Armenia: A country update. In Proceedings of the World Geothermal Congress, Kyushu-Tohoku, Japan, 28 May–10 June 2000; pp. 71–75. [Google Scholar]
- Philip, H.; Cisternas, A.; Gvishiani, A.; Gorshukov, A. The Caucasus: An actual example of the initial stages of continental collision. Tectonophysics 1989, 161, 1–21. [Google Scholar] [CrossRef]
- Reilinger, R.E.; McClusky, S.C.; Oral, M.B.; King, R.W.; Toksoz, M.N.; Barka, A.A.; Kinik, I.; Lenk, O.; Sanli, I. Global Positioning System measurements of present-day crustal movements in the Arabia-Africa-Eurasia plate collision zone. J. Geophys. Res. 1997, 102, 9983–9999. [Google Scholar] [CrossRef]
- Mkrtchyan, S. Geology of Armenian SSR; Publishing House of AS of ASSR: Yerevan, Armenia, 1969. (In Russian) [Google Scholar]
- Panosyan, H.; Birkeland, N.K. Microbial diversity in an Armenian geothermal spring assessed by molecular and culture-based methods. J. Basic Microbiol. 2014, 54, 1240–1250. [Google Scholar] [CrossRef]
- Panosyan, H.H. Thermophilic bacilli isolated from Armenian geothermal springs and their potential for production of hydrolytic enzymes. Int J. Biotech. Bioeng. 2017, 3, 239–244. [Google Scholar] [CrossRef][Green Version]
- Panosyan, H.; Margaryan, A.; Birkeland, N.K. Geothermal springs in Armenia and Nagorno-Karabakh: Potential sources of hydrolase-producing thermophilic bacilli. Extremophiles 2020, 24, 519–536. [Google Scholar] [CrossRef] [PubMed]
- Hovhannisyan, P.; Turabyan, A.; Panosyan, H.; Trchounian, A. Thermostable amylase production bacilli isolated from Armenian geothermal springs. Biol. J. Armenia 2016, 68, 6–15. [Google Scholar]
- Hovhannisyan, P.; Panosyan, H.; Trchounian, A.; Birkeland, N.K. Amplification and cloning of an alpha-amylase gene from Anoxybacillus flavithermus K103 isolated from an Armenian geothermal spring. In Proceedings of the 7th Congress of European Microbiologists (The FEMS 2017), Valencia, Spain, 9–13 July 2017. [Google Scholar]
- Poghosyan, L. Prokaryotic Diversity in an Armenian Geothermal Spring using Metagenomics, Anaerobic Cultivation and Genome Sequencing. Master’s Thesis, University of Bergen, Bergen, Norway, 2015. [Google Scholar]
- Shahinyan, G.; Margaryan, A.A.; Panosyan, H.H. Trchounian, A.H. Identification and sequence analysis of novel lipase encoding novel thermophilic bacilli isolated from Armenian geothermal springs. BMC Microbiol. 2017, 17, 103. [Google Scholar] [CrossRef]
- Vardanyan, G.; Margaryan, A.; Panosyan, H. Isolation and characterization of lipase-producting bacilli from Tatev geothermal spring (Armenia). In Collection of Scientific articles of YSU SSS: Materials of the Scientific Session Dedicated to the 95th Anniversary of YSU; Yerevan State University: Yerevan, Armenia, 2015; pp. 33–36. [Google Scholar]
- Panosyan, H.; Di Donato, P.; Poli, A.; Nicolaus, B. Production and characterization of exopolysaccharides by Geobacillus thermodenitrificans ArzA-6 and Geobacillus toebii ArzA-8 strains isolated from an Armenian geothermal spring. Extremophiles 2018, 22, 725–737. [Google Scholar] [CrossRef]
- Saghatelyan, A.; Poghosyan, L.; Panosyan, H.; Birkeland, N.K. Draft genome sequence of Thermus scotoductus strain K1, isolated from a geothermal spring in Karvachar, Nagorno Karabakh. Genome Announc. 2015, 3, e01346-15. [Google Scholar] [CrossRef]
- Saghatelyan, A.; Panosyan, H.; Trchounian, A.; Birkeland, N.K. Characteristics of DNA polymerase I from an extreme thermophile, Thermus scotoductus strain K1. Microbiol. Open 2021, 10, e1149. [Google Scholar] [CrossRef]
- Panosyan, H. Thermoactinomycetes isolated from geothermal springs in Armenia capable of producing extracellular hydrolases. Environ. Sustain. 2019, 2, 219–226. [Google Scholar] [CrossRef]
- Islam, T.; Larsen, Ø.; Torsvik, V.; Øvreås, L.; Panosyan, H.; Murrell, C.; Birkeland, N.K.; Bodrossy, L. Novel methanotrophs of the Family Methylococcaceae from different geographical regions and habitats. Microorganisms 2015, 3, 484–499. [Google Scholar] [CrossRef] [PubMed]
- Poghosyan, L.; Birkeland, N.K.; Panosyan, H. Diversity of thermophilic anaerobes in the geothermal spring Jermuk in Armenia. In Proceedings of the International Scientific Workshop on Trends in Microbiology and Microbial Biotechnology, Yerevan, Armenia, 5–8 October 2014; p. 83. [Google Scholar]
- Edwards, T.A.; Calica, N.A.; Huang, D.A.; Manoharan, N.; Hou, W.; Huang, L.; Panosyan, H.; Dong, H.; Hedlund, B.P. Cultivation and characterization of thermophilic Nitrospira species from geothermal springs in the U.S. Great Basin, China, and Armenia. FEMS Microbiol. Ecol. 2013, 85, 283–292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hedlund, B.P.; Dodsworth, J.A.; Cole, J.K.; Panosyan, H.H. An integrated study reveals diverse methanogens, Thaumarchaeota, and yet-uncultivated archaeal lineages in Armenian hot springs. Antonie Van Leeuwenhoek 2013, 104, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Paronyan, A.K.h. Ekologia, Biologicheskie Osobennosti Fototrofnikh Bakterii Armenii i Perspektiwi ikh Ispolzovanija. (Ecology, Biological Pecularities of Phototrophic Bacteria of Armenia and Prospects its Application). Ph.D. Disertation, Institute of Microbiology NAS of RA, Yerevan, Armenia, 2003. [Google Scholar]
- Paronyan, A. Ecophisiological charachteristics of phototrophic bacteria Rhodopseudomonas palustris isolated from mineral geothermal Jermuk. Biolog. J. Armenia 2007, 59, 73–77. (In Russian) [Google Scholar]
- Saghatelyan, A.; Panosyan, H. Study of bacterial diversity of Karvachar geothermal spring using culture-independent methods. In Collection of Scientific Articles of YSU SSS: Materials of the Scientific Session Dedicated to the 95th Anniversary of YSU; Yerevan State University: Yerevan, Armenia, 2015; pp. 13–18. [Google Scholar]
- Panosyan, H. Bacterial profiles of Karvachar hot spring identified by combination of different molecular approaches. Proc. Yerevan State Univ. Chem. Biol. 2020, 54, 147–153. [Google Scholar]
- Saghatelyan, A.; Panosyan, H.; Trchounian, A.; Birkeland, N.K. Investigation of microbial diversity of geothermal springs in Nagorno-Karabakh based on metagenomic and culture-based approaches. In Proceedings of the International Scientific Workshop on Trends in Microbiology and Microbial Biotechnology, Yerevan, Armenia, 5–8 October 2014; p. 38. [Google Scholar]
- Panosyan, H.H.; Margaryan, A.A.; Trchounian, A.H. Denaturing gradient gel electrophoresis (DGGE) profiles of the partial 16S rRNA genes defined bacterial population inhabiting in Armenian geothermal springs. Biolog. J. Armenia 2017, 68, 102–109. [Google Scholar]
- Kampfer, P.; Schulze, R.; Jackel, U.; Malik, K.A.; Amann, R.; Spring, S. Hydrogenophaga defluvii sp. nov. and Hydrogenophaga atypica sp. nov., isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 2005, 55, 341–344. [Google Scholar] [CrossRef]
- Kellermann, C.; Griebler, C. Thiobacillus thiophilus sp. nov., a chemolithoautotrophic, thiosulfate-oxidizing bacterium isolated from contaminated aquifer sediments. Int. J. Syst. Evol. Microbiol. 2009, 59, 583–588. [Google Scholar] [CrossRef]
- Kodama, Y.; Watanabe, K. Sulfuricurvum kujiense gen. nov., sp. nov., a facultatively anaerobic, chemolithoautotrophic, sulfur-oxidizing bacterium isolated from an underground crude-oil storage cavity. Int. J. Syst. Evol. Microbiol. 2004, 54, 2297–2300. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Z.; Belchik, S.M.; Edwards, M.J.; Liu, C.; Kennedy, D.W.; Merkley, E.D.; Lipton, M.D.; Butt, J.N.; Richardson, D.J.; et al. Identification and characterization of MtoA: A decaheme c-type cytochrome of the neutrophilic Fe (II)-oxidizing bacterium Sideroxydans lithotrophicus ES-1. Front. Microbiol. 2012, 3, 37. [Google Scholar] [CrossRef]
- Lee, S.Y.; Lee, M.H.; Oh, T.K.; Yoon, J.H. Lutibacter aestuarii sp. nov., isolated from a tidal flat sediment, and emended description of the genus Lutibacter Choi and Cho. Int. J. Syst. Evol. Microbiol. 2006, 62, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef] [PubMed]
- Panosyan, H.H.; Azaryan, A.S.; Birkeland, N.K. Microbiome structure of Karvachar (Nagorno-Karabakh) geothermal spring. In Proceedings of the 17th International Symposium on Microbial Ecology ISME17, Leipzig, Germany, 12–17 August 2018; p. 748. [Google Scholar]
- Wang, S.; Hou, W.; Dong, H.; Jiang, H.; Huang, L.; Wu, G.; Zhang, C.; Song, Z.; Zhang, Y.; Ren, H.; et al. Control of temperature on microbial community structure in hot springs of the Tibetan Plateau. PLoS ONE 2013, 8, e62901. [Google Scholar] [CrossRef]
- Poddar, A.; Das, S.K. Microbiological studies of hot springs in India: A review. Arch. Microbiol. 2017. [Google Scholar] [CrossRef]
- Cox, A.; Shock, E.L.; Havig, J.R. The transition to microbial photosynthesis in hot spring ecosystems. Chem. Geol. 2011, 280, 344–351. [Google Scholar] [CrossRef]
- Ochsenreiter, T.; Selezi, D.; Quaiser, A.; Bonch-Osmolovskaya, L.; Schleper, C. Diversity and abundance of Crenarchaeota in terrestrial habitats studied by 16S RNA surveys and real time PCR. Environ. Microbiol. 2003, 5, 787–797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.L.; Ye, Q.; Huang, Z.; Li, W.; Chen, J.; Song, Z.; Zhao, W.; Bagwell, C.; Inskeep, W.P.; Ross, C.; et al. Global occurrence of archaeal amoA genes in terrestrial hot springs. Appl. Environ. Microbiol. 2008, 74, 6417–6426. [Google Scholar] [CrossRef]
- Li, H.; Yang, Q.; Li, J.; Gao, H.; Li, P.; Zhou, H. The impact of temperature on microbial diversity and AOA activity in the Tengchong geothermal field, China. Sci. Rep. 2015, 5, 17056. [Google Scholar] [CrossRef]
- Chan, C.S.; Chan, K.G.; Ee, R.; Hong, K.W.; Urbieta, M.S.; Donati, E.R.; Shamsir, M.S.; Goh, K.M. Effects of Physiochemical Factors on Prokaryotic Biodiversity in Malaysian Circumneutral Hot Springs. Front. Microbiol. 2017, 8, 1252. [Google Scholar] [CrossRef]
- Badhai, J.; Ghosh, T.S.; Das, S.K. Taxonomic and functional characteristics of microbial communities and their correlation with physicochemical properties of four geothermal springs in Odisha, India. Front. Microbiol. 2015, 6, 1166. [Google Scholar] [CrossRef]
- Nakagawa, T.; Fukui, M. Phylogenetic characterization of microbial mats and streamers from a Japanese alkaline hot spring with a thermal gradient. J. Gen. Appl. Microbiol. 2002, 48, 211–222. [Google Scholar] [CrossRef]
- Derekova, A.; Mandeva, R.; Kambourova, M. Phylogenetic diversity of thermophilic carbohydrate degrading bacilli from Bulgarian hot springs. World J. Microbiol. Biotechnol. 2008, 24, 1697–1702. [Google Scholar] [CrossRef]
- Sahay, H.; Yadav, A.N.; Singh, A.K.; Singh, S.; Kaushik, R.; Saxena, A.K. Hot springs of Indian Himalayas: Potential sources of microbial diversity and thermostable hydrolytic enzymes. 3 Biotech 2017, 7, 118. [Google Scholar] [CrossRef]
- Adiguzel, A.; Ozkan, H.; Baris, O.; Inan, K.; Gulluce, M.; Sahin, F. Identification and characterization of thermophilic bacteria isolated from hot springs in Turkey. J. Microbiol. Methods. 2009, 79, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, R.J.; Grogan, D.W.; Taylor, J.W. Geographical barriers isolate endemic population of hyperthermophilic archaea. Science 2003, 301, 976–978. [Google Scholar] [CrossRef] [PubMed]
- Takacs-Vesbach, C.; Mitchell, K.; Jakson-Weaver, O.; Reysenbach, A.L. Volcanic calderas delineate biogeographi provinces among Yellowstone thermophiles. Environ. Microbiol. 2008, 10, 1681–1689. [Google Scholar] [CrossRef] [PubMed]
- Arya, M.; Joshi, G.K.; Gupta, A.K.; Kumar, A.; Raturi, A. Isolation and characterization of thermophilic bacterial strains from Soldhar (Tapovan) hot spring in Central Himalayan Region, India. Ann. Microbiol. 2015, 65, 1457–1464. [Google Scholar] [CrossRef]
- Yadav, P.; Korpole, S.; Prasad, G.S.; Sahni, G.; Maharjan, J.; Sreerama, L.; Bhattarai, T. Morphological, enzymatic screening, and phylogenetic analysis of thermophilic bacilli isolated from five hot springs of Myagdi. Nepal J. App. Biol. Biotech. 2018, 6, 1–8. [Google Scholar]
- Cihan, A.C.; Ozcan, B.; Tekin, N.; Cokmus, C. Phylogenetic diversity of isolates belonging to genera Geobacillus and Aeribacillus isolated from different geothermal regions of Turkey. World J. Microbiol. Biotechnol. 2011, 27, 2683–2696. [Google Scholar] [CrossRef]
- Gulecal-Pektas, Y.; Temel, M. A window to the subsurface: Microbial diversity in hot springs of a sulfidic cave (Kaklik, Turkey). Geomicrobiol. J. 2017, 34, 374–384. [Google Scholar] [CrossRef]
- Gulecal-Pektas, Y. Culture-Dependent and Culture-Independent Characterization of Microbial Communities in Hot springs of Bursa, Turkey. J. Pure Appl. Microbiol. 2016, 10, 1607–1612. [Google Scholar]
- Tomova, I.; Stoilova-Disheva, M.; Lyutskanova, D.; Pascual, J.; Petrov, P.; Kamburova, M. Phylogenetic analysis of the bacterial community in a geothermal spring, Rupi Basin, Bulgaria. World J. Microbiol. Biotechnol. 2010, 26, 2019–2028. [Google Scholar] [CrossRef]
- Mizerakis, P.; Stathopoulou, P.; Tsiamis, G.; Baeshen, M.N.; Mahyoub, J.A.; Elazzazy, A.M.; Bellou, S.; Sakoulogeorga, E.; Triantaphyllidou, I.-E.; Mazioti, T.; et al. Bacterial diversity of the outflows of a Polichnitos (Lesvos, Greece) hot spring, laboratory studies of a Cyanobacterium sp. strain and potential medical applications. Ann. Microbiol. 2017, 67, 643–654. [Google Scholar] [CrossRef]
- Sharma, N.; Kumar, J.; Abedin, M.M.; Sahoo, D.; Pandey, A.; Rai, A.K.; Singh, S.P. Metagenomics revealing molecular profiling of community structure and metabolic pathways in natural hot springs of the Sikkim Himalaya. BMC Microbiol. 2020, 20, 1–17. [Google Scholar] [CrossRef]
- Mehetre, G.T.; Paranjpe, A.S.; Dastager, S.G.; Dharne, M.S. Complete metagenome sequencing based bacterial diversity and functional insights from basaltic hot spring of Unkeshwar, Maharashtra, India. Genom. Data 2016, 7, 140–143. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.K.; Bisht, S.S.; de Mandal, S.; Kumar, N.S. Bacterial and archeal community composition in hot springs from Indo-Burma region, North-east India. AMB Express 2016, 6, 1–12. [Google Scholar] [CrossRef]
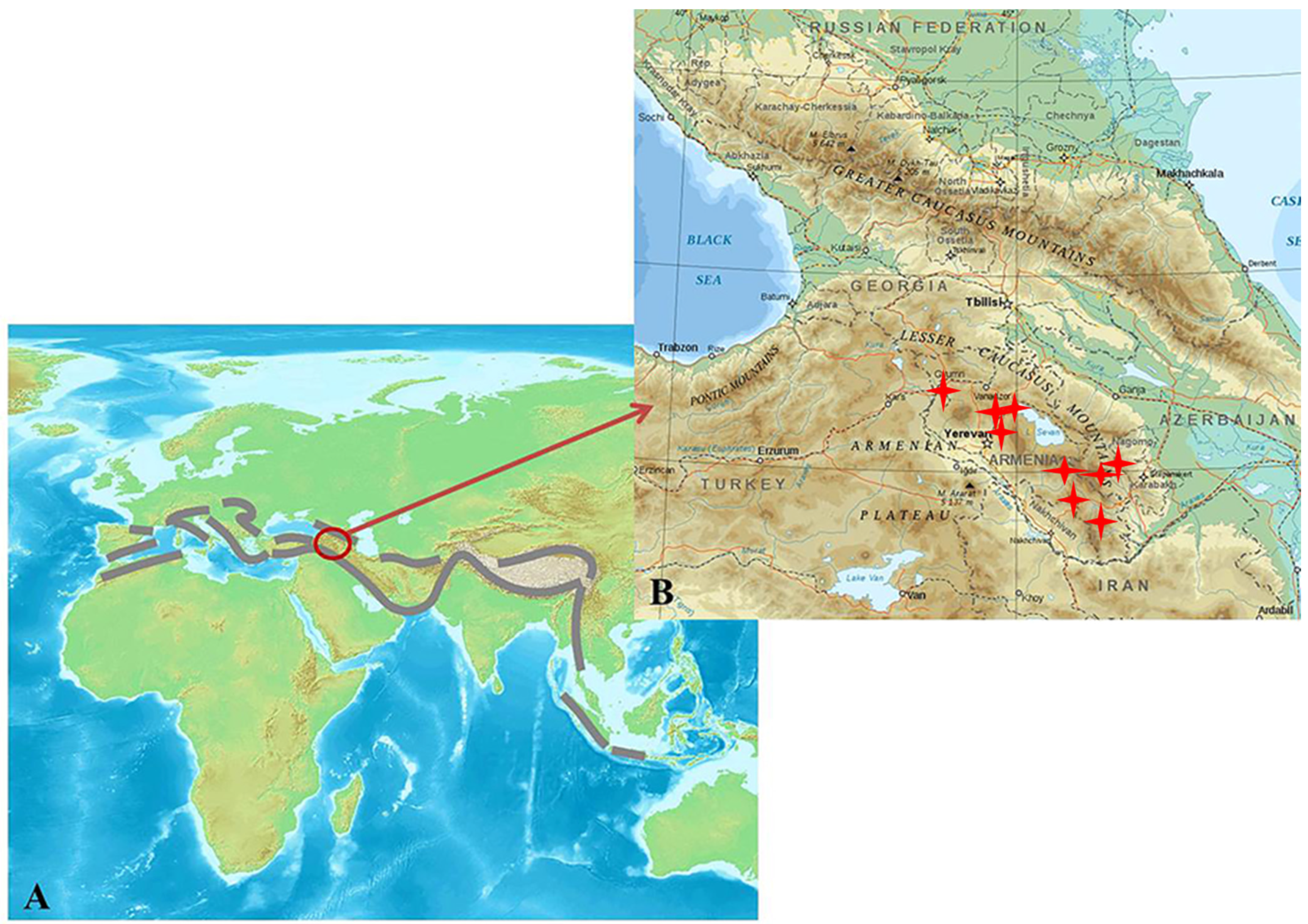
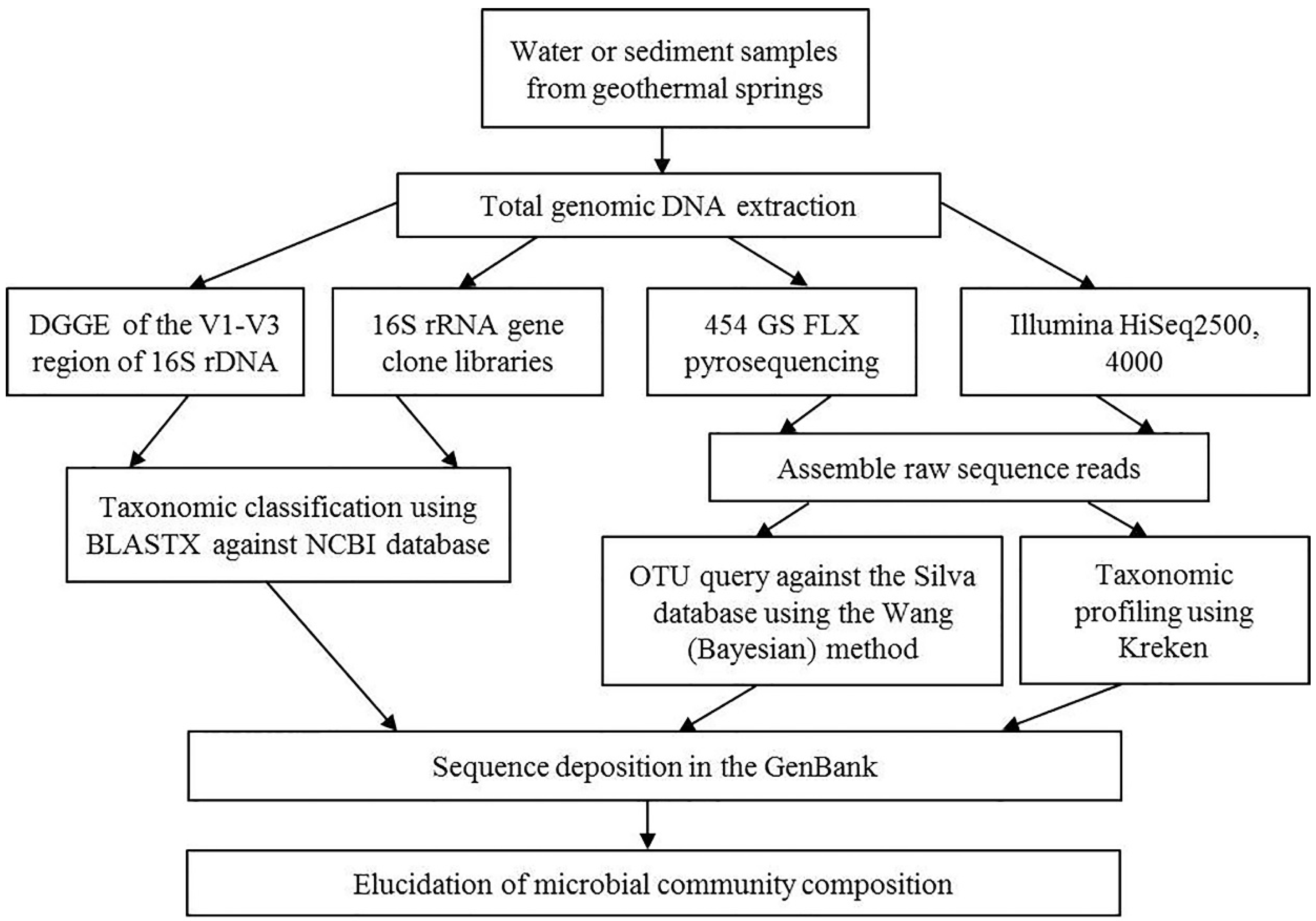
| Thermal Mineral Spring | Spring Location | Altitude, m Above Sea Level | pH | Conductivity, μS/cm | Temperature of Water in the Outlet, T, °C | Description | Photographs |
|---|---|---|---|---|---|---|---|
| Armenia | |||||||
| Akhurik | 40°44′34.04″ N 43°46′53.95″ E | 1490 | 6.5 | 2490 | 30 | Hydrocarbonate-sulphate sodium-magnesium type of spring. Slightly degassing. Sand at the bottom. |  |
| Arzakan | 40°27′36.10″ N 44°36′17.76″ E | 1490 | 7.2 | 4378.3 | 44 | Hydrocarbonate sodium class of mineral spring with a high concentration of dissolved minerals (of which > 20% is HCO3− and > 20% is Na+). Slightly degassing. Silicate sand at the bottom. |  |
| Bjni | 40°45’94.44” N 44°64’86.11” E | 1610 | 6.2–7.0 | 4138.3 | 30–37 | Chloride-hydrocarbonate sodium type of spring. Sand at the bottom. |  |
| Hankavan | 40°63′26.50″ N 44°48′46.00″ E | 1900 | 7.0–7.2 | 6722.9 | 42–44 | Hydrocarbonate-chloride sodium spring. Vigorously degassing. Silicate sand at the bottom. |  |
| Jermuk | 39°96′63.90″ N 45°68′52.80″ E | 2080 | 7.5 | 4340 | > 53 | Carbon hydro-sulphate-sodium water source. Sand at the bottom. |  |
| Tatev | 39°23′76.00″ N 46°15′48.00″ E | 960 | 6.0 | 1920 | 27.5 | Carbon-bicarbonate calcium water sources. Many bubbling sources and no visible outflow. Clays and sands at the bottom. Source was left in its natural form; no trace of human intervention was found. |  |
| Uytc (Uz) | 39°31′00″ N 46°03′09″ E | 1600 | 6.23 | 2700 | 25.8 | Hydrocarbonate-chloride-sulphate sodium source. Sand at the bottom. |  |
| Nagorno-Karabakh | |||||||
| Karvachar | 40°17′41.00″ N 46°27′50.00″ E | 1584 | 7.3 | 4600 | 70 | Hydrocarbonate-sulphate sodium source. Clear water and fine clay at the bottom. |  |
| Zuar | 40°02′47.60″ N 46°14′09.30″ E | 1520 | 7.0 | 4300 | 42 | Hydrocarbonate-sulphate sodium source. Clay and sand at the bottom. |  |
| Geothermal Spring | Bacterial and Archaeal Genera | Species | References |
|---|---|---|---|
| Armenia | |||
| Akhurik | Bacillus, Brevibacillus, Thermoactinomyces, Rhodobacter, Thiospirillum, Methylocaldum | B. licheniformis, B. pumilus, B. murimartini, B. psychrosaccharolyticus, B. borstelensis, Thermoactinomyces sp., R. sulfidophilus, T. jenense, Methylocaldum sp. | [35,39,44,45,49] |
| Arzakan | Bacillus, Paenibacillus, Geobacillus, Parageobacillus, Anoxybacillus, Rhodobacter, Rhodopseudomonas, Thiocapsa, Nitrospira, Arcobacter, Methanoculleus | B. licheniformis, B. simplex, Paenibacillus sp., G. thermodenitrificans, G. stearothermophilus, P. toebii, P. caldoxylosilyticus, A. rupiensis, R. shpaeroides, R. palustris, T. roseopersicina, N. calida, N. moscoviensis, Arcobacter sp., Methanoculleus sp. | [33,35,36,37,49] |
| Bjni | Bacillus, Ureibacillus, Parageobacillus, Anoxybacillus, Rhodobacter Rhodopseudomonas, Thiocapsa | B. licheniformis, U. thermosphaericus, P. toebii, Anoxybacillus sp., R. shpaeroides, R. palustris, T. roseopersicina | [35,49] |
| Hankavan | Bacillus, Brevibacillus, Geobacillus, Parageobacillus, Anoxybacillus | B. licheniformis,B. thermoruber, G. stearothermophilus, P. toebii, Anoxybacillus sp. | [35] |
| Jermuk | Bacillus, Parageobacillus, Geobacillus, Anoxybacillus, Desulfomicrobium, Desulfovibrio, Treponema, Rhodobacter, Rhodopseudomonas, Thiospirillum, Nitrospira | B. licheniformis, P. caldoxylosilyticus, Geobacillus sp., A. gonensis, A. kestanbolensis, A. flavithermus, D. thermophilum, D. psychrotolerans, Treponema sp., R. capsulatus, R. shpaeroides, R. palustris, T. jenense, N. calida, N. moscoviensis | [35,38,47,49] |
| Tatev | Bacillus, Geobacillus, Anoxybacillus, Parageobacillus, Thermoactinomyces | B. aerius, Geobacillus sp., Anoxybacillus sp., P. toebii, T. vulgaris | [35,44] |
| Uyts | Bacillus, Ureibacillus, Aeribacillus, Anoxybacillus, Geobacillus | B. licheniformis, B. glycinifermentans, U. terrenus, A. pallidus, Anoxybacillus sp., Geobacillus sp. | [35] |
| Nagorno-Karabakh | |||
| Karvachar | Bacillus, Aeribacillus, Anoxybacillus, Thermus | B. licheniformis, B. simplex, A. pallidus, A. suryakundensis, A. flavithermus, T. scotoductus | [35,42,43] |
| Zuar | Anoxybacillus, Geobacillus | A.rupiensis, Geobacillus sp. | [35] |
| Genome Features | Strains | ||
|---|---|---|---|
| Anoxybacillus sp. K1 | T. scotoductus K1 | Treponema sp. J25 | |
| GenBank accession | MQAD00000000 | LJJR01000000 | PTQW00000000 |
| Contigs | 48 | 55 | 72 |
| Base pairs | 2,722,200 | 2,379,636 | 3,180,620 |
| GC % | 41.6 | 65.2 | 49.6 |
| rRNAs (5S, 16S, 23S) | 5, 4, 1 | 3, 1, 3 | 1, 1, 1 |
| tRNAs | 64 | 48 | 44 |
| Genes (total) | 2883 | 2529 | 2744 |
| CRISPR arrays | 2 | 2 | 3 |
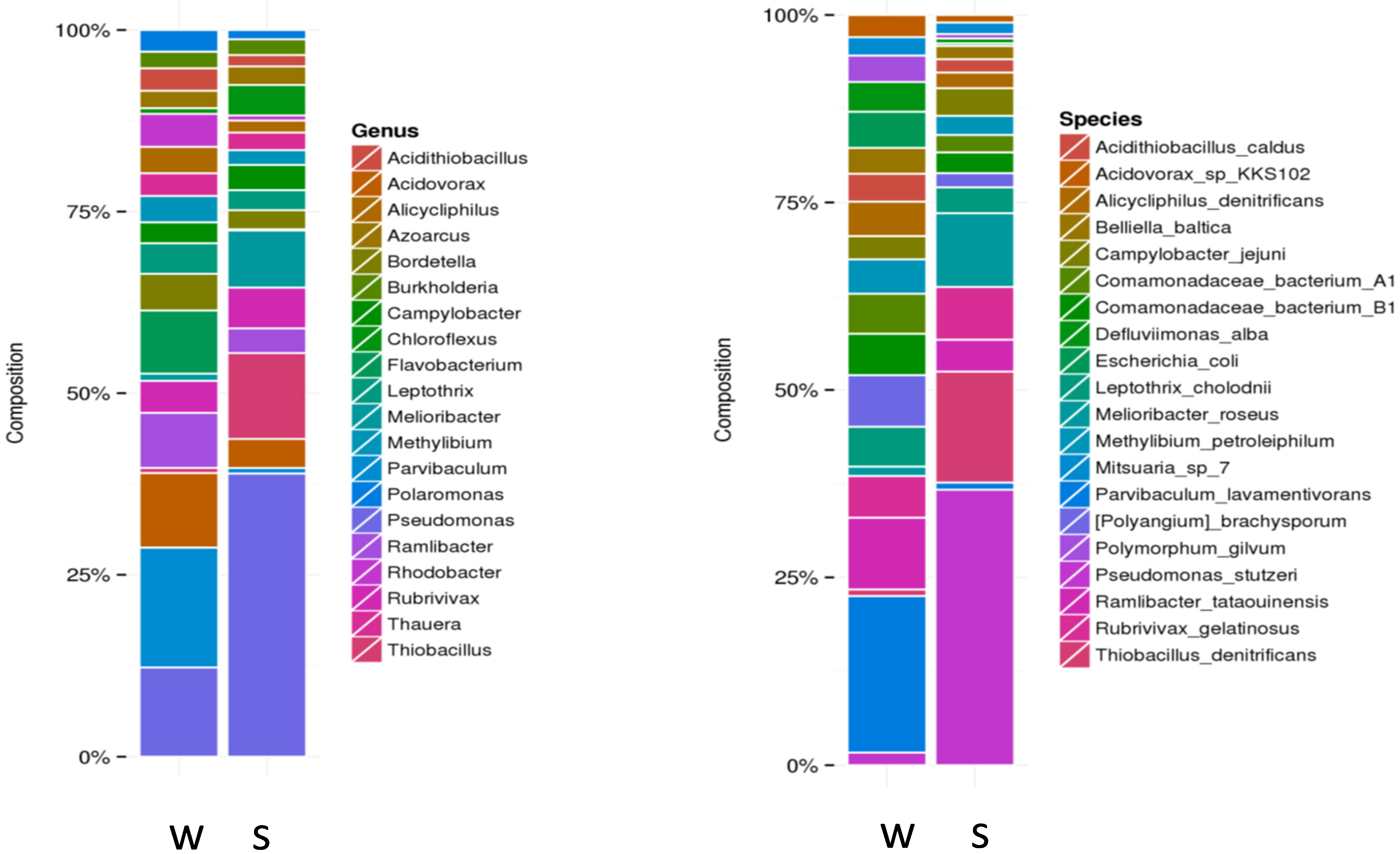
| Geothermal Spring | Approach | Dominant Bacterial and Archaeal Phyla | Accession Number | References |
|---|---|---|---|---|
| Armenia | ||||
| Arzakan | Shotgun pyrosequencing of the V4 region on the 454 GS FLX platform | Cyanobacteria, Proteobacteria, Bacteroidetes, Chloroflexi, Spirochaeta, Euryarchaeota, Crenarchaeota (the as yet uncultivated group, MCG) | SRR747863 | [48] |
| Bacterial 16S rRNA gene library | Bacteroidetes, Cyanobacteria, Betaproteobacteria, Gammaproteobacteria, Epsilonproteobacteria, Firmicutes, Alphaproteobacteria | JQ929026–JQ929037 | [33] | |
| Archaeal 16S rRNA gene library | Euryarchaeota, AOA Thaumarchaeota ‘‘Ca. Nitrososphaera gargensis’’, as yet uncultivated Crenarchaeota (MCG and DHVC1 groups) | KC682067–KC682083 | [48] | |
| DGGE | Beta-, Epsilon-, and Gammaproteobacteria, Bacteroidetes, Cyanobacteria | JX456536–JX456538 | [33,34] | |
| Jermuk | Shotgun pyrosequencing of the V4 region on the 454 GS FLX platform | Proteobacteria, Bacteroidetes, Synergistetes Euryarchaeota, as yet uncultivated Crenarchaeota (MCG and DHVC1 groups) | SRR747864 | [48] |
| Illumina HiSeq2500 paired-end sequencing | Proteobacteria, Firmicutes, Bacteroidetes, candidate division WS6, candidate phylum Ignavibacteria, Euryarchaeota, Crenarchaeota, Thaumarchaeota | - | [38] | |
| Archaeal 16S rRNA gene library | Euryarchaeota, AOA Thaumarchaeota ‘‘Ca. Nitrososphaera gargensis’’, as yet uncultivated Crenarchaeota (MCG group) | KC682084–KC682097 | [48] | |
| DGGE | Epsilonproteobacteria, Bacteroidetes, Spirochaetes, Ignavibacteriae, Firmicutes | [34] | ||
| Nagorno-Karabakh | ||||
| Karvachar | Bacterial 16S rRNA gene library | Proteobacteria, Cyanobacteria, Bacteroidetes, Chloroflexi, Verrumicrobia, Planctomycetes | - | [51,52] |
| DGGE | Bacteroidetes, Firmicutes | - | [34,51] | |
| Whole-metagenome shotgun sequencing using the Illumina Hiseq 4000 platform | Actinobacteria; Alpha-, Beta-, Delta-, Epsilon-, and Gammaproteobacteria; Bacteroidetes/Clorobi; Firmicutes; Clamydiaae; Cyanobacteria/Melainabacteria; Fusobacteria; Synergistia | - | [51] | |
| Zuar | Bacterial 16S rRNA gene library | Proteobacteria, Firmicutes, Bacteroidets, Cyanobacteria, Tenericutes, as yet unclassified phylotypes | - | [53] |
| Hot Spring | T (°C)/pH | Main Ions in Water | Dominant or Major Bacterial Phyla * | Approach | Reference |
|---|---|---|---|---|---|
| Karvachar, Nagorno-Karabakh | 70/7.3 | Anions: HCO3−, SO42− Cations: Na+ | Proteobacteria Bacteroidetes Ignavibacteriae Actinobacteria Chloroflexi Deinococcus–Thermus | Illumina HiSeq 4000 | [61] |
| Kaklik, Turkey | 35.4/6.77 | Anions: Cl−, SO42− Cations: Mg2+, Ca2+, Fe | Proteobacteria Bacteroidetes Verrucomicrobia Firmicutes Actinobacteria Nitrospirae Acidobacteria | 454 pyrosequencing | [79] |
| Orhaneli, Bursa, Turkey | 68/7.8 | Anions: Cl−, NO3−, PO43− Cations: Na+, NH3-N, K+, Mg2+, Ca2+ | Proteobacteria Chloroflexi Bacteroidetes Firmicutes | 454 pyrosequencing | [80] |
| Rupi Basin, Bulgaria | 79/8.6 | Anions: Cl−, SO42−, HCO3−, HS− Cations: Na+, K+, Ca2+ | Proteobacteria Hydrogenobacter Deinococcus–Thermus Cyanobacteria Thermotoga Cytophaga | Clone library | [81] |
| Polichnitos, Greece | 80/7.5 | Anions: Cl−, SO42−, NO3− Cations: Na+, K+, Mg2+, Ca2+, NH4+ | Proteobacteria Cyanobacteria Firmicutes | Clone library | [82] |
| Polok, Sikkim Himalaya, India | 62/8.0 | Nd | Proteobacteria Firmicutes Chloroflexi Deinococcus–Thermus Aquificae Bacteroidetes | Illumina MiSeq 2500 | [83] |
| Unkeshwar, Maharashtra, India | 50–60/7.3 | Anions: PO43-, SO42- | Actinobacteria Verrucomicrobia Bacteriodes Deinococcus–Thermus Firmicutes | Illumina HiSeq 2500 | [84] |
| Jakrem, Meghalaya, India | 46/9.0–10.0 | Anions: SO42−, NO3−, Cl− Cations: Mg2+, Na+, K+, Ca2+, Fe | Firmicutes Chloroflexi Proteobacteria | Illumina MiSeq | [85] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saghatelyan, A.; Margaryan, A.; Panosyan, H.; Birkeland, N.-K. Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review. Microorganisms 2021, 9, 1473. https://doi.org/10.3390/microorganisms9071473
Saghatelyan A, Margaryan A, Panosyan H, Birkeland N-K. Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review. Microorganisms. 2021; 9(7):1473. https://doi.org/10.3390/microorganisms9071473
Chicago/Turabian StyleSaghatelyan, Ani, Armine Margaryan, Hovik Panosyan, and Nils-Kåre Birkeland. 2021. "Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review" Microorganisms 9, no. 7: 1473. https://doi.org/10.3390/microorganisms9071473
APA StyleSaghatelyan, A., Margaryan, A., Panosyan, H., & Birkeland, N.-K. (2021). Microbial Diversity of Terrestrial Geothermal Springs in Armenia and Nagorno-Karabakh: A Review. Microorganisms, 9(7), 1473. https://doi.org/10.3390/microorganisms9071473






