Multiple Potential Plant Growth Promotion Activities of Endemic Streptomyces spp. from Moroccan Sugar Beet Fields with Their Inhibitory Activities against Fusarium spp.
Abstract
1. Introduction
2. Materials and Methods
2.1. Actinomycete Strains
2.2. Actinomycete Abilities to Use Orthoclase as Sole Potassium Source
2.3. Quantitative Estimation of the Amount of Solubilized Potassium by the Selected Actinomycete Strains
2.4. Assay for Indole Acetic Acid (IAA) Production
2.5. Isolation of Fungi from Symptomatic Sugar Beet Roots
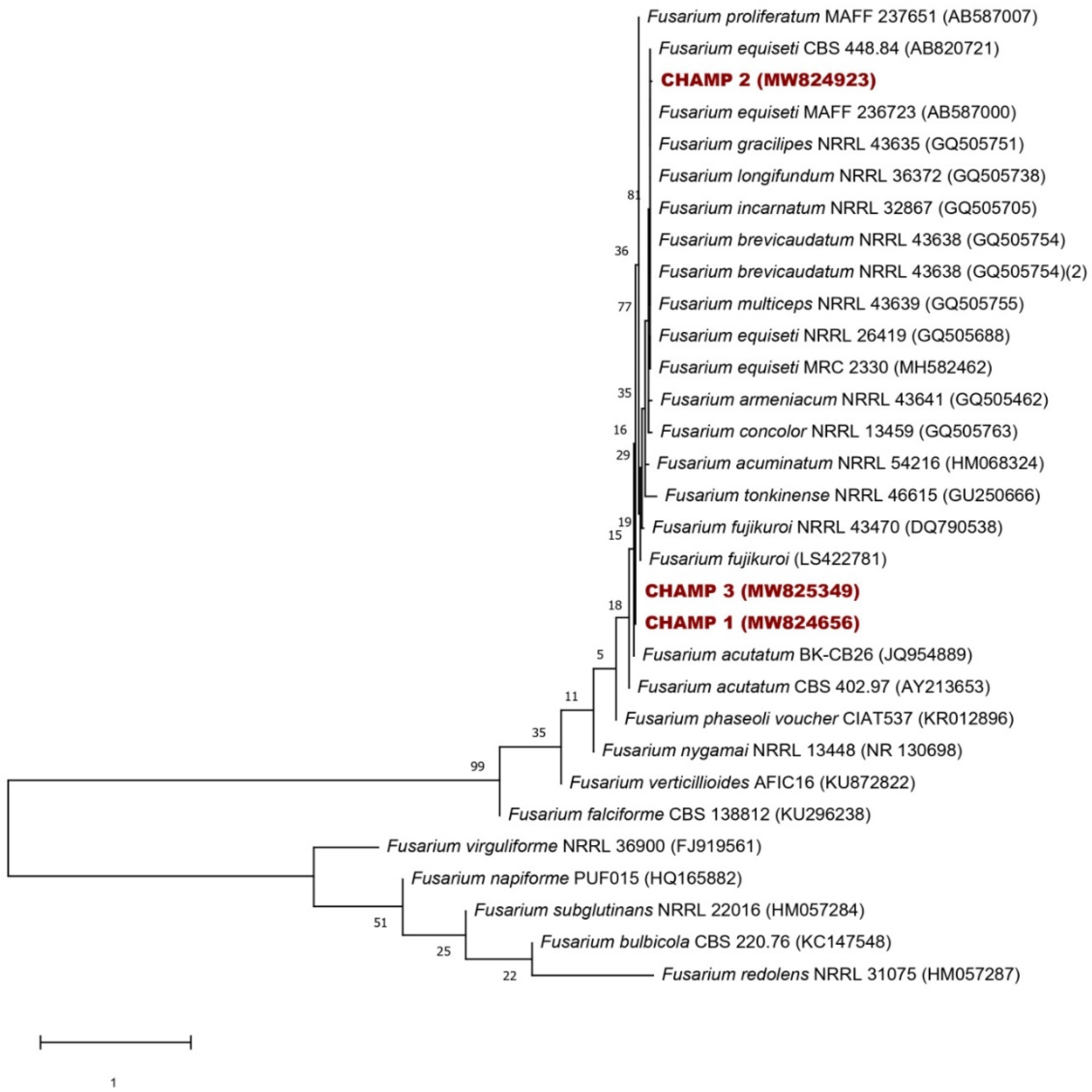
2.6. Molecular Determination of Isolated Fungi (DNA Extraction and ITS Amplification)
2.7. In Vitro Antagonistic Activity Assays
2.8. Statistical Analysis and Detection of the Most Promising Strains for Simultaneous Biofertilizer and Biocontrol Applications
3. Results
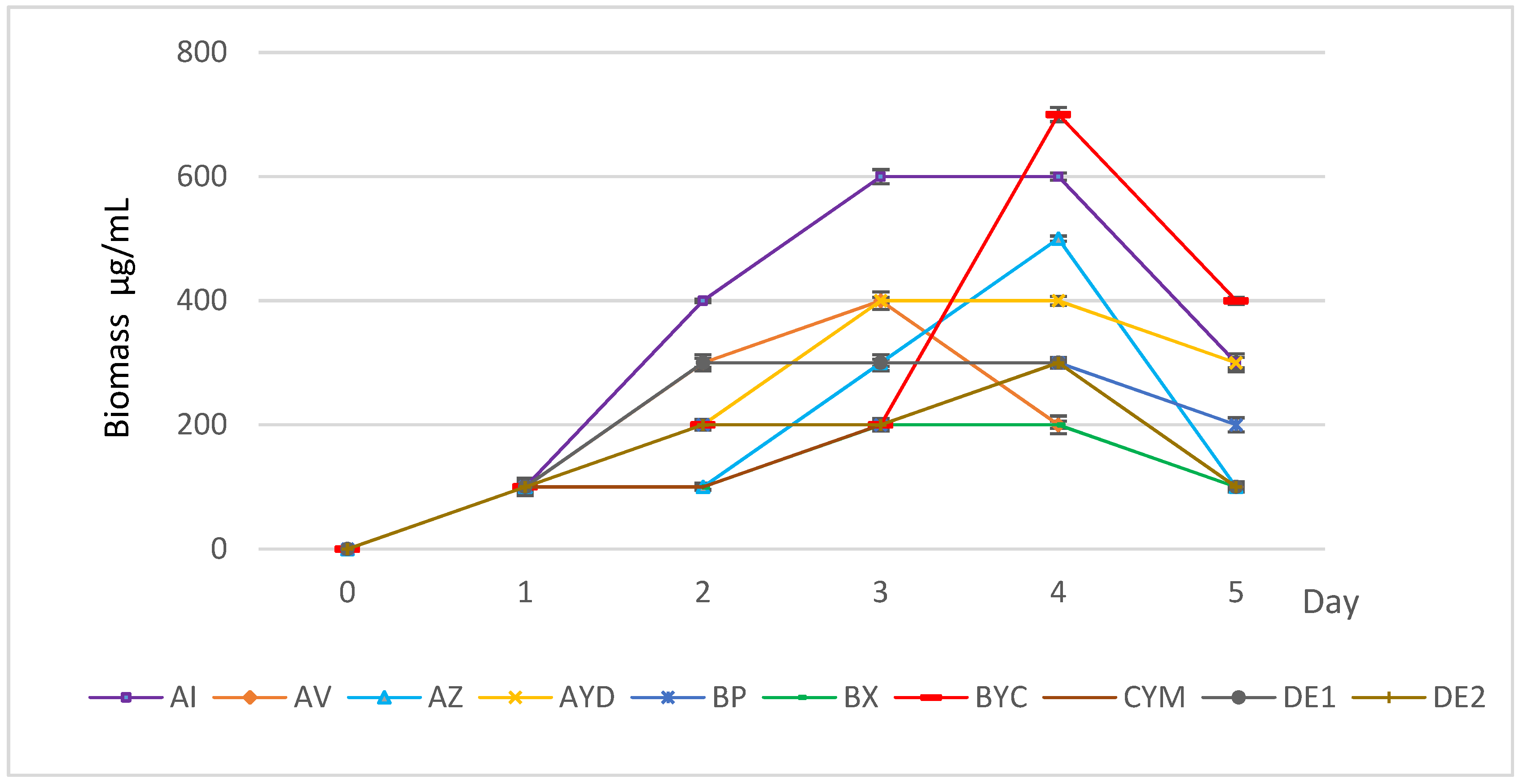
3.1. Growth Kinetics of the Selected Actinomycete Strains in AMM + Orthoclase
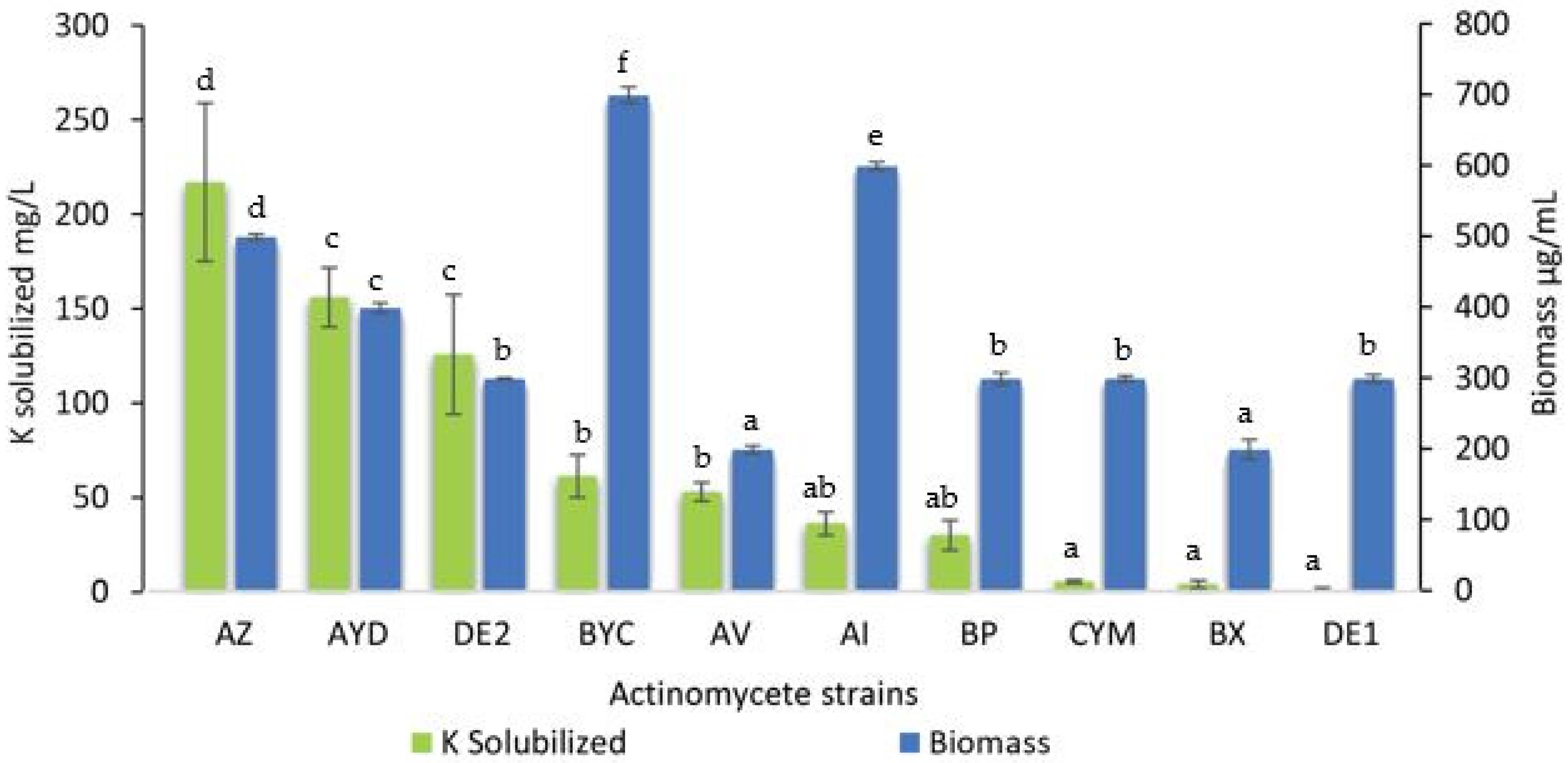
3.2. Estimation of the Amount Soluble Potassium Released from Orthoclase by the Selected Actinomycete Strains
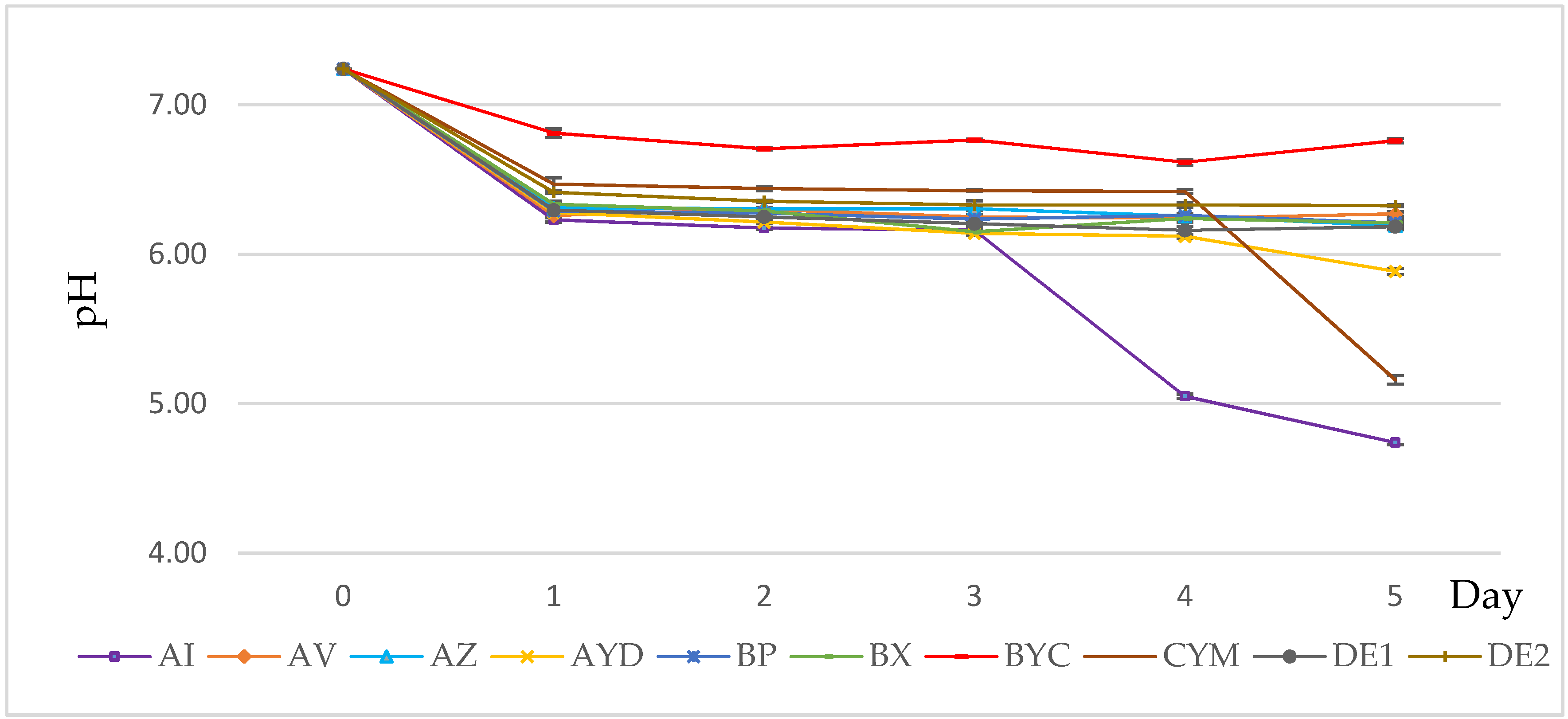
3.3. PH Evolution of the Growth Medium
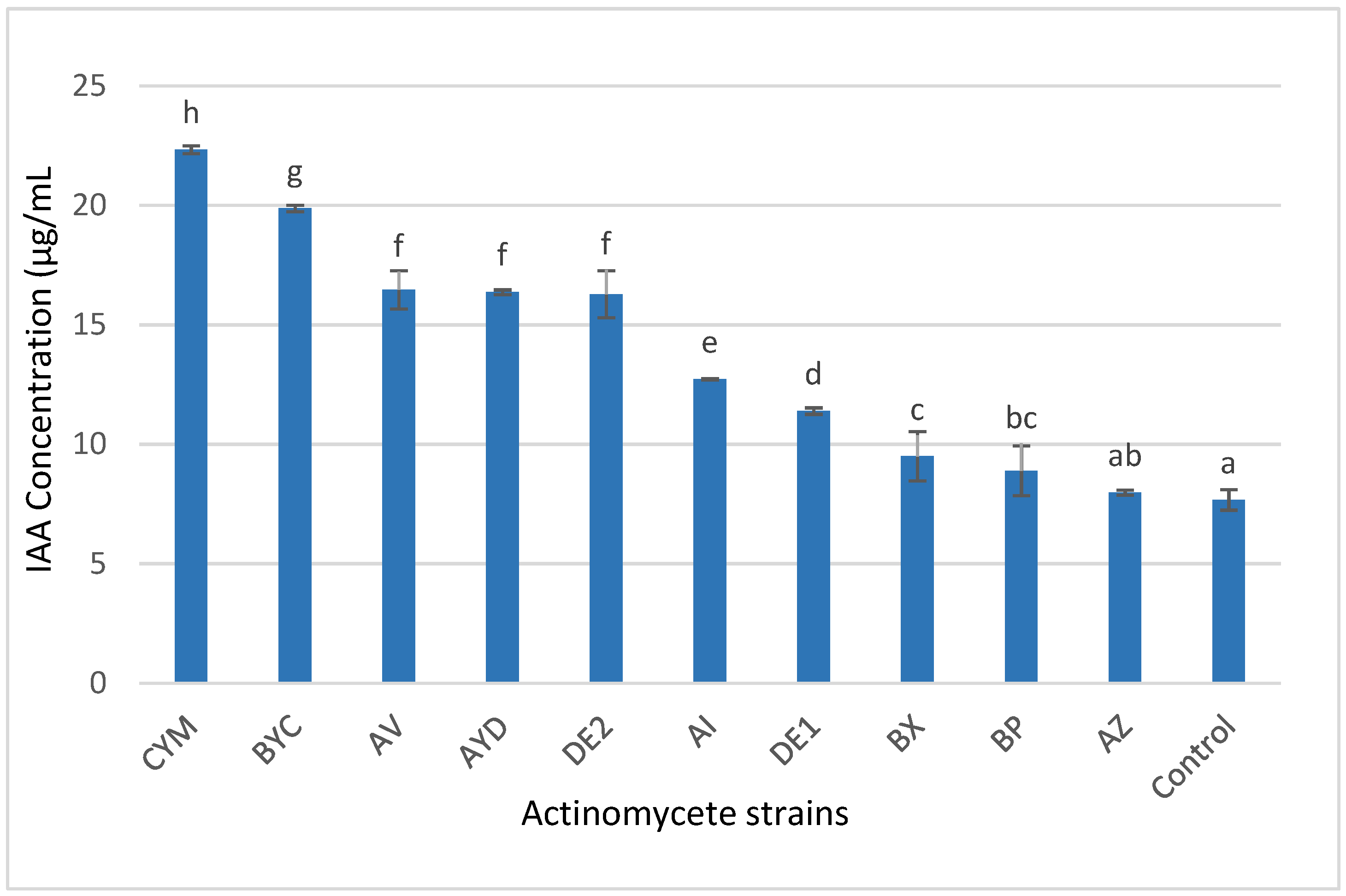
3.4. Indole Acetic Acid (IAA) Production
3.5. Identification of the Isolated Fungi from the Field
3.6. In Vitro Antagonistic Activity Assays
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lateef, E.; Abd El-salam, M.; Farrag, A.; Amin, G. Fertilizer Inputs Impact Of Different Bio-Solid Sources On Sugar Beet Yield In Sandy Soil. Int. J. Agric. For. Life Sci. 2019, 3, 106–114. [Google Scholar]
- Al Jbawi, E.; Al Raei, A.F.; Ali, A.A.A.; Al Zubi, H. Genotype-Environment Interaction Study in Sugar Beet (Beta vulgaris L.). Int. J. Environ. 2013, 5, 9–19. [Google Scholar] [CrossRef]
- FAOSTAT Food and Agricultural Organization Statistics. Available online: http://www.fao.org/faostat/en/#data/QC (accessed on 15 April 2021).
- ORMVAT Office Régional de Mise en Valeur Agricole de Tadla. Available online: www.ormvat.ma (accessed on 3 February 2021).
- Redani, L. Competitivite, Valorisation des Ressources et Objectifs de Sécurité Alimentaire pour la Filière Sucrière au Maroc. Ph.D Thesis, University of Liège, Gembloux, Belgium, 2015. [Google Scholar]
- Cakmak, I. The role of potassium in alleviating detrimental effects of abiotic stresses in plants. J. Plant Nutr. Soil. Sci. 2005, 168, 521–530. [Google Scholar] [CrossRef]
- Aksu, G.; Altay, H. The Effects of Potassium Applications on Drought Stress in Sugar Beet. Sugar Tech. 2020, 22, 1092–1102. [Google Scholar] [CrossRef]
- Barlog, P.; Grzebisz, W.; Peplinski, K.; Szczepaniak, W. Sugar Beet Response to Balanced Nitrogen Fertilization with Phosphorus and Potassium. Bulg. J. Agric. Sci. 2013, 19, 1311–1318. [Google Scholar]
- Hanafy, E.; El-Bana, A.; Yasin, M.; El-Naggar, N. Impact of Planting Density, Nitrogen and Potassium Fertilizer Levels on Yield and Quality of Sugar Beet. Zagazig J. Agric. Res. 2019, 46, 2133–2143. [Google Scholar] [CrossRef][Green Version]
- Abdel-Motagally, F.M.; Attia, K.K. Response of Sugar Beet Plants to Nitrogen and Potassium Fertilization in Sandy Calcareous Soil. Int. J. Agric. Biol. 2016, 11, 695–700. [Google Scholar]
- Sparks, D.L.; Huang, P.M. Physical Chemistry of Soil Potassium. In Potassium in Agriculture; Mounson, R.D., Ed.; American Society of Agronomy, Crop Science Society of America and Soil Science Society of America: Madison, WI, USA, 1985; pp. 201–276. [Google Scholar]
- Sugumaran, P.; Janarthanam, B. Solubilization of Potassium Containing Minerals by Bacteria and Their Effect on Plant Growth. World J. Agric. Sci. 2007, 3, 350–355. [Google Scholar]
- Li, Y.; Li, Q.; Guan, G.; Chen, S. Phosphate solubilizing bacteria stimulate wheat rhizosphere and endosphere biological nitrogen fixation by improving phosphorus content. PeerJ 2020, 8, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Z.; Xu, Z.; Fu, X. Effects of phosphate-solubilizing bacteria and N2-fixing bacteria on nutrient uptake, plant growth, and bioactive compound accumulation in Cyclocarya paliurus (Batal.) Iljinskaja. Forests 2019, 10, 772. [Google Scholar] [CrossRef]
- Baghel, V.; Thakur, J.K.; Yadav, S.S.; Manna, M.C.; Mandal, A.; Shirale, A.O.; Sharma, P.; Sinha, N.K.; Mohanty, M.; Singh, A.B.; et al. Phosphorus and Potassium Solubilization From Rock Minerals by Endophytic burkholderia sp. Strain FDN2-1 in Soil and Shift in Diversity of Bacterial Endophytes of Corn Root Tissue with Crop Growth Stage. Geomicrobiol. J. 2020, 1–14. [Google Scholar] [CrossRef]
- Ben-laouane, R.; Baslam, M.; Ait-el-mokhtar, M.; Anli, M.; Boutasknit, A.; Ait-Rahou, Y.; Toubali, S.; Mitsui, T. Potential of Native Arbuscular Mycorrhizal Fungi, Rhizobia, and/or Green Compost as Alfalfa (Medicago sativa) Enhancers under Salinity. Microorganisms 2020, 8, 1695. [Google Scholar] [CrossRef]
- Sattar, A.; Naveed, M.; Ali, M.; Zahir, Z.A.; Nadeem, S.M.; Yaseen, M.; Singh, S.V.; Farooq, M.; Singh, R.; Rahman, M.; et al. Perspectives of potassium solubilizing microbes in sustainable food production system: A review. Appl. Soil. Ecol. 2018, 133, 146–159. [Google Scholar] [CrossRef]
- Meena, V.S.; Maurya, B.R.; Verma, J.P. Does a rhizospheric microorganism enhance K+ availability in agricultural soils? Microbiol. Res. 2014, 169, 337–347. [Google Scholar] [CrossRef]
- Ali, A.M.; Awad, M.Y.M.; Hegab, S.A.; Abd, A.M.; Gawad, E.; Eissa, M.A.; Ali, A.M.; Awad, M.Y.M.; Hegab, S.A.; Abd, A.M. Effect of potassium solubilizing bacteria (Bacillus cereus) on growth and yield of potato. J. Plant Nutr. 2020, 44, 411–420. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, X.; Chen, W.; Huang, Q. Isolation and identification of three potassium- solubilizing bacteria from rape rhizospheric soil and their effects on ryegrass. Geomicrobiol. J. 2017, 34, 873–880. [Google Scholar] [CrossRef]
- Sun, F.; Ou, Q.; Wang, N.; Ou, Y.; Li, N.; Peng, C. Isolation and identification of potassium-solubilizing bacteria from Mikania micrantha rhizospheric soil and their effect on M. micrantha plants. Glob. Ecol. Conserv. 2020, 23, 1–27. [Google Scholar] [CrossRef]
- Saha, M.; Maurya, B.R.; Meena, S.; Bahadur, I.; Kumar, A. Identification and characterization of potassium solubilizing bacteria (KSB) from Indo-Gangetic Plains of India. Biocatal. Agric. Biotechnol. 2016, 7, 202–209. [Google Scholar] [CrossRef]
- Boubekri, K.; Soumare, A.; Mardad, I.; Lyamlouli, K.; Hafidi, M.; Ouhdouch, Y.; Kouisni, L. The Screening of Potassium- and Phosphate-Solubilizing Actinobacteria and the Assessment of Their Ability to Promote Wheat Growth Parameters. Microorganisms 2021, 9, 470. [Google Scholar] [CrossRef]
- Hillnhütter, C.; Mahlein, A.; Sikora, R.A.; Oerke, E. Field Crops Research Remote sensing to detect plant stress induced by Heterodera schachtii and Rhizoctonia solani in sugar beet fields. F. Crop. Res. J. 2011, 122, 70–77. [Google Scholar] [CrossRef]
- Wolfgang, A.; Zachow, C.; Müller, H.; Grand, A.; Temme, N.; Tilcher, R.; Berg, G. Understanding the Impact of Cultivar, Seed Origin, and Substrate on Bacterial Diversity of the Sugar Beet Rhizosphere and Suppression of Soil-Borne Pathogens. Front. Plant Sci. 2020, 11, 1–15. [Google Scholar] [CrossRef]
- De Lucchi, C.; Piergiorgio, S.; Linda, H.; Mitch, M.; Lee, P.; De, B.M.; Chiara, B.; Marco, B.; Sella, L.; Concheri, G. Molecular markers for improving control of soil-borne pathogen Fusarium oxysporum in sugar beet. Euphytica 2017, 213, 1–14. [Google Scholar] [CrossRef]
- Harveson, R.M.; Rush, C.M. Genetic Variation among Fusarium oxysporum Isolates from Sugar Beet as Determined by Vegetative Compatibility. Plant Dis. 1997, 81, 85–88. [Google Scholar] [CrossRef]
- Harveson, R.M.; Rush, C.M. Characterization of Fusarium Root Rot Isolates from Sugar Beet by Growth and Virulence at Different Temperatures and Irrigation Regimes. Plant Dis. 1998, 82, 1039–1042. [Google Scholar] [CrossRef]
- Hanson, L.E.; Hill, A.L.; Jacobsen, B.J.; Panella, L. Response of Sugarbeet Lines to Isolates of Fusarium oxysporum f. sp. Betae from the United States. J. Sugar Beet Res. 2009, 46, 11–26. [Google Scholar] [CrossRef]
- Jacobsen, B.J. Root Rot Diseases of Sugar Beet. Proc. Nat. Sci. Matica Srp. Novi. Sad. 2006, 9–19. [Google Scholar] [CrossRef]
- Errakhi, R.; Lebrihi, A.; Barakate, M. In vitro and in vivo antagonism of actinomycetes isolated from Moroccan rhizospherical soils against Sclerotium rolfsii: A causal agent of root rot on sugar beet (Beta vulgaris L.). J. Appl. Microbiol. 2009, 107, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Djebaili, R.; Pellegrini, M.; Smati, M.; Del Gallo, M.; Kitouni, M. Actinomycete Strains Isolated from Saline Soils: Plant-Growth-Promoting Traits and Inoculation Effects on Solanum lycopersicum. Sustainability 2020, 12, 4617. [Google Scholar] [CrossRef]
- Yoolong, S.; Kruasuwan, W.; Phạm, H.T.; Jaemsaeng, R.; Jantasuriyarat, C.; Thamchaipenet, A. Modulation of salt tolerance in Thai jasmine rice (Oryza sativa L. venezuelae cv KDLM105) by Streptomyces ATCC 10712 expressing ACC deaminase. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Gebauer, L.; Bouffaud, M.; Ganther, M.; Yim, B.; Vetterlein, D.; Smalla, K.; Buscot, F.; Heintz-buschart, A.; Tarkka, M.T. Soil Texture, Sampling Depth and Root Hairs Shape the Structure of ACC Deaminase Bacterial Community Composition in Maize Rhizosphere. Front. Microbiol. 2021, 12, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Elbendary, A.A.; Hessain, A.M.; El, M.D.; Seida, A.A.; Moussa, I.M.; Mubarak, A.S.; Kabli, S.A.; Hemeg, H.A.; Kamal, J.; Jakee, E. Isolation of antimicrobial producing Actinobacteria from soil samples. Saudi J. Biol. Sci. 2017, 25, 44–46. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Glick, B.R.; Babalola, O.O. Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol. 2017, 33, 1–16. [Google Scholar] [CrossRef]
- Hamdali, H.; Bouizgarne, B.; Hafidi, M.; Lebrihi, A.; Virolle, M.J.; Ouhdouch, Y. Screening for rock phosphate solubilizing Actinomycetes from Moroccan phosphate mines. Appl. Soil. Ecol. 2008, 38, 12–19. [Google Scholar] [CrossRef]
- Hamdali, H.; Lebrihi, A.; Monje, M.C.; Benharref, A.; Hafidi, M.; Ouhdouch, Y.; Virolle, M.J. A Molecule of the Viridomycin Family Originating from a Streptomyces griseus-Related Strain Has the Ability to Solubilize Rock Phosphate and to Inhibit Microbial Growth. Antibiotics 2021, 10, 72. [Google Scholar] [CrossRef] [PubMed]
- Evangelista-Martínez, Z. Isolation and characterization of soil Streptomyces species as potential biological control agents against fungal plant pathogens. World J. Microbiol. Biotechnol. 2014, 30, 1639–1647. [Google Scholar] [CrossRef]
- Aallam, Y.; Dhiba, D.; Lemriss, S.; Souiri, A.; Karray, F.; El Rasafi, T.; Saïdi, N.; Haddioui, A.; El Kabbaj, S.; Virolle, M.J.; et al. Isolation and characterization of phosphate solubilizing Streptomyces sp. Endemic from sugar beet fields of the beni-mellal region in morocco. Microorganisms 2021, 9, 914. [Google Scholar] [CrossRef] [PubMed]
- Meena, V.S.; Maurya, B.R.; Verma, J.P.; Aeron, A.; Kumar, A.; Kim, K.; Bajpai, V.K. Potassium solubilizing rhizobacteria (KSR): Isolation, identification, and K-release dynamics from waste mica. Ecol. Eng. 2015, 81, 340–347. [Google Scholar] [CrossRef]
- Hu, X.; Chen, J.; Guo, J.; Hangzhou, X. Two Phosphate- and Potassium-solubilizing Bacteria Isolated from Tianmu Mountain, Zhejiang, China. World J. Microbiol. Biotechnol. 2006, 22, 983–990. [Google Scholar] [CrossRef]
- Liu, D.F.; Lian, B.; Wang, B. Solubilization of potassium containing minerals by high temperature resistant Streptomyces sp. isolated from earthworm’s gut. Acta Geochim. 2016, 35, 262–270. [Google Scholar] [CrossRef]
- Sachdev, D.P.; Chaudhari, H.G.; Kasture, V.M.; Dhavale, D.D.; Chopade, B.A. Isolation and characterization of indole acetic acid (IAA) producing Klebsiella pneumonia strains from rhizosphere of wheat (Triticum aestivum) and their effect on plant growth. Indian J. Exp. Biol. 2009, 47, 993–1000. [Google Scholar]
- Loper, J.E.; Schroth, M.N. Influence of Bacterial Sources of Indole3-acetic Acid on Root Elongation of Sugar Beet. Physiol. Biochem. 1986, 76, 386–389. [Google Scholar] [CrossRef]
- Barakat, A.; Ennaji, W.; Krimissa, S.; Bouzaid, M. Heavy metal contamination and ecological-health risk evaluation in peri-urban wastewater-irrigated soils of Beni-Mellal city (Morocco). Int. J. Environ. Health Res. 2019, 30, 372–387. [Google Scholar] [CrossRef]
- Chenaoui, M.; Amar, M.; Benkhemmar, O.; El Aissami, A.; Arahou, M.; Rhazi, L. Isolation and characterization of fungi from sugar beet roots samples collected from Morocco. J. Mater. Environ. Sci. 2017, 8, 3962–3967. [Google Scholar]
- Leaw, S.; Chang, H.; Sun, H.; Barton, R.; Bouchara, J.; Chang, T. Identification d’espèces de levure médicalement importantes par analyse de séquence des régions d’espaceur transcrites internes. J. Clin. Microbiol. 2006, 44, 693–699. [Google Scholar] [CrossRef]
- Vainio, E.J.; Hantula, J. Direct analysis of wood-inhabiting fungi using denaturing gradient gel electrophoresis of amplified ribosomal DNA. Myco Res. 2000, 104, 927–936. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Phylogenies and the Comparative Method. Am. Nat. 1985, 125, 1–15. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.W.; Kirby, M.; Sherris, J.C.; Turck, M. Antibiotic susceptibility testing by a standardized single disk method. Am. J. Clin. Pathol. 1966, 45, 493–496. [Google Scholar] [CrossRef]
- Hamdali, H.; Hafidi, M.; Joe, M.; Ouhdouch, Y. Growth promotion and protection against damping-off of wheat by two rock phosphate solubilizing actinomycetes in a P-deficient soil under greenhouse conditions. Appl. Soil Ecol. 2008, 40, 510–517. [Google Scholar] [CrossRef]
- Karimi, E.; Safaie, N.; Shams-Baksh, M.; Mahmoudi, B. Bacillus amyloliquefaciens SB14 from rhizosphere alleviates Rhizoctonia damping-off disease on sugar beet. Microbiol. Res. 2016, 192, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Prasad, M.; Srinivasan, R.; Chaudhary, M.; Choudhary, M.; Jat, L.K. Plant Growth Promoting Rhizobacteria (PGPR) for Sustainable Agriculture: Perspectives and Challenges. In PGPR Amelioration in Sustainable Agriculture; Singh, K.A., Kumar, A., Singh, K.P., Eds.; Woodhead: Sawston, UK, 2019; pp. 129–157. [Google Scholar] [CrossRef]
- Han, D.; Wang, L.; Luo, Y. Isolation, identification, and the growth promoting effects of two antagonistic actinomycete strains from the rhizosphere of Mikania micrantha Kunth. Microbiol. Res. 2018, 208, 1–11. [Google Scholar] [CrossRef]
- Reyes-castillo, A.; Gerding, M.; Oyarzúa, P.; Zagal, E.; Gerding, J. Plant growth-promoting rhizobacteria able to improve NPK availability: Selection, identification and effects on tomato growth. Chil. J. Agric. Res. 2019, 79, 473–485. [Google Scholar] [CrossRef]
- Etesami, H.; Emami, S.; Alikhani, H.A. Potassium solubilizing bacteria (KSB): Mechanisms, promotion of plant growth, and future prospects—A review. J. Soil Sci. Plant Nutr. 2017, 17, 897–911. [Google Scholar] [CrossRef]
- Yuan, Z.S.; Liu, F.; Zhang, G.F. Characteristics and biodiversity of endophytic phosphorus- and potassium solubilizing bacteria in moso bamboo (Phyllostachys edulis). Acta Biol. Hung. 2015, 66, 449–459. [Google Scholar] [CrossRef]
- Dominguez-Nunez, J.A.; Benito, B.; Berrocal-Lobo, M.; Albanesi, A. Mycorrhizal Fungi: Role in the Solubilization of Potassium. In Potassium Solubilizing Microorganisms for Sustainable Agriculture; Meena, V., Maurya, B., Verma, J., Meena, R., Eds.; Springer: New Delhi, India, 2016; pp. 77–98. [Google Scholar] [CrossRef]
- Bashir, Z.; Zargar, M.Y.; Vishwakarma, D.K. Potassium-solubilizing Microorganisms for Sustainable Agriculture. In Applied Agricultural Practices for Mitigating Climate Change; Kumar, R., Singh, P.V., Jhajharia, D., Mirabbasi, R., Eds.; Taylor & Francis: New York, NY, USA, 2019; pp. 17–28. [Google Scholar]
- Bagyalakshmi, B.; Ponmurugan, P.; Balamurugan, A. Potassium solubilization, plant growth promoting substances by potassium solubilizing bacteria (KSB) from southern Indian Tea plantation soil. Biocatal. Agric. Biotechnol. 2017, 12, 116–124. [Google Scholar] [CrossRef]
- Liu, W.; Xu, X.; Wu, X.; Yang, Q.; Luo, Y.; Christie, P. Decomposition of silicate minerals by Bacillus mucilaginosus in liquid culture. Environ. Geochem. Health 2006, 28, 133–140. [Google Scholar] [CrossRef]
- Sheng, X.F.; He, L.Y. Solubilization of potassium-bearing minerals by a wild-type strain of Bacillus edaphicus and its mutants and increased potassium uptake by wheat. Can. J. Microbiol. 2006, 72, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Shelobolina, E.; Xu, H.; Konishi, H.; Kukkadapu, R.; Wu, T.; Blöthe, M.; Roden, E. Microbial Lithotrophic Oxidation of Structural Fe (II) in Biotite. Appl. Environ. Microbiol. 2012, 78, 5746–5752. [Google Scholar] [CrossRef]
- Dhiman, S.; Dubey, R.C.; Baliyan, N.; Kumar, S.; Maheshwari, D.K. Application of potassium—Solubilising Proteus mirabilis MG738216 inhabiting cattle dung in improving nutrient use efficiency of Foeniculum vulgare Mill. Environ. Sustain. 2019, 2, 401–409. [Google Scholar] [CrossRef]
- Reddy, K.R.K.; Jyothi, G.; Sowjanya, C.; Kusumanjali, K.; Malathi, N.; Reddy, K.R.N. Plant Growth-Promoting Actinomycetes: Mass Production, Delivery systems, and commercialization. In Plant Growth Promoting Actinobacteria; Subramaniam, G., Arumugam, S., Rajendran, V.G., Eds.; Springer: Singapore, 2016; pp. 1–298. [Google Scholar] [CrossRef]
- Getha, K.; Vikineswary, S.; Wong, W.H.; Seki, T.; Ward, A.; Goodfellow, M. Evaluation of Streptomyces sp. strain g10 for suppression of Fusarium wilt and rhizosphere colonization in pot-grown banana plantlets. J. Ind. Microbiol. Biotechnol. 2005, 32, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Du, C.; Tian, C. Suppression of Fusarium oxysporum and induced resistance of plants involved in the biocontrol of Cucumber Fusarium Wilt by Streptomyces bikiniensis HD-087. World J. Microbiol. Biotechnol. 2012, 28, 2919–2927. [Google Scholar] [CrossRef]
- Anitha, A.; Rabeeth, M. Control of Fusarium Wilt of Tomato by Bioformulation of Streptomyces griseus in Green House Condition. Afr. J. Basic. Appl. Sci. 2009, 1, 9–14. [Google Scholar]
- Yang, Y.; Zhang, S.W.; Li, K.T. Antagonistic activity and mechanism of an isolated Streptomyces corchorusii stain AUH-1 against phytopathogenic fungi. World J. Microbiol. Biotechnol. 2019, 35, 1–9. [Google Scholar] [CrossRef]
- Li, Y.; He, F.; Lai, H.; Xue, Q. Mechanism of in vitro antagonism of phytopathogenic Scelrotium rolfsii by actinomycetes. Eur. J. Plant Pathol. 2017, 149, 299–311. [Google Scholar] [CrossRef]
- Soares, A.C.F.; Sousa, C.D.S.; Garrido, M.D.S.; Perez, J.O.; De Almeida, N.S. Soil streptomycetes with in vitro activity against the yam pathogens Curvularia eragrostides and Colletotrichum gloeosporioides. Braz. J. Microbiol. 2006, 37, 456–461. [Google Scholar] [CrossRef]
- Li, Y.; Guo, Q.; He, F.; Li, Y.; Xue, Q.; Lai, H. Biocontrol of Root Diseases and Growth Promotion of the Tuberous Plant Aconitum carmichaelii Induced by Actinomycetes Are Related to Shifts in the Rhizosphere Microbiota. Microb. Ecol. 2020, 79, 134–147. [Google Scholar] [CrossRef]
- Kurth, F.; Mailänder, S.; Bönn, M.; Feldhahn, L.; Herrmann, S.; Große, I.; Buscot, F.; Schrey, S.D.; Tarkka, M.T. Streptomyces—Induced resistance against oak powdery mildew involves host plant responses in defence, photosynthesis and secondary metabolism pathways. Mol. Plant Microbe Interact. 2014, 27, 891–900. [Google Scholar] [CrossRef] [PubMed]
- Musa, Z.; Ma, J.; Egamberdieva, D.; Abdelshafy, M.O.A.; Abaydulla, G.; Liu, Y.; Li, W.J.; Li, L. Diversity and Antimicrobial Potential of Cultivable Endophytic Actinobacteria Associated With the Medicinal Plant Thymus roseus. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.P.; Gupta, R.; Gaur, R.; Srivastava, A.K. Antagonistic Actinomycetes Mediated Resistance in Solanum lycopersicon Mill. Against Rhizoctonia solani Kühn. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2015, 87, 789–798. [Google Scholar] [CrossRef]

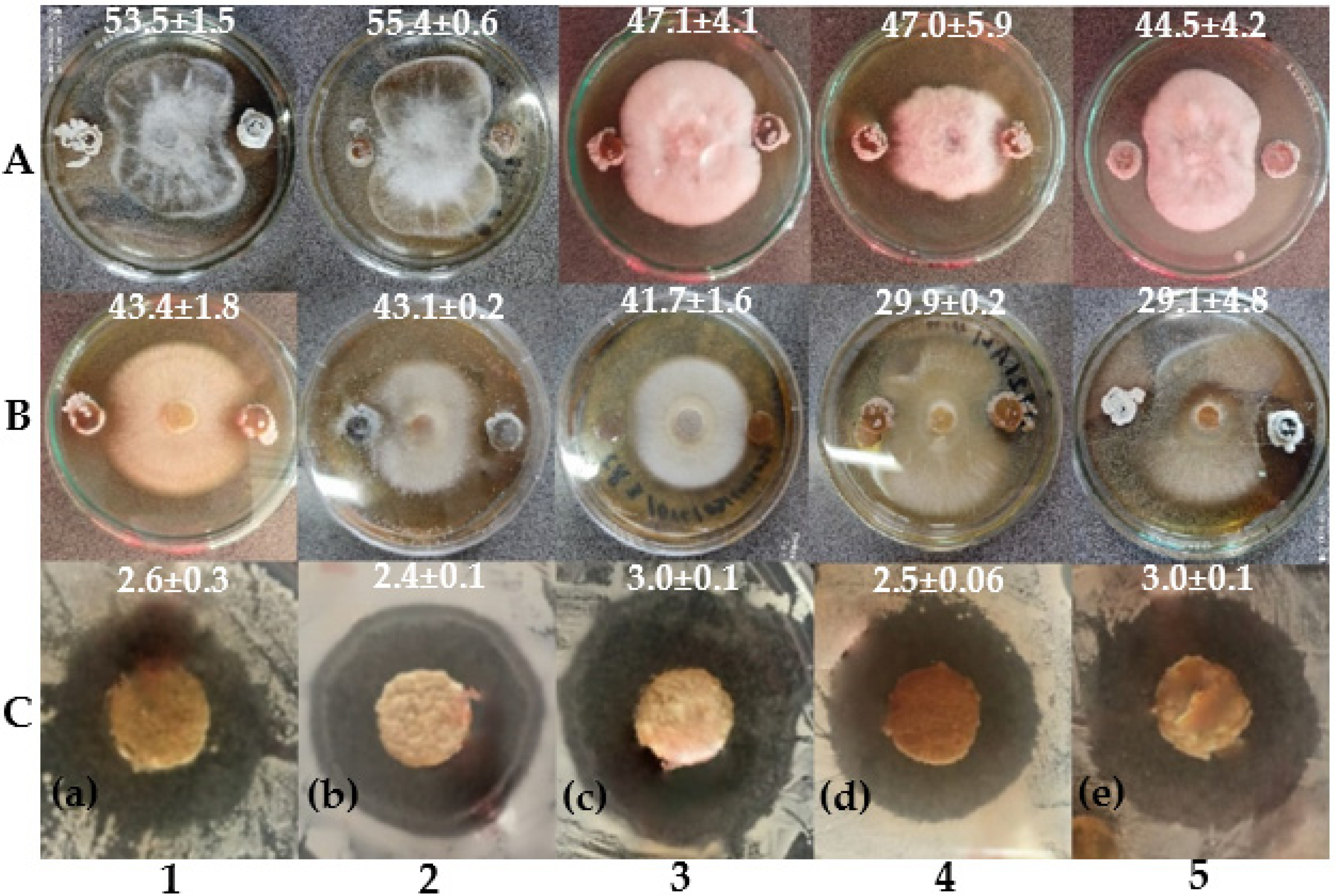
| Fungi | Bacteria Gram+ | Bacteria Gram– | Yeast | |||||
|---|---|---|---|---|---|---|---|---|
| Streptomyces Strains | CHAMP 1 | CHAMP 2 | CHAMP 3 | S. aureus CCMM/B2 | Salmonella sp. CCMM/B17 | K. pneumonia E40 | E. coli CCMM/B4 | C. albicans CCMM/L11 |
| Percentage of Inhibition (PI ± SD mm) | Zone of Inhibition ± SD cm | |||||||
| CYM | 41.3 ± 8.8 d | 35.4 ± 3.5 de | 30.7 ± 5.7 b | 2.8 ± 0.1 e | 0 | 2.7 ± 0.1 bc | 0 | 2.9 ± 0.1 d |
| BP | 24.2 ± 1.2 c | 8.2 ± 7.5 a | 23.8 ± 5.3 ab | 1.3 ± 0.0 a | 0 | 3.0 ± 0.1 d | 0 | 1.3 ± 0.1 a |
| DE1 | 20.4 ± 4.2 bc | 13.7 ± 7.3 ab | 23.0 ± 4.3 ab | 2.5 ± 0.2 cd | 0 | 2.8 ± 0.2 c | 0 | 2.6 ± 0.1 c |
| AZ | 7.6 ± 3.4 a | 19.2 ± 11 abc | 18.5 ± 2.6 ab | 2.3 ± 0.2 c | 0 | 2.2 ± 0.1 a | 0 | 1.7 ± 0.5 ab |
| BX | 12.7 ± 3.3 ab | 7.6 ± 0.7 a | 6.9 ± 0.4 a | 2.2 ± 0.4 c | 0 | 2.3 ± 0.1 a | 0 | 2.3 ± 0.3 c |
| Streptomyces Strains | Solubilization of RP *,ψ | Solubilization of TCP * | Potassium Solubilization ψ | Antifungal Activity ψ | Antibacterial Activity | Production of IAA ψ | Production of Siderophores * | Final Score |
|---|---|---|---|---|---|---|---|---|
| BYC | 20 | 9 | 14 | 16 | 4 | 18 | 9 | 90 |
| AYD | 16 | 8 | 18 | 20 | 3 | 14 | 8 | 87 |
| DE2 | 10 | 10 | 16 | 18 | 7 | 12 | 10 | 83 |
| AZ | 14 | 7 | 20 | 4 | 6 | 2 | 7 | 60 |
| AI | 12 | 6 | 10 | 14 | 1 | 10 | 6 | 59 |
| CYM | 8 | 2 | 6 | 10 | 10 | 20 | 2 | 58 |
| AV | 4 | 4 | 12 | 12 | 5 | 16 | 4 | 57 |
| BP | 18 | 5 | 8 | 8 | 9 | 4 | 5 | 57 |
| DE1 | 6 | 3 | 2 | 6 | 8 | 8 | 3 | 36 |
| BX | 2 | 1 | 4 | 2 | 2 | 6 | 1 | 18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aallam, Y.; Maliki, B.E.; Dhiba, D.; Lemriss, S.; Souiri, A.; Haddioui, A.; Tarkka, M.; Hamdali, H. Multiple Potential Plant Growth Promotion Activities of Endemic Streptomyces spp. from Moroccan Sugar Beet Fields with Their Inhibitory Activities against Fusarium spp. Microorganisms 2021, 9, 1429. https://doi.org/10.3390/microorganisms9071429
Aallam Y, Maliki BE, Dhiba D, Lemriss S, Souiri A, Haddioui A, Tarkka M, Hamdali H. Multiple Potential Plant Growth Promotion Activities of Endemic Streptomyces spp. from Moroccan Sugar Beet Fields with Their Inhibitory Activities against Fusarium spp. Microorganisms. 2021; 9(7):1429. https://doi.org/10.3390/microorganisms9071429
Chicago/Turabian StyleAallam, Yassine, Bouchra El Maliki, Driss Dhiba, Sanaa Lemriss, Amal Souiri, Abdelmajid Haddioui, Mika Tarkka, and Hanane Hamdali. 2021. "Multiple Potential Plant Growth Promotion Activities of Endemic Streptomyces spp. from Moroccan Sugar Beet Fields with Their Inhibitory Activities against Fusarium spp." Microorganisms 9, no. 7: 1429. https://doi.org/10.3390/microorganisms9071429
APA StyleAallam, Y., Maliki, B. E., Dhiba, D., Lemriss, S., Souiri, A., Haddioui, A., Tarkka, M., & Hamdali, H. (2021). Multiple Potential Plant Growth Promotion Activities of Endemic Streptomyces spp. from Moroccan Sugar Beet Fields with Their Inhibitory Activities against Fusarium spp. Microorganisms, 9(7), 1429. https://doi.org/10.3390/microorganisms9071429







