Abstract
Several parasite species are shared between humans and pigs. We explored the application of next-generation sequencing-based metabarcoding supplemented with real-time PCR to fecal DNAs from 259 samples from 116 pigs in Denmark to detect and differentiate single-celled intestinal parasites of zoonotic relevance. Enterocytozoon bieneusi, Balantioides coli, and Giardia duodenalis were observed in 34/37 (92%), 148/259 (57%), and 86/259 (33%) samples, respectively. Entamoeba polecki ST1, E. polecki ST3, and Entamoeba hartmanni were detected in 104/259 (40%), 161/259 (62%), and 8/259 (3%) samples, respectively. Metabarcoding and real-time PCR detected Cryptosporidium in 90/259 (35%) and 239/259 (92%) of the samples, respectively, with Cryptosporidium suis and Cryptosporidium scrofarum observed in nearly equal proportions. Blastocystis subtypes 1, 3, 5, and 15 were found in 72 (28%), 6 (2%), 176 (68%), and 36 (14%) of 259 samples, respectively. Iodamoeba was identified in 1/259 samples (<1%), while none of 37 tested samples was positive for Dientamoeba fragilis. Our results illustrate how metabarcoding exemplifies a ‘one-fits-many’ approach to detecting intestinal single-celled parasites in feces supplemented with real-time PCR for selected parasites. Using metabarcoding with pathogen-specific assays may help detect emerging and previously underdetected pathogens and further elucidate the role of micro-eukaryotic parasites in human and animal health and disease.
1. Introduction
Several parasites can be hosted by both human and non-human hosts. Zoonotic intestinal parasitic genera taking a toll on both human and animal health include Giardia and Cryptosporidium [1]. Meanwhile, a few others can be observed with varying frequency in both humans and larger mammals, such as pigs, including Enterocytozoon, Balantioides, Blastocystis, Dientamoeba, and Entamoeba [2,3,4,5,6,7,8,9], the public health and animal health significance and epidemiology of which remain unclear.
The screening of human fecal samples for parasites of clinical and epidemiological relevance is increasingly being assisted by DNA-based tools such as targeted conventional and real-time PCRs [10] and commercial solutions offering multiplex PCR assays. Traditionally, routine parasitological testing of samples from domestic animals (including pigs) has relied mainly on coprological approaches using concentration techniques (e.g., flotation and sedimentation) [11], while molecular methods are increasingly used. The results from the current routine approaches often indicate only the presence/absence of parasites, with little or no information on genetic diversity (such as species, lineages, genotypes, subtypes, etc.) due to their high morphological uniformity (e.g., Blastocystis or Giardia) or due to the use of pathogen-specific targeted assays.
Meanwhile, next-generation sequencing (NGS)-assisted assays have been introduced as a comprehensive, ‘one-fits-many’ approach to detecting and differentiating parasites at a given taxonomic level. As an example, we previously introduced a metabarcoding assay relying on amplicon-based NGS of nuclear ribosomal genes from bacteria, fungi, and parasites with automated software-based annotation of sequences to genus and—oftentimes—species/sub-species level. This method has proven useful for screening various matrices such as skin samples, cornea scrapings, sewage samples, and human stool samples for parasites and other non-viral organisms, the relevance of which depends on the focus and scope of the investigation [12,13,14,15].
Based on available literature, shotgun sequencing was recently applied to a minor collection of fecal DNA from pigs for parasite detection and differentiation [16], and amplicon-based NGS of ribosomal DNA in fecal samples from pigs has been attempted once [17] with no reporting of, for example, Giardia and Cryptosporidium. The aim of this study was therefore to explore the applicability of the metabarcoding approach supplemented by specific real-time PCR tests for testing samples from pigs for intestinal parasitic protists, with a focus on protists that can be hosted by both pigs and humans. We discuss the benefits and challenges of the approach, and the results provide insight into the genetic diversity and host specificity of the detected genera.
2. Materials and Methods
2.1. Samples
A total of 273 fecal DNAs from 120 pigs were available from previous studies [18,19,20,21]. The 5–12 week-old pigs had been sampled in the weaning units of four conventional pig farms in different regions in Denmark [20]. Herd 1 (n = 34 samples) and Herd 5 (n = 33) samples were from Jutland, while Herd 3 (n = 28 samples) and Herd 4 (n = 178 samples) were from Zealand. The sampling procedure is described in the study by Græsbøll et al., 2017 [18], and the herd numbers applied in the present study are the same as in the study by Græsbøll et al. The samples were collected either at defecation or per rectum. A minor proportion (13%) of the pigs had received tetracycline prior to sampling (8/34 from Herd 1; 15/28 from Herd 3; 12/178 from Herd 5, and 0/33 from Herd 5). However, it was not a goal in this study to investigate the impact of tetracycline on parasite positivity.
2.2. DNA Extraction
All samples were subject to DNA extraction as previously described [19]. Briefly, total DNA was extracted using the Maxwell® 16 LEV Blood DNA Kit (Promega Corporation, Madison, WI, USA). The samples were homogenized in a cell and tissue disruptor (Tissuelyser II, Qiagen, Hilden, Germany) with 5-mm stainless steel beads (Qiagen, Hilden, Germany) and bead-beaten for 2 min in a 25 mg/mL lysozyme buffer (Sigma-Aldrich, Søborg, Denmark A/S). Next, they were transferred to Maxwell® extraction kit cartridges, and DNA was extracted according to manufacturer’s instructions. DNA concentration and purity were evaluated by the 260/280 nm-ratio using the NanoDropND-1000 spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). The extracted DNAs were diluted to 40 ng/μL in nuclease-free water (Qiagen, Hilden, Germany) and stored at −20 °C until further processing.
2.3. Detection and Differentiation of Parasitic Genera by Metabarcoding
The DNAs were processed by the metabarcoding assay [12,13,14,15,22]. This method involves PCR-based amplification (PCR 1) of ribosomal genes using one set of primers targeting 16S and three sets of primers targeting 18S (Table 1). The G3 and G6 primers target the hyper-variable regions V3–V4 of the 18S, and G4 primers target V3–V5.

Table 1.
Primers used in the metabarcoding assay.
An adaptor PCR (PCR 2) was performed, and DNA concentration was quantified using the Quant-ITTM High-Sensitivity dsDNA Assay Kit (Thermo Fisher Scientific, Hvidovre, Denmark). PCR2 products were pooled in equimolar amounts across samples. Undesirable DNA amplicons were removed from the pooled amplicon library (PAL) by Agencourt AMPure XP bead (Beckman Coulter) purification. The resulting AMPure beads-purified PAL (bPAL) was diluted to its final concentration of 11.5 pM DNA in 0.001 N NaOH and used for sequencing on the Illumina MiSeq desktop sequencer (Illumina Inc., San Diego, CA, USA). The library was sequenced with the 500-cycle MiSeq Reagent Kit V2 in a 2 × 250 nt setup (Illumina Inc., San Diego, CA, USA).
Raw reads were assigned to taxon by the “BION” package (http://box.com/bion, accessed on 28 May, 2021), and involved quality trimming, read pairing, and chimera filtering before taxonomic classification of sequences. Sequences from the three 18s rRNA targets were queried against the SILVA database in combination with an in-house database.
2.4. Real-Time PCR for Cryptosporidium, Dientamoeba, Enterocytozoon, and Giardia
Based on previous experience [15], the sensitivity of the metabarcoding assay in terms of detecting Giardia, Dientamoeba, and microsporidia was expected to be low; therefore, specific molecular assays were applied for these.
We applied the duplex real-time PCR for Cryptosporidium spp. and Giardia duodenalis that is used in the Laboratory of Parasitology, Statens Serum Institut to all the DNAs. The method has been applied in One Health context previously [23], and the primers used are listed in Table 2. This was done to ensure detection of Giardia, and to add support to the data output on Cryptosporidium generated by metabarcoding.
The 25-µL reaction mixture contained 0.2 µL IMMOLASE™ DNA Polymerase (Bioline, Denmark), 2.5 µL 10 × ImmoBuffer (Bioline), 2.5 µL 10× dUTP (1×), 2.5 µL 50 mM MgCl2 (5 mM), 0.5 µL ROX Reference Dye (Invitrogen) (diluted 1:30), 5 µL 50% glycerol (5%), 1.25 µL (1 µM) of each primer, 0.125 µL (0.075 µM) of each probe, 0,25 µL IPC (Internal Process Control DNA), 5 µL of DNA eluate, and water sufficient to reach the total volume of 25 µL. Negative (water) and positive (G. duodenalis and C. parvum DNA) controls and inhibition controls were included in each run on the Applied Biosystems QuantStudio™ 5 Real-Time PCR System (Thermo Fisher Scientific, Hvidovre, Denmark). PCR cycling conditions were as follows: 10 min at 95 °C (initial denaturation), 50 cycles of 15 s at 95 °C, and 60 s at 60 °C. PCR products were analyzed with the QuantStudioTM Design&Analysis Software v1.5.1 (Thermo Fisher Scientific, Hvidovre, Denmark). Samples were considered positive, if they exhibited a sigmoid function with a threshold cycle value (Ct-value) ≤ 42.
Similarly, we screened a subset of the samples (n = 37; chosen by convenience sampling) for Dientamoeba fragilis [24] and Enterocytozoon bieneusi [25] using the same PCR conditions as described above. The primers used are listed in Table 2.
Conventional PCR followed by Sanger sequencing was applied to a minor selection of samples that were real-time PCR-positive but metabarcoding-negative for Cryptosporidium for test result confirmation; the primers published by Xiao et al. [26] were used.
Genotyping of E. bieneusi was performed using the method by Buckholt et al., 2002 [27].

Table 2.
Oligos used in the present study for real-time PCR-based detection of selected parasites.
Table 2.
Oligos used in the present study for real-time PCR-based detection of selected parasites.
| Parasite (Target Gene) | Oligonucleotides (Primer/Probe) | Primer and Probe Sequences | PCR Product Size (bp) | Reference |
|---|---|---|---|---|
| Cryptosporidium spp. | Cryptosporidium CRY F3 | 5′-CTA CAC TGA TGC ATC CAT CRA GT-3′ | 78 | Present study |
| (18S) | Cryptosporidium CRY R3 | 5′-CCC ATC ACG ATG CAT AYT CAA AA-3′ | ||
| Cryptosporidium CRY P | VIC-TCC TGT TTC GAA GGA AAT GGG TAA TC-MGB | |||
| Dientamoeba fragilis | DF-124f | 5′-CAA CGG ATG TCT TGG CTC TTT A-3′ | 97 | [24] |
| (ITS1-5.8S-ITS2) | Df-221r | 5′-TGC ATT CAA AGA TCG AAC TTA TCA C-3′ | ||
| Probe Df-172 | 6-Fam CAA TTC TAG CCG CTT AT-MGBNFQ | |||
| Enterocytozoon bieneusi | EblTS-89F | 5′-TGT GTA GGC GTG AGA GTG TAT CTG-3′ | 103 | [25] |
| (ITS) | EblTS-191r | 5′-CAT CCA ACC ATC ACG TAC CAA TC-3′ | ||
| Probe EblTS-114rev T | FAM-CAC TGC ACC CAC ATC CCT CAC CCT T-BHQ-1 | |||
| Giardia duodenalis | Giardia-80F | 5′-GAC GGC TCA GGA CAA CGG TT-3′ | 62 | [28] |
| (18S) | Giardia-127R | 5′-TTG CCA GCG GTG TCC G-3′ | ||
| Giardia-105T | FAM-CCC GCG GCG GTC CCT GCT AG-BHQ-1 |
2.5. Analysis of DNA Sequence Read Outputs
Samples with 1000 reads or fewer mapping to eukaryotes were excluded from the analysis, as such low read counts most likely reflected PCR inhibition and would not reflect the true distribution of eukaryotic organisms in the sample.
The highest sequence read output for Cryptosporidium was obtained by the G3 primers; for Balantioides, the sequence output generated by the G4 primer pair was used (Table 3). For Blastocystis, Entamoeba, and Iodamoeba, the G6 primer pair produced the largest sequence read output.

Table 3.
DNA sequence read yield obtained per genus by metabarcoding of 259 fecal DNAs from pigs from four different herds in Denmark.
For each sample positive for parasites (except for Balantioides and Blastocystis ST15 for which a different approach was used), DNA sequences were downloaded in fasta format for each relevant genus from the BION server and collated in sample-specific files. DNA sequences were submitted to multiple sequence alignment using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/; accessed on 28 May, 2021), and each alignment was inspected by eye to identify sequence variation that could indicate genetic diversity and not PCR or sequencing error. For each major cluster of near-identical sequences (sporadic single nucleotide polymorphism [SNPs] were commonly noticed and thought to be due to PCR and sequencing-introduced errors), a consensus sequence was generated, recorded, and queried in the NCBI Database (https://blast.ncbi.nlm.nih.gov/Blast.cgi; accessed on 28 May 2021). Sequences are available at GITHUB (https://github.com/Entamoeba/pig-study-2020; accessed on 28 May 2021). Specifically, for Balantioides, sequence reads from a variety of positive samples were pooled with a view to identifying major lineages. This approach was applied to Balantioides because of (1) a large amount of minor genetic diversity per sample and (2) the fact that most of the sequences had gaps in the middle (the PCR product generated by the G4 primers is larger than those generated for the G3 and G6 primers, and sequencing using ILLUMINA may not provide sufficient coverage over the middle part of the amplified fragment). For sequences reflecting Blastocystis ST15, examples of sequences were pooled and uploaded as documentation (GITHUB). It should be noted that for those samples potentially positive for ST15, the number of ST15-associated reads was limited (typically between 1 and 50). Even the pooled sequences did not align very well and none of the consensus sequences that could be generated were 100% similar to sequences in GenBank.
For Balantioides, Blastocystis, and Entamoeba, samples for which <40 sequence reads were observed per genus were deemed negative for that particular genus. This arbitrary threshold was chosen in order to avoid erroneous classification of a sample with very few sequence reads as positive, since these sequence reads might reflect spill-over of ID tags from highly positive samples. Since the average number of Cryptosporidium-specific reads was generally noticeably lower in comparison, samples with Cryptosporidium-specific sequence reads >0 were scored as positive.
3. Results
3.1. DNA Sequence Read Yield in the Metabarcoding Assay
The total number of sequence reads obtained by all primer sets that could be mapped was 19,777,284 (Table 4), with a median combined number of Archaea, Prokaryotes, and Eukaryotes reads of 72,180 per sample (interquartile range, 50,922–87,416). Fourteen of the samples (5%) had very few reads representing eukaryotic DNA (~1000 reads or fewer, indicating PCR inhibition), so these samples were excluded from the sample set. Hence, the overall number of samples included for data analysis was 259 from 116 pigs.

Table 4.
Total number of DNA sequence reads generated by the metabarcoding assay according to taxonomic group across all primers sets.
Data on DNA sequence read yield obtained per genus by the metabarcoding assay are also listed in Table 4.
3.2. Parasites Detected by Metabarcoding and Real-Time PCR
The genera of potential zoonotic relevance detected by metabarcoding were Balantioides, Blastocystis, Cryptosporidium, Entamoeba, and Iodamoeba. Meanwhile, Dientamoeba, Enterocytozoon, and Giardia were not detected by the metabarcoding assay in any of the samples (Table 4), supporting the low sensitivity of the assay for these genera.
With regard to real-time PCR results, 86 (33%) samples tested positive for Giardia. Meanwhile, the number of samples positive for Cryptosporidium was 239 (92%), which was a substantially higher positivity rate than that obtained by the metabarcoding assay (35%). The real-time PCR cycle threshold values for the Cryptosporidium-positive samples ranged from 22 to 40, with a median (interquartile range [IQR]) of 32 (30–34). Given this IQR, most of the samples identified as positive by the real-time PCR assay could be considered weakly positive samples. For the 113 samples that had a Ct value ≥ 33, 15 (13%) were positive by the metabarcoding assay. Meanwhile, for the 126 samples with a Ct value < 33, 72 (57%) were identified as Cryptosporidium-positive by metabarcoding; these observations clearly support a higher sensitivity of the real-time PCR assay. Six samples with Ct values < 30 that were negative by the metabarcoding assay were subjected to conventional PCR followed by Sanger sequencing. In all six cases, sequencing of the PCR products revealed C. scrofarum.
While none of the subset samples were positive for D. fragilis, 34 (92%) of the 37 samples were positive for E. bieneusi. One genotype of E. bieneusi was observed by nested PCR and Sanger sequencing, namely EbpA.
An overview of the parasite positivity rate according to herd is provided in Figure 1. Comparing the herds, it appeared that the positivity rate was generally lower in Herd 5 than in the other three herds, whereas samples from Herd 1 exhibited very high positivity rates overall. In samples from Herd 4, Cryptosporidium was quite common compared with the other herds as observed by the microbiome assay (Figure 1); however, real-time data revealed that the positivity rate was high across all herds. For Giardia, the situation was different, with 6% of samples positive from Herd 5, compared with 47% in samples from Herd 1.
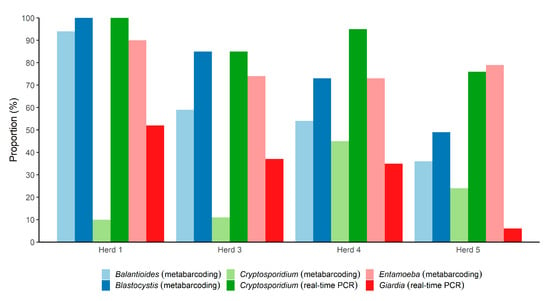
Figure 1.
Overview of the proportions of the 259 fecal DNAs from pigs from four different herds in Denmark positive for five parasitic genera, according to herd. For Balantioides, Blastocystis, and Entamoeba, only metabarcoding was used. For Giardia and Cryptosporidium, both metabarcoding and real-time PCR were used (Giardia failed to be detected by the metabarcoding assay).
3.3. Differentiation of Species and Subtypes Detected in the Samples by Metabarcoding
A detailed presentation of the species and subtypes identified in the samples by metabarcoding is provided in Table 5. The sequences obtained for the organisms below are available on GITHUB (https://github.com/Entamoeba/pig-study-2020; accessed on 28 May 2021).

Table 5.
Parasitic species and subtypes of Balantioides, Cryptosporidium, Entamoeba, and Blastocystis identified by cluster analysis of metabarcoding data obtained from 259 fecal DNAs from pigs from four different herds in Denmark.
3.3.1. Cryptosporidium
By metabarcoding, C. suis and C. scrofarum were identified in 54/259 (21%) and 46/259 (18%) of the samples, respectively; no other species of Cryptosporidium were observed. Eight samples were positive for both C. suis and C. scrofarum.
No intra-species DNA sequence variation was observed for Cryptosporidium; all C. suis sequences were identical to AF108861 [29], while all C. scrofarum sequences were identical to e.g., JX424840 [30]. All samples with Cryptosporidium-specific reads were positive for Cryptosporidium by real-time PCR. Overall, the Ct values of the C. suis-positive samples were lower than those of the C. scrofarum-positive samples. Meanwhile, the crude read counts generated by the metabarcoding assay appeared similar for the two species.
3.3.2. Giardia
The metabarcoding assay did not detect Giardia in any of the samples, while the real-time PCR-based analysis of the samples identified Giardia in 86 samples (33%).
3.3.3. Entamoeba
About three out of four samples were positive for species of Entamoeba, in particular E. polecki, which was found in 193 samples (75%). E. polecki ST1 was detected in 104 samples (40%), while E. polecki ST3 was found in 161 samples (62%). Both subtypes were observed in 37.4% of the E. polecki-positive samples. No intra-subtype diversity was observed in any of the two subtypes.
E. hartmanni was detected in 8 (3%) samples, and all E. hartmanni consensus sequences exhibited 100% similarity to GenBank accession numbers FR686375 and FR686374, which are sequences of E. hartmanni found in human stool in Sweden [31].
3.3.4. Iodamoeba
Among the 259 samples, one tested positive for Iodamoeba bütschlii. The results for the two remaining primer sets (G3 and G4) were also checked for Iodamoeba-specific sequences; however, no evidence of other positive samples was observed. The file with the sequence output exemplifies the extreme variability seen among Iodamoeba-specific SSU rRNA genes that can be present in a fecal sample, and the consensus sequence ’731IOa_RL2′ (https://github.com/Entamoeba/pig-study-2020; accessed on 28 May, 2021) was generated from only a few of the reads and showed 100% similarity to Iodamoeba sp. RL2.
3.3.5. Balantioides
Balantioides was detected in 57% of the samples. The positivity rate of the parasite differed substantially across the herds (Figure 1). While a positivity rate of 93.5% was observed among samples from Herd 1, it was 36.4% in samples from Herd 5; the positivity rates in samples from Herd 3 and Herd 4 were 59% and 54%, respectively.
Two different sequence types were identified: One had a 100% match to MK801486 from a pig sampled in Germany [16] and 99.79% to several DNA sequences from domestic pigs sampled in the Czech republic (JQ07324,-23,-21-04) and in Cameroon (JQ07334) [4]; we refer to this sequence as ‘sequence type I’ and it was present in 64.2% of the Balantioides-positive samples. The other sequence type (‘sequence type II’) shared 100% identify to, for example, MK801495 from a pig in Germany [16] and GQ903678 from a pig sampled in the Philippines [32] and present in the remaining Balantioides-positive samples (35.8%).
3.3.6. Blastocystis
Blastocystis was observed in 193/259 (75%) samples. Sixty-eight percent of all tested samples were positive for ST5 (Table 5), and ST5 was observed in 91% of all Blastocystis-positive samples (Figure 2). ST1 was seen in 28% of the samples, ST15 in 14%, and ST3 in 2% of the samples. ST15 was typically observed admixed with ST5 (81% of the ST15-positive samples were positive for ST5) (Figure 2). Both ST1 and ST5 were seen in 62 samples (32% of all Blastocystis-positive samples); both ST1 and ST15 were observed in 29 samples (15% of all Blastocystis-positive samples), and both subtypes 1 and 3 were seen in one sample (0.5% of all Blastocystis-positive samples). One sample was found to be positive for subtypes 1, 3, and 5, and another sample was positive for subtypes, 3, 5, and 15.
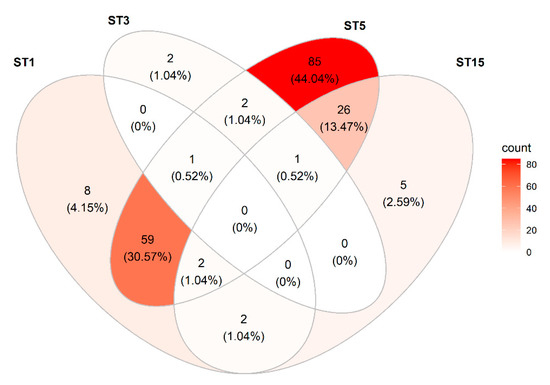
Figure 2.
Venn diagram showing the distribution and co-occurrence of Blastocystis sp. subtypes detected in the study of fecal DNAs from pigs from four different herds in Denmark.
All samples from Herd 1 were positive for Blastocystis sp. ST5, and some were positive for other subtypes as well: typically, ST1, but also a few ST15.
For ST5, several different sequence types were observed (ST5a through ST5f; Table 6), although ST5a and ST5c were strikingly similar and maybe the same, and this was also the case for ST5d and ST5f; these variants differed by only one or two SNPs. Similarly, for ST1 and ST3, two different sequence types were observed for each (Table 6).

Table 6.
Sequence types of Blastocystis subtypes 1, 3, 5, and 15 observed in the present study.
For ST3, two different sequence types were observed; one sample was positive for both types, while the other ST3-positive samples had only one of the two types.
3.4. Examples of Detected Polyparasitism
Most samples tested positive for multiple parasitic species. For instance, 164 of 193 (85.0%) of the Blastocystis-positive samples were positive for E. polecki, and 108 of these 164 Blastocystis- and Entamoeba-positive samples (65.9%) were positive for Balantioides. One hundred and six (41%) of the samples were positive for Balantioides, Blastocystis, and Entamoeba. Forty-four (17%) samples were positive for Cryptosporidium, Giardia, Balantioides, Blastocystis, and Entamoeba.
Of the 54 C. suis-positive samples, 40 (74.1%) and 37 (68.5%) were positive for E. polecki and Blastocystis, respectively, while 36 (78.3%) and 40 (87.0%) of the 46 C. scrofarum-positive samples were positive for the two, respectively.
The single sample in which Iodamoeba was observed was also positive for C. suis, C. scrofarum, E. polecki ST1, E. polecki ST3, Blastocystis ST5, and Blastocystis ST15.
4. Discussion
The amplicon-based NGS-based approach to detecting and differentiating parasites used in this study has been applied in a number of studies involving human, animal, and environmental samples, including corneal scrapings, fecal samples, and sewage samples [14,15,22,33], and with different foci from clinically relevant opportunistic parasites to foodborne parasites [34]. This ‘one-fits-many’ approach could be cost-effective for screening DNAs from large numbers of samples for DNA from parasites, fungi, and bacteria, although some limitations have been identified [15,34].
In the present study, we screened for intestinal protists previously reported in pigs, focusing on those with zoonotic potential. Of the species detected, E. hartmanni, Blastocystis sp. ST1 and ST3, and I. bütschlii are all common in humans. E. polecki is rarely reported in humans [35], and the same is true for Blastocystis sp. ST5 (see below), C. suis [36], and Balantioides coli [6], although the distribution of reported human cases of balantidiosis differs substantially according to geographical region [6]. Cryptosporidium scrofarum, previously referred to as Cryptosporidium pig genotype II, has been reported in at least one human case [37].
Our metabarcoding approach was supplemented by real-time PCR for Dientamoeba, Enterocytozoon, Giardia, and Cryptosporidium in order to increase test sensitivity for these parasites. Interestingly, Giardia was not identified in any of the pig samples subject to shotgun sequencing by Wylezich et al. [16]. That team recently published an evaluation of their method [38] and mentioned the low recovery rate for Giardia as a limitation, which means that Giardia may have been overlooked in their study of pigs [16], and thus, it is worth noticing that Giardia may generally be difficult to detect not only by the metabarcoding methods but also by a metagenomics approach. To this end, Ramayo-Caldas et al. [17] used amplicon-based NGS relying on detection and differentiation of ITS and 18S rRNA fragments and mentioned neither Giardia nor Cryptosporidium among their findings. In the study by Parfrey and colleagues [39], one set of primers was used to amplify eukaryotic DNA in human stool samples, and also here, Giardia- and Cryptosporidium-specific DNA was not detected. The authors speculated that the primers might not be suited for the detection of Giardia and that DNA extraction might not have been optimal for the detection of Cryptosporidium. However, there was no baseline or reference data (e.g., microscopy data) indicating whether any of the samples included in that study were positive for these two parasites.
Although this study was not designed to evaluate the sensitivity of the metabarcoding assay, it is worth noting that the sensitivity of the assay appears to differ according to genus. While Enterocytozoon and Giardia were not detected at all by the metabarcoding assay, Cryptosporidium was identified in some samples but to a much lesser extent than by real-time PCR. Interestingly, Cryptosporidium was readily detected in samples with Cryptosporidium amplicon-specific Ct values lower than 33. The reason for the relatively high positivity rate in samples from Herd 4 by the metabarcoding assay might reflect the fact that 54/71 (76%) samples with Cryptosporidium-specific Ct values by real-time between 22 and 30 were from this herd, and so these would be more likely to contain sufficient Cryptosporidium-specific DNA detectable by the metabarcoding assay. We successfully sequenced Cryptosporidium-specific SSU rDNA from all six samples that were real-time PCR-positive with Ct values < 30 and metabarcoding-negative, which confirms the higher sensitivity of the real-time PCR assay compared with the metabarcoding assay.
Cryptosporidium parvum, which has previously been reported in pigs in Sweden [40] but not in Denmark [41], was not detected in the present study.
The Ct values of the C. suis-positive samples were significantly lower than those of the C. scrofarum-positive samples. This may reflect (i) a higher infection intensity for C. suis than for C. scrofarum, (ii) a difference between the two species in terms of oocyst resistance towards DNA extraction, (iii) genetic polymorphism in the primer-probe annealing sites, resulting in preferential amplification/probe-based detection of C. suis over the C. scrofarum, and/or (iv) variation in copy number of the SSU rRNA gene between these two species. Of these scenarios, the first one might be of particular relevance here, since an earlier study from Denmark showed that pigs infected with Cryptosporidium scrofarum excreted fewer oocysts compared with pigs infected with Cryptosporidium suis [42], adding support to the observations in the present study.
Enterocytozoon bieneusi is a common finding in pigs all over the world, and multiple genotypes have been observed in this host type [9], some of which are zoonotic. For example, Sak and colleagues [43] detected E. bieneusi in 74/79 pigs (94%) in the Czech Republic. In that study, nested PCR was used for screening, and most of the E. bieneusi-positive pigs (70/74, 95%) had the zoonotic genotype EbpA, which is the one identified in the present study. In the present study, only EbpA was identified, and although we only tested a minor subset of samples, the subset included samples from all four pig herds, and thus our finding suggests that this may be a common genotype in pig herds in Denmark. Enterocytozoon bieneusi has been a rare observation in human fecal samples in Denmark. However, it should be mentioned that there was an E. bieneusi outbreak in 2020 in a company based in the Copenhagen metropolitan area (Michlmayr et al., in preparation); in this outbreak, Genotype C was identified. The same genotype was also the cause of an outbreak in Sweden in 2009 [44]. To date, no evidence of an animal reservoir for Genotype C has been identified [45].
Dientamoeba fragilis is very common in the human population in Denmark [46,47]. Data from Italy suggested that pigs may be common and natural hosts of D. fragilis [5,48]. Since we did not detect this parasite in any of the samples, we were not able to produce evidence supporting those findings.
Other parasites (Blastocystis, Entamoeba, and Balantioides) were detected in most samples, and the current version of the metabarcoding assay therefore appears to be particularly useful for detecting these.
Blastocystis is very common in the human population, including in Denmark [49,50]. The observation that Blastocystis sp. ST1 is so common in samples from pigs from Denmark suggests a potential for zoonotic transmission, given the fact that ST1 is one of the most common subtypes found in the human population in Denmark [49,50]. A previous study on Blastocystis in pigs in Denmark revealed only ST5 and ST3, with a clear preponderance of ST5 [51]. Meanwhile, Navarro et al. found ST1 to predominate among pigs sampled in Spain [52], and ST1 has also been found to be common in pigs in Thailand [53] and goats in Malaysia [54]. Apart from ST1, ST5 appears to be the most common subtype in domestic pigs [55,56], and this subtype has only rarely been observed in human fecal samples [57]. It should be noted, however, that the two sequence types displayed in Table 6 would both be scored as ST1 allele 4 according to the terminology by Stensvold et al. [58], and so the allele system, which is currently commonly used, might not be sufficiently discriminative to separate ST1 strains of uncertain host specificity. Blastocystis ST15 was described for the first time in 2013 by Alfellani et al. [59] in a camel and a gibbon. Since then, this subtype was found to be common in wild boars sampled in Italy and it was also found in a couple of domestic pigs [60]. In the study by Wylezich et al. [16], 14/41 samples were positive for ST15. Hence, just like ST5, ST15 appears to be a parasite of mainly suids.
The intra-subtype variation accounted for in Table 6 shows the extent of within-subtype variation in Blastocystis and the discriminatory ability of the metabarcoding assay.
Of particular note is the finding of E. hartmanni, which has not previously been reported in pigs. Kessel reported Entamoeba cysts with 1–4 nuclei in pigs, and the cysts were between 5 and 12 µm, which would include the E. hartmanni cyst size range. These were referred to as E. dysenteriae-like, which is a synonym of E. histolytica, and at that time, E. hartmanni was referred to as ‘small-race E. histolytica’. Burrows indicated that E. histolytica might frequently have been mixed up with E. hartmanni [61,62]. In a study specifically investigating the distribution of Entamoeba spp. in pigs in China by DNA-based methods, Li et al. tested for E. histolytica, E. suis, and E. polecki, but not for E. hartmanni. No cases of E. histolytica were found [63].
E. polecki was divided into four specific subtypes by Verweij et al. in 2001 [64], all of which have been isolated from human stool, but which appear to differ in terms of host reservoir specificity. Of these four subtypes, ST1 and ST3 appear to be common in pigs [16,63,65,66]. Meanwhile ST4 has only been found in humans so far, whereas ST2 has been found in non-human primates as well. In the present study, subtypes 1 and 3 were found in 41% and 63% of the samples, respectively, and none of the remaining two E. polecki subtypes were found. It should be noted that even though both subtypes of E. polecki appear to be very common in pigs on a general basis and even though these subtypes may be found in human stool samples, the extent of human infection/colonization appears limited [35].
Of note, there was no evidence of Entamoeba suis-specific DNA in the samples. E. suis was observed in 81.1% samples from pigs sampled in West Java, Indonesia [66], while Ji et al. observed a considerable lower positive rate (13.0%) for E. suis in pigs sampled in south-eastern China [65]. To our knowledge, E. suis is yet to be reported and confirmed at DNA-level in Europe.
The zoonotic ciliate Balantioides coli was detected in a total of 57% of samples in this study, with the proportion of positives varying between herds and ranging between 36.4% and 93.5%. This is consistent with the results from prevalence studies from other countries [16,17,67,68]. In the present study, based on the SSU rDNA marker, two types of B. coli sequences were detected, for which we introduced a designation: sequence types I and II. Similar results were revealed in the previous molecular-phylogenetic study focused on the genetic diversity of B. coli in pigs and primates [4], in which two clusters of B. coli were revealed using SSU rDNA sequence analysis; one group corresponding to sequence type I showed a broader host specificity and included strains from domestic pigs from different localities (Philippines, Spain, Central African Republic, Czech Republic, and Kenya), wild boar (Czech Republic), and captive apes (chimpanzees and gorillas from different zoological gardens and rehabilitation centers). The second group, which corresponds to sequence type II was dominated by B. coli sequences from domestic pigs sampled in Czech Republic, Madagascar, and Cameroon. Both sequence variants of B. coli were detected in a later large-scale study in domestic pigs in Germany [16]. Unfortunately, these data cannot be compared with data from other studies on molecular detection of B. coli and epidemiology because they address its genetic diversity using hypervariable marker ITS1-5.8rRNA-ITS2 [69,70]. Although several variants of B. coli (A0–A2, B0, B1) have been described based on the primary and secondary structure of the ITS markers, it is unclear whether this may suggest host specificity or transmission due to the presence of more variants in one isolate, even within a single B. coli cell, also in different host species [71]. Despite this, Pomajbíková et al. [4] showed that two clusters based on the SSU sequences in B. coli (corresponding to the two sequence types I and II in this study) were supported also by ITS DNA sequence analysis. Our present results suggest that the SSU rDNA marker may be sensitive enough to distinguish intraspecific variability of B. coli, and information on sequence types might assist in studies of zoonotic transmission, especially studies involving pig farms or slaughterhouses and the personnel in these places. A recent study from Argentina confirmed 100% genetic identity between isolates obtained from pigs and humans using the SSU marker, which may suggest zoonotic transmission [72].
It might be surprising that only one sample was positive for Iodamoeba. In the study by Wylezich et al. [16], 13/41 (32%) pigs tested positive for Iodamoeba, and the authors identified RL2 in all the Iodamoeba-positive samples. RL2 has previously been identified in pig feces as well as in human stool [73]. Iodamoeba-specific PCR could be used to confirm this relatively low positivity rate.
A number of pigs received tetracycline treatment prior to the sampling. However, our aim was not to investigate the effect of tetracycline on parasite positivity. Suffice to say that since 13% of the pigs had received antibiotics prior to sampling, this might to a minor extent account for one or more of the differences observed in parasite positivity rate.
Using NGS-based metabarcoding in addition to pathogen-specific assays may be useful for detection of emerging and previously underdetected pathogens in a One Health setting, across host species. We observed extensive examples of polyparasitism in this study, which highlights the relevance of using assays such as the metabarcoding assay used in the present study, not only when the aim is to screen for multiple parasites but also when multiple taxa (species, subtypes) of parasites are expected. It should be noted that little data is available with regard to the limit of detection of the metabarcoding assay for most of the parasites studied here. Meanwhile, the assay appears to be a suitable method for generating baseline data on parasite occurrence and diversity in various hosts.
Author Contributions
Conceptualization, C.R.S., Ø.A., and H.V.N.; methodology, C.R.S., K.J.-P., E.M., Ø.A., K.W.T., R.F.P., L.O.A., and R.P.K.D.B.; software, C.R.S. and L.O.A.; validation, C.R.S., K.J.-P., K.W.T., Ø.A., and L.O.A.; formal analysis, C.R.S., K.J.-P., P.J., and R.P.K.D.B.; investigation, C.R.S., K.J.-P., and P.J.; resources, C.R.S., Ø.A., and H.V.N., data curation, C.R.S., K.W.T., Ø.A., and L.O.A.; writing—original draft preparation, C.R.S. and K.J.-P.; writing—review and editing, C.R.S. P.J., K.W.T., R.P.K.D.B., L.O.A., and H.V.N.; visualization, C.R.S. and R.P.K.D.B.; supervision, C.R.S.; project administration, C.R.S., K.W.T., Ø.A., and H.V.N.; funding acquisition, C.R.S., P.J., L.O.A., and H.V.N. All authors have read and agreed to the published version of the manuscript.
Funding
C.R.S.: P.J.: L.O.A., R.F.P., and H.V.N. are part of the PARADISE consortium, supported by funding from the European Union’s Horizon 2020 Research and Innovation Programme under grant agreement No 773830: One Health European Joint Programme.
Institutional Review Board Statement
The swine samples were collected in connection with a clinical trial. The clinical trial was approved by the Danish Medicines Agency (license no. 2011090862/2012053751), and the participating herd owners signed a written owner informed consent explaining the scope of the field trial.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are available at https://github.com/Entamoeba/pig-study-2020 (accessed on 28 May 2021).
Acknowledgments
We thank the laboratories in the Department of Bacteria, Parasites, and Fungi for excellent technical assistance. Marianne Lebbad is thanked for commenting on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Robertson, L.J.; Lalle, M.; Paulsen, P. Why we need a European focus on foodborne parasites. Exp. Parasitol. 2020, 214, 107900. [Google Scholar] [CrossRef]
- Lhotská, Z.; Jirků, M.; Hložková, O.; Brožová, K.; Jirsová, D.; Stensvold, C.R.; Kolísko, M.; Jirků Pomajbíková, K. A Study on the Prevalence and Subtype Diversity of the Intestinal Protist Blastocystis sp. in a Gut-Healthy Human Population in the Czech Republic. Front. Cell Infect. Microbiol. 2020, 10, 544335. [Google Scholar] [CrossRef]
- Schuster, F.L.; Ramirez-Avila, L. Current world status of Balantidium coli. Clin. Microbiol. Rev. 2008, 21, 626–638. [Google Scholar] [CrossRef] [PubMed]
- Pomajbikova, K.; Obornik, M.; Horak, A.; Petrzelkova, K.J.; Grim, J.N.; Levecke, B.; Todd, A.; Mulama, M.; Kiyang, J.; Modry, D. Novel insights into the genetic diversity of Balantidium and Balantidium-like cyst-forming ciliates. PLoS Negl. Trop. Dis. 2013, 7, e2140. [Google Scholar] [CrossRef] [PubMed]
- Cacciò, S.M.; Sannella, A.R.; Manuali, E.; Tosini, F.; Sensi, M.; Crotti, D.; Pozio, E. Pigs as natural hosts of Dientamoeba fragilis genotypes found in humans. Emerg. Infect. Dis. 2012, 18, 838–841. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.; Ijaz, M.; Ayyub, R.M.; Ghaffar, A.; Ghauri, H.N.; Aziz, M.U.; Ali, S.; Altaf, M.; Awais, M.; Naveed, M.; et al. Balantidium coli in domestic animals: An emerging protozoan pathogen of zoonotic significance. Acta Trop. 2020, 203, 105298. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.G.; Kaffashian, F.; Tawari, B.; Windsor, J.J.; Twigg-Flesner, A.; Davies-Morel, M.C.; Blessmann, J.; Ebert, F.; Peschel, B.; Le Van, A.; et al. New insights into the phylogeny of Entamoeba species provided by analysis of four new small-subunit rRNA genes. Int. J. Syst. Evol. Microbiol. 2006, 56, 2235–2239. [Google Scholar] [CrossRef]
- Dashti, A.; Rivero-Juarez, A.; Santin, M.; Lopez-Lopez, P.; Caballero-Gomez, J.; Frias-Casas, M.; Koster, P.C.; Bailo, B.; Calero-Bernal, R.; Briz, V.; et al. Enterocytozoon bieneusi (Microsporidia): Identification of novel genotypes and evidence of transmission between sympatric wild boars (Sus scrofa ferus) and Iberian pigs (Sus scrofa domesticus) in Southern Spain. Kransbound. Emerg. Dis. 2020, 67, 2869–2880. [Google Scholar] [CrossRef]
- Li, W.; Feng, Y.; Xiao, L. Diagnosis and molecular typing of Enterocytozoon bieneusi: The significant role of domestic animals in transmission of human microsporidiosis. Res. Vet. Sci. 2020, 133, 251–261. [Google Scholar] [CrossRef]
- Verweij, J.J.; Stensvold, C.R. Molecular Testing for Clinical Diagnosis and Epidemiological Investigations of Intestinal Parasitic Infections. Clin. Microbiol. Rev. 2014, 27, 371–418. [Google Scholar] [CrossRef]
- FAO. Epidemiology, diagnosis and control of helminth parasites of swine. In FAO Animal Health Manual; FAO: Rome, Italy, 1998. [Google Scholar]
- Krogsgaard, L.R.; Andersen, L.O.; Johannesen, T.B.; Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. Characteristics of the bacterial microbiome in association with common intestinal parasites in irritable bowel syndrome. Clin. Transl. Gastroenterol. 2018, 9, 161. [Google Scholar] [CrossRef]
- Ring, H.C.; Thorsen, J.; Saunte, D.M.; Lilje, B.; Bay, L.; Riis, P.T.; Larsen, N.; Andersen, L.O.; Nielsen, H.V.; Miller, I.M.; et al. The Follicular Skin Microbiome in Patients With Hidradenitis Suppurativa and Healthy Controls. JAMA Dermatol. 2017, 153, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Holmgaard, D.B.; Barnadas, C.; Mirbarati, S.H.; Andersen, L.O.; Nielsen, H.V.; Stensvold, C.R. Detection and identification of Acanthamoeba and other non-viral causes of infectious keratitis in corneal scrapings by real-time PCR and next-generation sequencing-based 16S-18S gene analysis. J. Clin. Microbiol. 2020, 59. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Lebbad, M.; Hansen, A.; Beser, J.; Belkessa, S.; O’Brien Andersen, L.; Clark, C.G. Differentiation of Blastocystis and parasitic archamoebids encountered in untreated wastewater samples by amplicon-based next-generation sequencing. Parasite Epidemiol. Control 2020, 9, e00131. [Google Scholar] [CrossRef] [PubMed]
- Wylezich, C.; Belka, A.; Hanke, D.; Beer, M.; Blome, S.; Höper, D. Metagenomics for broad and improved parasite detection: A proof-of-concept study using swine faecal samples. Int. J. Parasitol. 2019, 49, 769–777. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Prenafeta-Boldu, F.; Zingaretti, L.M.; Gonzalez-Rodriguez, O.; Dalmau, A.; Quintanilla, R.; Ballester, M. Gut eukaryotic communities in pigs: Diversity, composition and host genetics contribution. Anim. Microbiome 2020, 2, 18. [Google Scholar] [CrossRef]
- Græsbøll, K.; Damborg, P.; Mellerup, A.; Herrero-Fresno, A.; Larsen, I.; Holm, A.; Nielsen, J.P.; Christiansen, L.E.; Angen, Ø.; Ahmed, S.; et al. Effect of Tetracycline Dose and Treatment Mode on Selection of Resistant Coliform Bacteria in Nursery Pigs. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef]
- Græsbøll, K.; Larsen, I.; Clasen, J.; Birkegård, A.C.; Nielsen, J.P.; Christiansen, L.E.; Olsen, J.E.; Angen, Ø.; Folkesson, A. Effect of tetracycline treatment regimens on antibiotic resistance gene selection over time in nursery pigs. BMC Microbiol. 2019, 19, 269. [Google Scholar] [CrossRef]
- Larsen, I.; Hjulsager, C.K.; Holm, A.; Olsen, J.E.; Nielsen, S.S.; Nielsen, J.P. A randomised clinical trial on the efficacy of oxytetracycline dose through water medication of nursery pigs on diarrhoea, faecal shedding of Lawsonia intracellularis and average daily weight gain. Prev. Vet. Med. 2016, 123, 52–59. [Google Scholar] [CrossRef] [PubMed]
- Larsen, I.; Nielsen, S.S.; Olsen, J.E.; Nielsen, J.P. The efficacy of oxytetracycline treatment at batch, pen and individual level on Lawsonia intracellularis infection in nursery pigs in a randomised clinical trial. Prev. Vet. Med. 2016, 124, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Fredensborg, B.L.; Fossdal í Kálvalíð, I.; Johannesen, T.B.; Stensvold, C.R.; Nielsen, H.V.; Kapel, C.M.O. Parasites modulate the gut-microbiome in insects: A proof-of-concept study. PLoS ONE 2020, 15, e0227561. [Google Scholar] [CrossRef]
- Thomas-Lopez, D.; Muller, L.; Vestergaard, L.S.; Christoffersen, M.; Andersen, A.M.; Jokelainen, P.; Agerholm, J.S.; Stensvold, C.R. Veterinary Students Have a Higher Risk of Contracting Cryptosporidiosis when Calves with High Fecal Cryptosporidium Loads Are Used for Fetotomy Exercises. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Verweij, J.J.; Mulder, B.; Poell, B.; van Middelkoop, D.; Brienen, E.A.; van Lieshout, L. Real-time PCR for the detection of Dientamoeba fragilis in fecal samples. Mol. Cell. Probes 2007, 21, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Verweij, J.J.; Ten Hove, R.; Brienen, E.A.; van Lieshout, L. Multiplex detection of Enterocytozoon bieneusi and Encephalitozoon spp. in fecal samples using real-time PCR. Diagn. Microbiol. Infect. Dis. 2007, 57, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Escalante, L.; Yang, C.; Sulaiman, I.; Escalante, A.A.; Montali, R.J.; Fayer, R.; Lal, A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999, 65, 1578–1583. [Google Scholar] [CrossRef] [PubMed]
- Buckholt, M.A.; Lee, J.H.; Tzipori, S. Prevalence of Enterocytozoon bieneusi in swine: An 18-month survey at a slaughterhouse in Massachusetts. Appl. Environ. Microbiol. 2002, 68, 2595–2599. [Google Scholar] [CrossRef]
- Verweij, J.J.; Schinkel, J.; Laeijendecker, D.; van Rooyen, M.A.; van Lieshout, L.; Polderman, A.M. Real-time PCR for the detection of Giardia lamblia. Mol. Cell Probes 2003, 17, 223–225. [Google Scholar] [CrossRef]
- Morgan, U.M.; Monis, P.T.; Fayer, R.; Deplazes, P.; Thompson, R.C. Phylogenetic relationships among isolates of Cryptosporidium: Evidence for several new species. J. Parasitol. 1999, 85, 1126–1133. [Google Scholar] [CrossRef] [PubMed]
- Kváč, M.; Kestřánová, M.; Pinková, M.; Květoňová, D.; Kalinová, J.; Wagnerová, P.; Kotková, M.; Vítovec, J.; Ditrich, O.; McEvoy, J.; et al. Cryptosporidium scrofarum n. sp. (Apicomplexa: Cryptosporidiidae) in domestic pigs (Sus scrofa). Vet. Parasitol. 2013, 191, 218–227. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Lebbad, M.; Victory, E.L.; Verweij, J.J.; Tannich, E.; Alfellani, M.; Legarraga, P.; Clark, C.G. Increased sampling reveals novel lineages of Entamoeba: Consequences of genetic diversity and host specificity for taxonomy and molecular detection. Protist 2011, 162, 525–541. [Google Scholar] [CrossRef]
- Nilles-Bije, M.L.; Rivera, W.L. Ultrastructural and molecular characterization of Balantidium coli isolated in the Philippines. Parasitol. Res. 2010, 106, 387–394. [Google Scholar] [CrossRef]
- Stolzenbach, S.; Myhill, L.J.; Andersen, L.O.; Krych, L.; Mejer, H.; Williams, A.R.; Nejsum, P.; Stensvold, C.R.; Nielsen, D.S.; Thamsborg, S.M. Dietary Inulin and Trichuris suis Infection Promote Beneficial Bacteria Throughout the Porcine Gut. Front. Microbiol. 2020, 11, 312. [Google Scholar] [CrossRef]
- Stensvold, C.R.; O’Brien Andersen, L.; Jokelainen, P. Protocol for 18S rDNA-based amplicon sequencing for detection of relevant Foodborne Parasites. Zenodo 2021. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Winiecka-Krusnell, J.; Lier, T.; Lebbad, M. Evaluation of a PCR Method for Detection of Entamoeba polecki, with an Overview of Its Molecular Epidemiology. J. Clin. Microbiol. 2018, 56. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017, 8–9, 14–32. [Google Scholar] [CrossRef]
- Kvac, M.; Kvetonova, D.; Sak, B.; Ditrich, O. Cryptosporidium pig genotype II in immunocompetent man. Emerg. Infect. Dis. 2009, 15, 982–983. [Google Scholar] [CrossRef]
- Wylezich, C.; Caccio, S.M.; Walochnik, J.; Beer, M.; Höper, D. Untargeted metagenomics shows a reliable performance for synchronous detection of parasites. Parasitol. Res. 2020, 119, 2623–2629. [Google Scholar] [CrossRef] [PubMed]
- Parfrey, L.W.; Walters, W.A.; Lauber, C.L.; Clemente, J.C.; Berg-Lyons, D.; Teiling, C.; Kodira, C.; Mohiuddin, M.; Brunelle, J.; Driscoll, M.; et al. Communities of microbial eukaryotes in the mammalian gut within the context of environmental eukaryotic diversity. Front. Microbiol. 2014, 5, 298. [Google Scholar] [CrossRef] [PubMed]
- Pettersson, E.; Ahola, H.; Frossling, J.; Wallgren, P.; Troell, K. Detection and molecular characterisation of Cryptosporidium spp. in Swedish pigs. Acta Vet. Scand. 2020, 62, 40. [Google Scholar] [CrossRef]
- Langkjaer, R.B.; Vigre, H.; Enemark, H.L.; Maddox-Hyttel, C. Molecular and phylogenetic characterization of Cryptosporidium and Giardia from pigs and cattle in Denmark. Parasitology 2007, 134, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Petersen, H.H.; Jianmin, W.; Katakam, K.K.; Mejer, H.; Thamsborg, S.M.; Dalsgaard, A.; Olsen, A.; Enemark, H.L. Cryptosporidium and Giardia in Danish organic pig farms: Seasonal and age-related variation in prevalence, infection intensity and species/genotypes. Vet. Parasitol. 2015, 214, 29–39. [Google Scholar] [CrossRef]
- Sak, B.; Kvác, M.; Hanzlíková, D.; Cama, V. First report of Enterocytozoon bieneusi infection on a pig farm in the Czech Republic. Vet. Parasitol. 2008, 153, 220–224. [Google Scholar] [CrossRef]
- Decraene, V.; Lebbad, M.; Botero-Kleiven, S.; Gustavsson, A.M.; Löfdahl, M. First reported foodborne outbreak associated with microsporidia, Sweden, October 2009. Epidemiol. Infect. 2012, 140, 519–527. [Google Scholar] [CrossRef]
- Li, W.; Feng, Y.; Santin, M. Host Specificity of Enterocytozoon bieneusi and Public Health Implications. Trends Parasitol. 2019, 35, 436–451. [Google Scholar] [CrossRef]
- Jokelainen, P.; Hebbelstrup Jensen, B.; Andreassen, B.U.; Petersen, A.M.; Röser, D.; Krogfelt, K.A.; Nielsen, H.V.; Stensvold, C.R. Dientamoeba fragilis—A Commensal in Children in Danish Day Care Centers. J. Clin. Microbiol. 2017, 55, 1707–1713. [Google Scholar] [CrossRef]
- Roser, D.; Simonsen, J.; Nielsen, H.V.; Stensvold, C.R.; Molbak, K. Dientamoeba fragilis in Denmark: Epidemiological experience derived from four years of routine real-time PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1303–1310. [Google Scholar] [CrossRef] [PubMed]
- Crotti, D.; Sensi, M.; Crotti, S.; Grelloni, V.; Manuali, E. Dientamoeba fragilis in swine population: A preliminary investigation. Vet. Parasitol. 2007, 145, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Krogsgaard, L.R.; Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. The prevalence of intestinal parasites is not greater among individuals with irritable bowel syndrome: A population-based case-control study. Clin. Gastroenterol. Hepatol. 2015, 13, 507–513.e502. [Google Scholar] [CrossRef] [PubMed]
- Andersen, L.O.; Bonde, I.; Nielsen, H.B.; Stensvold, C.R. A retrospective metagenomics approach to studying Blastocystis. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; Alfellani, M.A.; Nørskov-Lauritsen, S.; Prip, K.; Victory, E.L.; Maddox, C.; Nielsen, H.V.; Clark, C.G. Subtype distribution of Blastocystis isolates from synanthropic and zoo animals and identification of a new subtype. Int. J. Parasitol. 2009, 39, 473–479. [Google Scholar] [CrossRef]
- Navarro, C.; Domínguez-Márquez, M.V.; Garijo-Toledo, M.M.; Vega-García, S.; Fernández-Barredo, S.; Pérez-Gracia, M.T.; García, A.; Borrás, R.; Gómez-Muñoz, M.T. High prevalence of Blastocystis sp. in pigs reared under intensive growing systems: Frequency of ribotypes and associated risk factors. Vet. Parasitol. 2008, 153, 347–358. [Google Scholar] [CrossRef]
- Thathaisong, U.; Worapong, J.; Mungthin, M.; Tan-Ariya, P.; Viputtigul, K.; Sudatis, A.; Noonai, A.; Leelayoova, S. Blastocystis isolates from a pig and a horse are closely related to Blastocystis hominis. J. Clin. Microbiol. 2003, 41, 967–975. [Google Scholar] [CrossRef]
- Tan, T.C.; Tan, P.C.; Sharma, R.; Sugnaseelan, S.; Suresh, K.G. Genetic diversity of caprine Blastocystis from Peninsular Malaysia. Parasitol. Res. 2013, 112, 85–89. [Google Scholar] [CrossRef]
- Rauff-Adedotun, A.A.; Mohd Zain, S.N.; Farah Haziqah, M.T. Current status of Blastocystis sp. in animals from Southeast Asia: A review. Parasitol. Res. 2020, 119, 3559–3570. [Google Scholar] [CrossRef]
- Danisova, O.; Valencakova, A. First detection of Blastocystis sp. in pigs in Slovakia and in Europe. Parasitol. Int. 2020, 81, 102235. [Google Scholar] [CrossRef]
- Jimenez, P.A.; Jaimes, J.E.; Ramirez, J.D. A summary of Blastocystis subtypes in North and South America. Parasit. Vectors 2019, 12, 376. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Alfellani, M.; Clark, C.G. Levels of genetic diversity vary dramatically between Blastocystis subtypes. Infect. Genet. Evol. 2012, 12, 263–273. [Google Scholar] [CrossRef]
- Alfellani, M.A.; Taner-Mulla, D.; Jacob, A.S.; Imeede, C.A.; Yoshikawa, H.; Stensvold, C.R.; Clark, C.G. Genetic diversity of Blastocystis in livestock and zoo animals. Protist 2013, 164, 497–509. [Google Scholar] [CrossRef]
- Russini, V.; Di Filippo, M.M.; Fanelli, R.; Polidori, M.; Berrilli, F.; Di Cave, D.; Novelletto, A.; Calderini, P. Characterization of prevalence and genetic subtypes of Blastocystis sp. in wild and domestic Suidae of central Italy aided by amplicon NGS. Vet. Parasitol. Reg. Stud. Rep. 2020, 22, 100472. [Google Scholar] [CrossRef] [PubMed]
- Burrows, R.B. Morphological differentiation of Entamoeba hartmanni and E. polecki from E. histolytica. Am. J. Trop. Med. Hyg. 1959, 8, 583–589. [Google Scholar] [CrossRef]
- Burrows, R.B. Microscopic Diagnosis of the Parasites of Man; Yale University Press: New Haven, CT, USA, 1965; pp. 118–119. [Google Scholar]
- Li, W.C.; Geng, J.Z.; Chen, C.; Qian, L.; Zhang, T.; Liu, J.L.; Luo, J.X.; Gu, Y.F. First report on the occurance of intestinal Entamoeba spp. in pigs in China. Acta Trop. 2018, 185, 385–390. [Google Scholar] [CrossRef] [PubMed]
- Verweij, J.J.; Polderman, A.M.; Clark, C.G. Genetic Variation among Human Isolates of Uninucleated Cyst-Producing Entamoeba Species. J. Clin. Microbiol. 2001, 39, 1644–1646. [Google Scholar] [CrossRef]
- Ji, T.; Cao, H.X.; Wu, R.; Cui, L.L.; Su, G.M.; Niu, C.; Zhang, N.; Wang, S.K.; Zhou, D.H. Prevalence and Genetic Identification of Three Entamoeba Species in Pigs in Southeastern China. Biomed Res. Int. 2019, 2824017. [Google Scholar] [CrossRef] [PubMed]
- Wardhana, A.H.; Sawitri, D.H.; Ekawasti, F.; Martindah, E.; Apritadewi, D.; Shibahara, T.; Kusumoto, M.; Tokoro, M.; Sasai, K.; Matsubayashi, M. Occurrence and genetic identifications of porcine Entamoeba, E. suis and E. polecki, at Tangerang in West Java, Indonesia. Parasitol. Res. 2020, 119, 2983–2990. [Google Scholar] [CrossRef]
- Schubnell, F.; von Ah, S.; Graage, R.; Sydler, T.; Sidler, X.; Hadorn, D.; Basso, W. Occurrence, clinical involvement and zoonotic potential of endoparasites infecting Swiss pigs. Parasitol. Int. 2016, 65, 618–624. [Google Scholar] [CrossRef] [PubMed]
- Roesel, K.; Dohoo, I.; Baumann, M.; Dione, M.; Grace, D.; Clausen, P.H. Prevalence and risk factors for gastrointestinal parasites in small-scale pig enterprises in Central and Eastern Uganda. Parasitol. Res. 2017, 116, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Da Silva Barbosa, A.; Ponce-Gordo, F.; Dib, L.V.; Antunes Uchoa, C.M.; Bastos, O.M.P.; Pissinatti, A.; Amendoeira, M.R.R. First molecular characterization of Balantioides coli (Malmsten, 1857) isolates maintained in vitro culture and from feces of captive animals, Rio de Janeiro, Brazil. Vet. Parasitol. Reg. Stud. Rep. 2017, 10, 102–113. [Google Scholar] [CrossRef]
- Li, Y.H.; Yao, Q.; Dong, H.P.; Wang, S.S.; Chen, R.R.; Song, J.K.; Yan, W.C.; Zhao, G.H. Molecular characterization of Balantioides coli in pigs from Shaanxi province, northwestern China. Parasitol. Res. 2020, 119, 3075–3081. [Google Scholar] [CrossRef]
- Ponce-Gordo, F.; Fonseca-Salamanca, F.; Martinez-Diaz, R.A. Genetic heterogeneity in internal transcribed spacer genes of Balantidium coli (Litostomatea, Ciliophora). Protist 2011, 162, 774–794. [Google Scholar] [CrossRef]
- Lopez Arias, L.; Guillemi, E.; Bordoni, N.; Farber, M.; Garbossa, G. Development of a PCR assay for identification of Neobalantidium coli () in Argentina. Vet. Parasitol. Reg. Stud. Rep. 2017, 10, 114–118. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Lebbad, M.; Clark, C.G. Last of the human protists: The phylogeny and genetic diversity of Iodamoeba. Mol. Biol Evol. 2012, 29, 39–42. [Google Scholar] [CrossRef][Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).