Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe
Abstract
1. Introduction
2. Material and Methods
2.1. Geographical Area of Research and Study Model
2.2. Faecal Sample Collection
2.3. Sample Processing and DNA Extraction
2.4. Cryptosporidium spp. Detection, and Subtyping
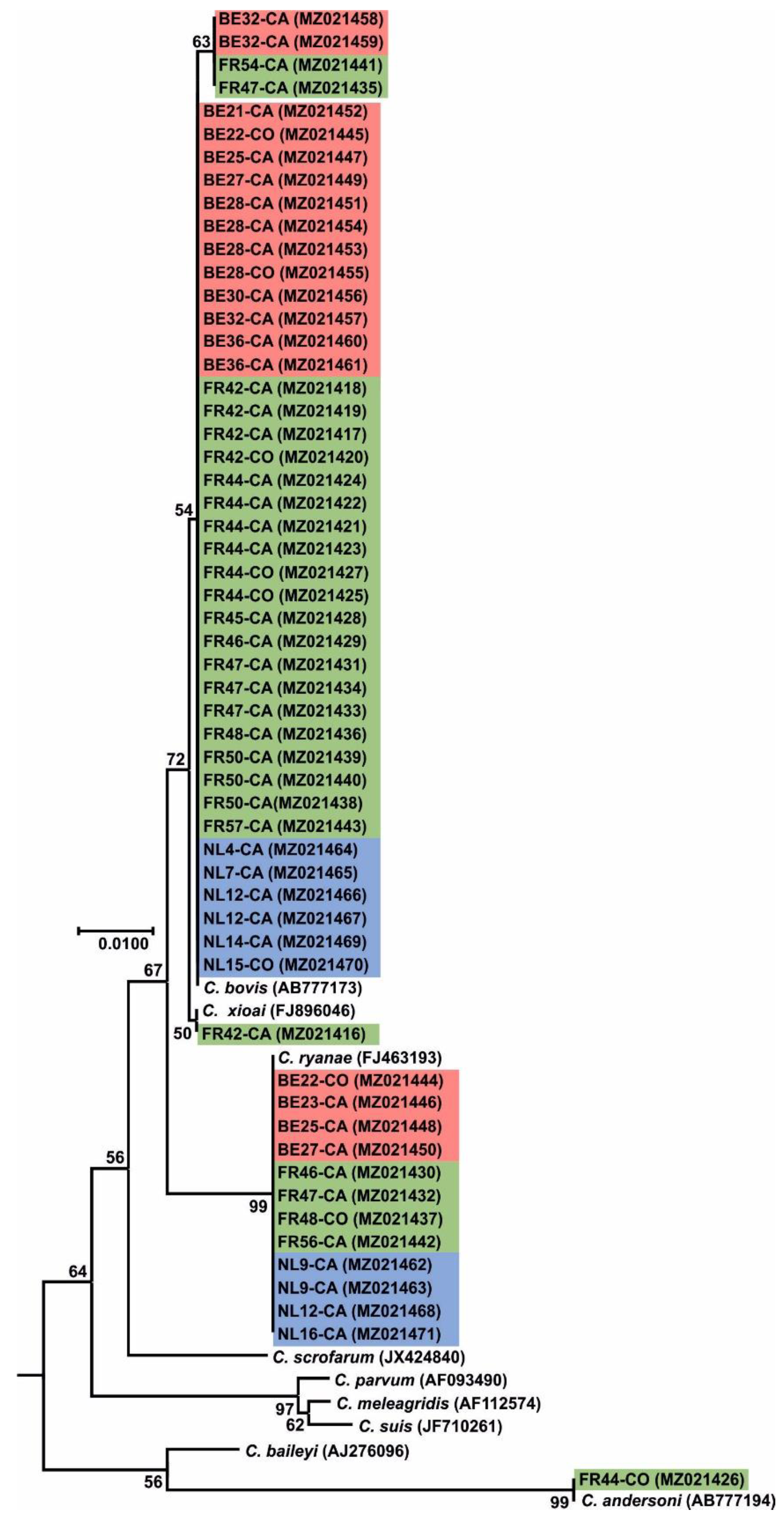
2.5. Sequencing and Phylogenetic Analysis
3. Results
3.1. Sampling Report
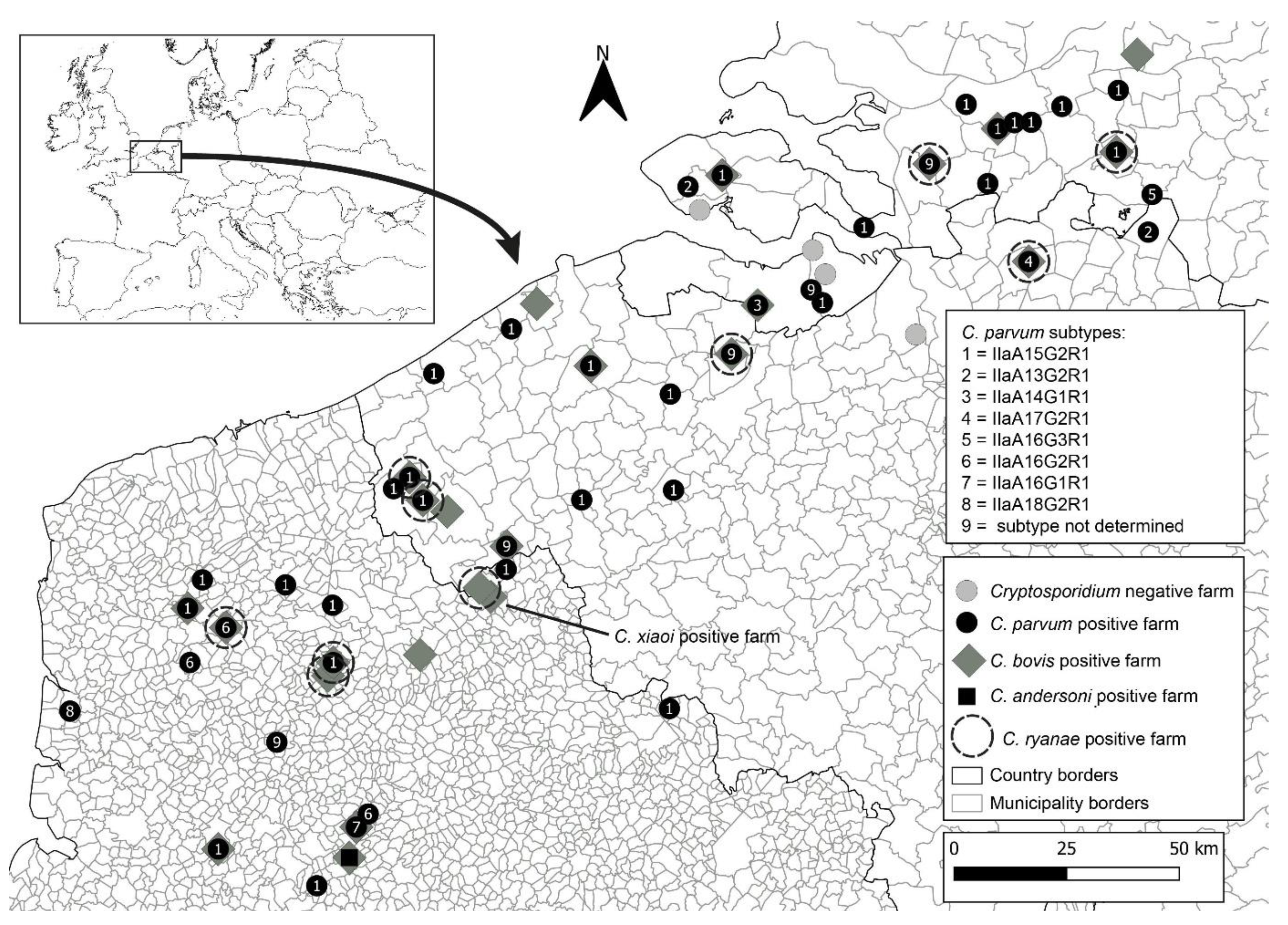
3.2. Cryptosporidium spp. Occurrence and Prevalence in Farms across Belgium, France, and the Netherlands
3.2.1. Belgium
3.2.2. France
3.2.3. The Netherlands
3.2.4. Cryptosporidium parvum Subtyping through gp60 Molecular Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tandel, J.; English, E.D.; Sateriale, A.; Gullicksrud, J.A.; Beiting, D.P.; Sullivan, M.C.; Pinkston, B.; Striepen, B. Life cycle progression and sexual development of the apicomplexan parasite Cryptosporidium Parvum. Nat. Microbiol. 2019, 4, 2226–2236. [Google Scholar] [CrossRef]
- Xiao, L. Molecular epidemiology of cryptosporidiosis: An update. Exp. Parasitol. 2010, 124, 80–89. [Google Scholar] [CrossRef]
- Feng, Y.; Ryan, U.M.; Xiao, L. Genetic Diversity and Population Structure of Cryptosporidium. Trends Parasitol. 2018, 34, 997–1011. [Google Scholar] [CrossRef] [PubMed]
- Checkley, W.; White, A.C.; Jaganath, D.; Arrowood, M.J.; Chalmers, R.M.; Chen, X.M.; Fayer, R.; Griffiths, J.K.; Guerrant, R.L.; Hedstrom, L.; et al. A review of the global burden, novel diagnostics, therapeutics, and vaccine targets for Cryptosporidium. Lancet Infect. Dis. 2015, 15, 85–94. [Google Scholar] [CrossRef]
- Ryan, U.; Fayer, R.; Xiao, L. Cryptosporidium species in humans and animals: Current understanding and research needs. Parasitology 2014, 141, 1667–1685. [Google Scholar] [CrossRef]
- Cacciò, S.M.; Chalmers, R.M. Human cryptosporidiosis in Europe. Clin. Microbiol. Infect. 2016, 22, 471–480. [Google Scholar] [CrossRef]
- Innes, E.A.; Chalmers, R.M.; Wells, B.; Pawlowic, M.C. A One Health Approach to Tackle Cryptosporidiosis. Trends Parasitol. 2020, 36, 290–303. [Google Scholar] [CrossRef]
- Bouzid, M.; Hunter, P.R.; Chalmers, R.M.; Tyler, K.M. Cryptosporidium Pathogenicity and Virulence. Clin. Microbiol. Rev. 2013, 26, 115–134. [Google Scholar] [CrossRef]
- Brainard, J.; Hooper, L.; McFarlane, S.; Hammer, C.C.; Hunter, P.R.; Tyler, K. Systematic review of modifiable risk factors shows little evidential support for most current practices in Cryptosporidium management in bovine calves. Parasitol. Res. 2020, 119, 3571–3584. [Google Scholar] [CrossRef] [PubMed]
- Shaw, H.J.; Innes, E.A.; Morrison, L.J.; Katzer, F.; Wells, B. Long-term production effects of clinical cryptosporidiosis in neonatal calves. Int. J. Parasitol. 2020, 50, 371–376. [Google Scholar] [CrossRef]
- Brook, E.; Hart, C.A.; French, N.; Christley, R. Prevalence and risk factors for Cryptosporidium spp. infection in young calves. Vet. Parasitol. 2008, 152, 46–52. [Google Scholar] [CrossRef] [PubMed]
- Brainard, J.; Hammer, C.C.; Hunter, P.R.; Katzer, F.; Hurle, G.; Tyler, K. Efficacy of halofuginone products to prevent or treat cryptosporidiosis in bovine calves: A systematic review and meta-analyses. Parasitology 2021, 148, 408–409. [Google Scholar] [CrossRef] [PubMed]
- Thomson, S.; Hamilton, C.A.; Hope, J.C.; Katzer, F.; Mabbott, N.A.; Morrison, L.J.; Innes, E.A. Bovine cryptosporidiosis: Impact, host-parasite interaction and control strategies. Vet. Res. 2017, 48, 42. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.; IJpelaar, J. Updated Estimates of the Costs Associated with Thirty Four Endemic Livestock Diseases in Great Britain: A Note. J. Agric. Econ. 2005, 56, 135–144. [Google Scholar] [CrossRef]
- Shaw, H.J. Digest Paper-cryptosporidiosis in calves, the economic impact and best control measures. Br. Cattle Breed. Club 2014, 69. Available online: https://www.cattlebreeders.org.uk/digests/73/papers/1109/ (accessed on 20 November 2021).
- Fayer, R.; Santín, M.; Trout, J.M.; Greiner, E. Prevalence of species and genotypes of Cryptosporidium found in 1-2-year-old dairy cattle in the eastern United States. Vet. Parasitol. 2006, 135, 105–112. [Google Scholar] [CrossRef]
- Fayer, R.; Santin, M.; Trout, J.M. Prevalence of Cryptosporidium species and genotypes in mature dairy cattle on farms in eastern United States compared with younger cattle from the same locations. Vet. Parasitol. 2007, 145, 260–266. [Google Scholar] [CrossRef]
- Santín, M.; Trout, J.M.; Fayer, R. A Longitudinal study of cryptosporidiosis in dairy cattle from birth to 2 years of age. Vet. Parasitol. 2008, 155, 15–23. [Google Scholar] [CrossRef]
- Santín, M.; Trout, J.M.; Xiao, L.; Zhou, L.; Greiner, E.; Fayer, R. Prevalence and age-related variation of Cryptosporidium species and genotypes in dairy calves. Vet. Parasitol. 2004, 122, 103–117. [Google Scholar] [CrossRef]
- Smith, R.P.; Clifton-Hadley, F.A.; Cheney, T.; Giles, M. Prevalence and molecular typing of Cryptosporidium in dairy cattle in England and Wales and examination of potential on-farm transmission routes. Vet. Parasitol. 2014, 204, 111–119. [Google Scholar] [CrossRef]
- Wang, R.; Zhao, G.; Gong, Y.; Zhang, L. Advances and Perspectives on the Epidemiology of Bovine Cryptosporidium in China in the Past 30 Years. Front. Microbiol. 2017, 8, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Cao, J.; Chang, Y.; Yu, F.; Zhang, S.; Wang, R.; Zhang, L. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Gansu, northwest China. Parasite 2020, 27, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santín, M.; Xiao, L. Cryptosporidium bovis n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos taurus). J. Parasitol. 2005, 91, 624–629. [Google Scholar] [CrossRef] [PubMed]
- Fayer, R.; Santín, M.; Trout, J.M. Cryptosporidium ryanae n. sp. (Apicomplexa: Cryptosporidiidae) in cattle (Bos Taurus). Vet. Parasitol. 2008, 156, 191–198. [Google Scholar] [CrossRef]
- Åberg, M.; Emanuelson, U.; Troell, K.; Björkman, C. Infection dynamics of Cryptosporidium bovis and Cryptosporidium ryanae in a Swedish dairy herd. Vet. Parasitol. 2019, 276S, 100010. [Google Scholar] [CrossRef]
- Izzo, M.M.; Kirkland, P.D.; Mohler, V.L.; Perkins, N.R.; Gunn, A.A.; House, J.K. Prevalence of major enteric pathogens in Australian dairy calves with diarrhoea. Aust. Vet. J. 2011, 89, 167–173. [Google Scholar] [CrossRef]
- Kváč, M.; Kouba, M.; Vítovec, J. Age-related and housing-dependence of Cryptosporidium infection of calves from dairy and beef herds in South Bohemia, Czech Republic. Vet. Parasitol. 2006, 137, 202–209. [Google Scholar] [CrossRef]
- Ryan, U.; Zahedi, A.; Paparini, A. Cryptosporidium in humans and animals—A one health approach to prophylaxis. Parasite Immunol. 2016, 38, 535–547. [Google Scholar] [CrossRef]
- Nydam, D.V.; Wade, S.E.; Schaaf, S.L.; Mohammed, H.O. Number of Cryptosporidium parvum oocysts or Giardla spp. cysts shed by dairy calves after natural infection. Am. J. Vet. Res. 2001, 62, 1612–1615. [Google Scholar] [CrossRef]
- Bushkin, G.G.; Motari, E.; Carpentieri, A. Evidence for a Structural Role for Acid-Fast Lipids in Oocyst Walls of Cryptosporidium, Toxoplasma, and Eimeria. MBio 2013, 4, 1–8. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Herman, L.; Hilbert, F.; et al. Public health risks associated with food-borne parasites. EFSA J. 2018, 16, e05495. [Google Scholar] [CrossRef]
- Robertson, L.J.; Campbell, A.T.; Smith, H.V. Survival of Cryptosporidium parvum Oocysts under Various Environmental Pressures. Appl. Environ. Microbiol. 1992, 58, 3494–3500. [Google Scholar] [CrossRef]
- Xiao, L.; Feng, Y. Molecular epidemiologic tools for waterborne pathogens Cryptosporidium spp. and Giardia duodenalis. Food Waterborne Parasitol. 2017, 8–9, 14–32. [Google Scholar] [CrossRef]
- Sulaiman, I.M.; Hira, P.R.; Zhou, L.; Al-Ali, F.M.; Al-Shelahi, F.A.; Shweiki, H.M.; Iqbal, J.; Khalid, N.; Xiao, L. Unique Endemicity of Cryptosporidiosis in Children in Kuwait. J. Clin. Microbiol. 2005, 43, 2805–2809. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, R.M.; Robinson, G.; Elwin, K.; Elson, R. Analysis of the Cryptosporidium spp. and gp60 subtypes linked to human outbreaks of cryptosporidiosis in England and Wales, 2009 to 2017. Parasites Vectors 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Follet, J.; Guyot, K.; Leruste, H.; Follet-Dumoulin, A.; Hammouma-Ghelboun, O.; Certad, G.; Dei-Cas, E.; Halama, P. Cryptosporidium infection in a veal calf cohort in France: Molecular characterization of species in a longitudinal study. Vet. Res. 2011, 42, 116. [Google Scholar] [CrossRef]
- Mammeri, M.; Chevillot, A.; Chenafi, I.; Julien, C.; Vallée, I.; Polack, B.; Follet, J.; Adjou, K.T. Molecular characterization of Cryptosporidium isolates from diarrheal dairy calves in France. Vet. Parasitol. Reg. Stud. Rep. 2019, 18, 100323. [Google Scholar] [CrossRef]
- Ngouanesavanh, T.; Guyot, K.; Certad, G.; Fichoux, Y.L.; Chartier, C.; Verdier, R.I.; Cailliez, J.C.; Camus, D.; Dei-Cas, E.; Bañuls, A.L. Cryptosporidium Population Genetics: Evidence of Clonality in Isolates from France and Haiti. J. Eukaryot. Microbiol. 2006, 53 (Suppl. 1), 33–36. [Google Scholar] [CrossRef] [PubMed]
- Rieux, A.; Chartier, C.; Pors, I.; Paraud, C. Dynamics of excretion and molecular characterization of Cryptosporidium isolates in pre-weaned French beef calves. Vet. Parasitol. 2013, 195, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Rieux, A.; Chartier, C.; Pors, I.; Delafosse, A.; Paraud, C. Molecular characterization of Cryptosporidium isolates from high-excreting young dairy calves in dairy cattle herds in Western France. Parasitol. Res. 2013, 112, 3423–3431. [Google Scholar] [CrossRef]
- Rieux, A.; Paraud, C.; Pors, I.; Chartier, C. Molecular characterization of Cryptosporidium isolates from beef calves under one month of age over three successive years in one herd in western France. Vet. Parasitol. 2014, 202, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Rieux, A.; Paraud, C.; Pors, I.; Chartier, C. Molecular characterization of Cryptosporidium isolates from pre-weaned calves in western France in relation to age. Vet. Parasitol. 2013, 197, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Geurden, T.; Berkvens, D.; Martens, C.; Casaert, S.; Vercruysse, J.; Claerebout, E. Molecular epidemiology with subtype analysis of Cryptosporidium in calves in Belgium. Parasitology 2007, 134, 1981–1987. [Google Scholar] [CrossRef] [PubMed]
- Wielinga, P.R.; de Vries, A.; van der Goot, T.H.; Mank, T.; Mars, M.H.; Kortbeek, L.M.; van der Giessen, J.W.B. Molecular epidemiology of Cryptosporidium in humans and cattle in The Netherlands. Int. J. Parasitol. 2008, 38, 809–817. [Google Scholar] [CrossRef]
- Ziegler, P.E.; Santucci, F.; Lindergard, G.; Nydam, D.V.; Wade, S.E.; Schaaf, S.L.; Chang, Y.-F.; Mohammed, H.O. Evaluation of polymerase chain reaction diagnosis of Cryptosporidium spp. in dairy cattle and wildlife. Vet. Ther. 2007, 8, 148–159. [Google Scholar] [PubMed]
- Alves, M.; Xiao, L.; Sulaiman, I.; Lal, A.A.; Matos, O.; Antunes, F. Subgenotype Analysis of Cryptosporidium Isolates from Humans, Cattle, and Zoo Ruminants in Portugal. J. Clin. Microbiol. 2003, 41, 2744–2747. [Google Scholar] [CrossRef]
- Guindon, S.; Gascuel, O. A Simple, Fast, and Accurate Algorithm to Estimate Large Phylogenies by Maximum Likelihood. Syst. Biol. 2003, 52, 696–704. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the Number of Nucleotide Substitutions When There Are Strong Transition-Transversion and G+C-Content Biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Santoro, A.; Dorbek-Kolin, E.; Jeremejeva, J.; Tummeleht, L.; Orro, T.; Jokelainen, P.; Lassen, B. Molecular epidemiology of Cryptosporidium spp. in calves in Estonia: High prevalence of Cryptosporidium parvum shedding and 10 subtypes identified. Parasitology 2019, 146, 261–267. [Google Scholar] [CrossRef]
- Díaz, P.; Navarro, E.; Remesar, S.; García-Dios, D.; Martínez-Calabuig, N.; Prieto, A.; López-Lorenzo, G.; López, C.M.; Panadero, R.; Fernández, G.; et al. The Age-Related Cryptosporidium Species Distribution in Asymptomatic Cattle from North-Western Spain. Animals 2021, 11, 256. [Google Scholar] [CrossRef]
- Kváč, M.; Hromadová, N.; Květoňová, D.; Rost, M.; Sak, B. Molecular characterization of Cryptosporidium spp. in pre-weaned dairy calves in the Czech Republic: Absence of C. ryanae and management-associated distribution of C. andersoni, C. bovis and C. parvum subtypes. Vet. Parasitol. 2011, 177, 378–382. [Google Scholar] [CrossRef]
- Ondráčková, Z.; Kváč, M.; Sak, B.; Květoňová, D.; Rost, M. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle in South Bohemia, the Czech Republic. Vet. Parasitol. 2009, 165, 141–144. [Google Scholar] [CrossRef]
- Díaz, P.; Varcasia, A.; Pipia, A.P.; Tamponi, C.; Sanna, G.; Prieto, A.; Ruiu, A.; Spissu, P.; Díez-Baños, P.; Morrondo, P.; et al. Molecular characterisation and risk factor analysis of Cryptosporidium spp. in calves from Italy. Parasitol. Res. 2018, 117, 3081–3090. [Google Scholar] [CrossRef]
- Holzhausen, I.; Lendner, M.; Göhring, F.; Steinhöfel, I.; Daugschies, A. Distribution of Cryptosporidium parvum gp60 subtypes in calf herds of Saxony, Germany. Parasitol. Res. 2019, 118, 1549–1558. [Google Scholar] [CrossRef]
- Delafosse, A.; Chartier, C.; Dupuy, M.C.; Dumoulin, M.; Pors, I.; Paraud, C. Cryptosporidium parvum infection and asociated risk factors in dairy calves in western France. Prev. Vet. Med. 2015, 118, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Lichtmannsperger, K.; Hinney, B.; Joachim, A.; Wittek, T. Molecular characterization of Giardia intestinalis and Cryptosporidium parvum from calves with diarrhoea in Austria and evaluation of point-of-care tests. Comp. Immunol. Microbiol. Infect. Dis. 2019, 66, 101333. [Google Scholar] [CrossRef] [PubMed]
- Thomson, S.; Innes, E.A.; Jonsson, N.N.; Katzer, F. Shedding of Cryptosporidium in calves and dams: Evidence of re-infection and shedding of different gp60 subtypes. Parasitology 2019, 146, 1404–1413. [Google Scholar] [CrossRef]
- Feng, Y.; Ortega, Y.; He, G.; Das, P.; Xu, M.; Zhang, X.; Fayer, R.; Gatei, W.; Cama, V.; Xiao, L. Wide geographic distribution of Cryptosporidium bovis and the deer-like genotype in bovines. Vet. Parasitol. 2007, 144, 1–9. [Google Scholar] [CrossRef]
- Huetink, R.E.C.; Van der Giessen, J.W.B.; Noordhuizen, J.P.T.M.; Ploeger, H.W. Epidemiology of Cryptosporidium spp. and Giardia duodenalis on a dairy farm. Vet. Parasitol. 2001, 102, 53–67. [Google Scholar] [CrossRef]
- Khan, S.M.; Debnath, C.; Pramanik, A.K.; Xiao, L.; Nozaki, T.; Ganguly, S. Molecular characterization and assessment of zoonotic transmission of Cryptosporidium from dairy cattle in West Bengal, India. Vet. Parasitol. 2010, 171, 41–47. [Google Scholar] [CrossRef]
- Maddox-Hyttel, C.; Langkjær, R.B.; Enemark, H.L.; Vigre, H. Cryptosporidium and Giardia in different age groups of Danish cattle and pigs-Occurrence and management associated risk factors. Vet. Parasitol. 2006, 141, 48–59. [Google Scholar] [CrossRef] [PubMed]
- Maikai, B.V.; Umoh, J.U.; Kwaga, J.K.P.; Lawal, I.A.; Maikai, V.A.; Cama, V.; Xiao, L. Molecular characterization of Cryptosporidium spp. in native breeds of cattle in Kaduna State, Nigeria. Vet. Parasitol. 2011, 178, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.B.; Sharma, R.; Kumar, H.; Banga, H.S.; Aulakh, R.S.; Gill, J.P.S.; Sharma, J.K. Prevalence of Cryptosporidium parvum infection in Punjab (India) and its association with diarrhea in neonatal dairy calves. Vet. Parasitol. 2006, 140, 162–165. [Google Scholar] [CrossRef] [PubMed]
- Kváč, M.; Vitovec, J. Prevalence and pathogenicity of Cryptosporidium andersoni in one herd of beef cattle. J. Vet. Med. Ser. B 2003, 50, 451–457. [Google Scholar] [CrossRef]
- Wells, B.; Shaw, H.; Hotchkiss, E.; Gilray, J.; Ayton, R.; Green, J.; Katzer, F.; Wells, A.; Innes, E. Prevalence, species identification and genotyping Cryptosporidium from livestock and deer in a catchment in the Cairngorms with a history of a contaminated public water supply. Parasites Vectors 2015, 8, 1–13. [Google Scholar] [CrossRef]
- Soba, B.; Logar, J. Genetic classification of Cryptosporidium isolates from humans and calves in Slovenia. Parasitology 2008, 135, 1263–1270. [Google Scholar] [CrossRef]
- Ayinmode, A.B.; Olakunle, F.B.; Xiao, L. Molecular characterization of Cryptosporidium spp. in native calves in Nigeria. Parasitol. Res. 2010, 107, 1019–1021. [Google Scholar] [CrossRef]
- Björkman, C.; Lindström, L.; Oweson, C.; Ahola, H.; Troell, K.; Axén, C. Cryptosporidium infections in suckler herd beef calves. Parasitology 2015, 142, 1108–1114. [Google Scholar] [CrossRef]
- Budu-Amoako, E.; Greenwood, S.J.; Dixon, B.R.; Barkema, H.W.; McClure, J.T. Giardia and Cryptosporidium on Dairy Farms and the Role these Farms May Play in Contaminating Water Sources in Prince Edward Island, Canada. J. Vet. Intern. Med. 2012, 26, 668–673. [Google Scholar] [CrossRef]
- Fan, Y.; Wang, T.; Koehler, A.V.; Hu, M.; Gasser, R.B. Molecular investigation of Cryptosporidium and Giardia in pre- and post-weaned calves in Hubei Province, China. Parasites Vectors 2017, 10, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, P.; Zhao, X.; Xu, H.; Wu, W.; Wang, Y.; Guo, Y.; Wang, L.; Feng, Y.; Xiao, L. Occurrence and molecular characterization of Cryptosporidium spp. and Enterocytozoon bieneusi in dairy cattle, beef cattle and water buffaloes in China. Vet. Parasitol. 2015, 207, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.; Yang, R.; McCarthy, S.; Gordon, C.; Hijjawi, N.; Ryan, U. Molecular characterization of Cryptosporidium and Giardia in pre-weaned calves in Western Australia and New South Wales. Vet. Parasitol. 2011, 176, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Silverlås, C.; Näslund, K.; Björkman, C.; Mattsson, J.G. Molecular characterisation of Cryptosporidium isolates from Swedish dairy cattle in relation to age, diarrhoea and region. Vet. Parasitol. 2010, 169, 289–295. [Google Scholar] [CrossRef]
- Silverlås, C.; Blanco-Penedo, I. Cryptosporidium spp. in calves and cows from organic and conventional dairy herds. Epidemiol. Infect. 2013, 141, 529–539. [Google Scholar] [CrossRef]
- Wang, R.; Wang, H.; Sun, Y.; Zhang, L.; Jian, F.; Qi, M.; Ning, C.; Xiao, L. Characteristics of Cryptosporidium Transmission in Preweaned Dairy Cattle in Henan, China. J. Clin. Microbiol. 2011, 49, 1077–1082. [Google Scholar] [CrossRef]
- Zhang, X.X.; Tan, Q.D.; Zhou, D.H.; Ni, X.T.; Liu, G.X.; Yang, Y.C.; Zhu, X.Q. Prevalence and molecular characterization of Cryptosporidium spp. in dairy cattle, northwest China. Parasitol. Res. 2015, 114, 2781–2787. [Google Scholar] [CrossRef]
- Santín, M. Clinical and subclinical infections with Cryptosporidium in animals. N. Z. Vet. J. 2013, 61, 1–10. [Google Scholar] [CrossRef]
- Fayer, R.; Santín, M. Cryptosporidium xiaoi n. sp. (Apicomplexa: Cryptosporidiidae) in sheep (Ovis aries). Vet. Parasitol. 2009, 164, 192–200. [Google Scholar] [CrossRef]
- Hijjawi, N.; Mukbel, R.; Yang, R.; Ryan, U. Genetic characterization of Cryptosporidium in animal and human isolates from Jordan. Vet. Parasitol. 2016, 228, 116–120. [Google Scholar] [CrossRef]
- Gong, C.; Cao, X.F.; Deng, L.; Li, W.; Huang, X.M.; Lan, J.C.; Xiao, Q.C.; Zhong, Z.J.; Feng, F.; Zhang, Y.; et al. Epidemiology of Cryptosporidium infection in cattle in China: A review. Parasite 2017, 24, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Mirhashemi, M.E.; Zintl, A.; Grant, T.; Lucy, F.; Mulcahy, G.; De Waal, T. Molecular epidemiology of Cryptosporidium species in livestock in Ireland. Vet. Parasitol. 2016, 216, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.; Sturbaum, G.D.; Hoover, P.J.; Sterling, C.R. Cryptosporidium parvum Mixed Genotypes Detected by PCR-Restriction Fragment Length Polymorphism Analysis. Appl. Environ. Microbiol. 2002, 68, 427–429. [Google Scholar] [CrossRef][Green Version]
- Tanriverdi, S.; Arslan, M.Ö.; Akiyoshi, D.E.; Tzipori, S.; Widmer, G. Identification of genotypically mixed Cryptosporidium parvum populations in humans and calves. Mol. Biochem. Parasitol. 2003, 130, 13–22. [Google Scholar] [CrossRef]
- Hadfield, S.J.; Robinson, G.; Elwin, K.; Chalmers, R.M. Detection and Differentiation of Cryptosporidium spp. in Human Clinical Samples by Use of Real-Time PCR. J. Clin. Microbiol. 2011, 49, 918–924. [Google Scholar] [CrossRef]
- Fayer, R.; Santín, M.; Dargatz, D. Species of Cryptosporidium detected in weaned cattle on cow-calf operations in the United States. Vet. Parasitol. 2010, 170, 187–192. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Ethelberg, S.; Hansen, L.; Sahar, S.; Voldstedlund, M.; Kemp, M.; Hartmeyer, G.N.; Otte, E.; Engsbro, A.L.; Nielsen, H.V.; et al. Cryptosporidium infections in Denmark, 2010–2014. Dan. Med. J. 2015, 62, 3–6. [Google Scholar]
- Alves, M.; Xiao, L.; Antunes, F.; Matos, O. Distribution of Cryptosporidium subtypes in humans and domestic and wild ruminants in Portugal. Parasitol. Res. 2006, 99, 287–292. [Google Scholar] [CrossRef]
- Aita, J.; Ichikawa-Seki, M.; Kinami, A.; Yaita, S.; Kumagai, Y.; Nishikawa, Y.; Itagaki, T. Molecular characterization of Cryptosporidium parvum detected in Japanese black and Holstein calves in Iwate Prefecture and Tanegashima Island, Kagoshima Prefecture, Japan. J. Vet. Med. Sci. 2015, 77, 997–999. [Google Scholar] [CrossRef][Green Version]
- Caffarena, R.D.; Meireles, M.V.; Carrasco-Letelier, L.; Picasso-Risso, C.; Santana, B.N.; Riet-Correa, F.; Giannitti, F. Dairy Calves in Uruguay Are Reservoirs of Zoonotic Subtypes of Cryptosporidium parvum and Pose a Potential Risk of Surface Water Contamination. Front. Vet. Sci. 2020, 7, 1–15. [Google Scholar] [CrossRef]
- Garcia-R, J.C.; Pita, A.B.; Velathanthiri, N.; French, N.P.; Hayman, D.T.S. Species and genotypes causing human cryptosporidiosis in New Zealand. Parasitol. Res. 2020, 119, 2317–2326. [Google Scholar] [CrossRef] [PubMed]
- Guy, R.A.; Yanta, C.A.; Muchaal, P.K.; Rankin, M.A.; Thivierge, K.; Lau, R.; Boggild, A.K. Molecular characterization of Cryptosporidium isolates from humans in Ontario, Canada. Parasites Vectors 2021, 14, 1–14. [Google Scholar] [CrossRef]
- Ma, D.W.; Lee, M.R.; Hong, S.H.; Cho, S.H.; Lee, S.E. Molecular Prevalence and Genotypes of Cryptosporidium parvum and Giardia duodenalis in Patients with Acute Diarrhea in Korea, 2013–2016. Korean J. Parasitol. 2019, 57, 531–536. [Google Scholar] [CrossRef]
- Quilez, J.; Torres, E.; Chalmers, R.M.; Robinson, G.; Del Cacho, E.; Sanchez-Acedo, C. Cryptosporidium species and subtype analysis from dairy calves in Spain. Parasitology 2008, 135, 1613–1620. [Google Scholar] [CrossRef] [PubMed]
- García-Presedo, I.; Pedraza-Díaz, S.; González-Warleta, M.; Mezo, M.; Gómez-Bautista, M.; Ortega-Mora, L.M.; Castro-Hermida, J.A. Presence of Cryptosporidium scrofarum, C. suis and C. parvum subtypes IIaA16G2R1 and IIaA13G1R1 in Eurasian wild boars (Sus scrofa). Vet. Parasitol. 2013, 196, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Hijjawi, N.; Zahedi, A.; Kazaleh, M.; Ryan, U. Prevalence of Cryptosporidium species and subtypes in paediatric oncology and non-oncology patients with diarrhoea in Jordan. Infect. Genet. Evol. 2017, 55, 127–130. [Google Scholar] [CrossRef]
- Hutter, J.A.; Dion, R.; Irace-Cima, A.; Fiset, M.; Guy, R.; Dixon, B.; Aguilar, J.L.; Trépanier, J.; Thivierge, K. Cryptosporidium spp.: Human incidence, molecular characterization and associated exposures in Québec, Canada (2016–2017). PLoS ONE 2020, 15, 1–13. [Google Scholar] [CrossRef]
- Benhouda, D.; Hakem, A.; Sannella, A.R.; Benhouda, A.; Cacciò, S.M. First molecular investigation of Cryptosporidium spp. in young calves in Algeria. Parasite 2017, 24, 1–6. [Google Scholar] [CrossRef]
- Taylan-Ozkan, A.; Yasa-Duru, S.; Usluca, S.; Lysen, C.; Ye, J.; Roellig, D.M.; Feng, Y.; Xiao, L. Cryptosporidium species and Cryptosporidium parvum subtypes in dairy calves and goat kids reared under traditional farming systems in Turkey. Exp. Parasitol. 2016, 170, 16–20. [Google Scholar] [CrossRef]
- Trotz-Williams, L.A.; Martin, D.S.; Gatei, W.; Cama, V.; Peregrine, A.S.; Martin, S.W.; Nydam, D.V.; Jamieson, F.; Xiao, L. Genotype andsSubtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol. Res. 2006, 99, 346–352. [Google Scholar] [CrossRef]
- Iqbal, A.; Lim, Y.A.L.; Surin, J.; Sim, B.L.H. High Diversity of Cryptosporidium Subgenotypes Identified in Malaysian HIV/AIDS Individuals Targeting gp60 Gene. PLoS ONE 2012, 7, e31139. [Google Scholar] [CrossRef] [PubMed]
- Kaupke, A.; Rzeżutka, A. Emergence of novel subtypes of Cryptosporidium parvum in calves in Poland. Parasitol. Res. 2015, 114, 4709–4716. [Google Scholar] [CrossRef] [PubMed]
- Hatalová, E.; Valenčáková, A.; Luptáková, L.; Špalková, M.; Kalinová, J.; Halánová, M.; Bednárová, V.; Gabzdilová, J.; Dedinská, K.; Ondriska, F.; et al. The first report of animal genotypes of Cryptosporidium parvum in immunosuppressed and immunocompetent humans in Slovakia. Transbound. Emerg. Dis. 2019, 66, 243–249. [Google Scholar] [CrossRef]
- Certad, G.; Dupouy-Camet, J.; Gantois, N.; Hammouma-Ghelboun, O.; Pottier, M.; Guyot, K.; Benamrouz, S.; Osman, M.; Delaire, B.; Creusy, C.; et al. Identification of Cryptosporidium Species in Fish from Lake Geneva (Lac Léman) in France. PLoS ONE 2015, 10, 1–13. [Google Scholar] [CrossRef]
- Certad, G.; Follet, J.; Gantois, N.; Hammouma-Ghelboun, O.; Guyot, K.; Benamrouz-Vanneste, S.; Fréalle, E.; Seesao, Y.; Delaire, B.; Creusy, C.; et al. Prevalence, Molecular Identification, and Risk Factors for Cryptosporidium Infection in Edible Marine Fish: A Survey across Sea Areas Surrounding France. Front. Microbiol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Thompson, H.P.; Dooley, J.S.G.; Kenny, J.; McCoy, M.; Lowery, C.J.; Moore, J.E.; Xiao, L. Genotypes and subtypes of Cryptosporidium spp. in neonatal calves in Northern Ireland. Parasitol. Res. 2007, 100, 619–624. [Google Scholar] [CrossRef]
- Danišová, O.; Valenčáková, A.; Petrincová, A. Detection and identification of six Cryptospordium species in livestock in Slovakia by amplification of SSU and GP60 Genes with the Use of PCR Analysis. Ann. Agric. Environ. Med. 2016, 23, 254–258. [Google Scholar] [CrossRef][Green Version]
- Kiani, H.; Haghighi, A.; Seyyedtabaei, S.J.; Azargashsb, E.; Zebardast, N.; Taghipour, N.; Rostami, A.; Xiao, L. Prevalence, Clinical Manifestations and Genotyping of Cryptosporidium spp. in Patients with Gastrointestinal Illnesses in Western Iran. Iran. J. Parasitol. 2017, 12, 169–176. [Google Scholar]
- Imre, K.; Lobo, L.M.; Matos, O.; Popescu, C.; Genchi, C.; Dǎrǎbuş, G. Molecular characterisation of Cryptosporidium isolates from pre-weaned calves in Romania: Is there an actual risk of zoonotic infections? Vet. Parasitol. 2011, 181, 321–324. [Google Scholar] [CrossRef] [PubMed]
- Plutzer, J.; Karanis, P. Genotype and subtype analyses of Cryptosporidium isolates from cattle in Hungary. Vet. Parasitol. 2007, 146, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Broglia, A.; Reckinger, S.; Cacció, S.M.; Nöckler, K. Distribution of Cryptosporidium parvum subtypes in calves in Germany. Vet. Parasitol. 2008, 154, 8–13. [Google Scholar] [CrossRef] [PubMed]
- Lassen, B.; Ståhl, M.; Enemark, H.L. Cryptosporidiosis—An occupational risk and a disregarded disease in Estonia. Acta Vet. Scand. 2014, 56, 36. [Google Scholar] [CrossRef] [PubMed]
- Kinross, P.; Beser, J.; Troell, K.; Silverlas, C.; Björkman, C.; Lebbad, M.; Winiecka-Krusnell, J.; Lindh, J.; Löfdahl, M. Cryptosporidium parvum infections in a cohort of veterinary students in Sweden. Epidemiol. Infect. 2015, 143, 2748–2756. [Google Scholar] [CrossRef]
- Baptista, R.P.; Cooper, G.W.; Kissinger, J.C. Challenges for Cryptosporidium Population Studies. Genes 2021, 12, 894. [Google Scholar] [CrossRef]
- Troell, K.; Hallström, B.; Divne, A.M.; Alsmark, C.; Arrighi, R.; Huss, M.; Beser, J.; Bertilsson, S. Cryptosporidium as a testbed for single cell genome characterization of unicellular eukaryotes. BMC Genom. 2016, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Morris, A.; Robinson, G.; Swain, M.T.; Chalmers, R.M. Direct Sequencing of Cryptosporidium in Stool Samples for Public Health. Front. Public Health 2019, 7, 360. [Google Scholar] [CrossRef]
- Grinberg, A.; Biggs, P.J.; Dukkipati, V.S.R.; George, T.T. Extensive intra-host genetic diversity uncovered in Cryptosporidium parvum using Next Generation Sequencing. Infect. Genet. Evol. 2013, 15, 18–24. [Google Scholar] [CrossRef]
- Zahedi, A.; Gofton, A.W.; Jian, F.; Paparini, A.; Oskam, C.; Ball, A.; Robertson, I.; Ryan, U. Next Generation Sequencing uncovers within-host differences in the genetic diversity of Cryptosporidium gp60 subtypes. Int. J. Parasitol. 2017, 47, 601–607. [Google Scholar] [CrossRef]

| Country | Farm ID | Number of Screened/Positive Animals | C. parvum | C. bovis | C. andersoni | C. ryanae | C. xiaoi | gp60 Subtypes of C. parvum (n) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calf | Cow | Calf | Cow | Calf | Cow | Calf | Cow | Calf | Cow | Calf | Cow | Calf | Cow | ||
| Belgium | BE1 | 7/3 | 7/0 | 3 | - | - | - | - | - | - | - | - | - | IiaA15G2R1 (3) | - |
| BE2 | 20/5 | 21/3 | 5 | 1 | - | 1 | - | - | - | 1 | - | - | IIaA15G2R1 (5) | IIaA15G2R1 (1) | |
| BE3 | 9/4 | 11/0 | 3 | - | - | - | - | - | 1 | - | - | - | IIaA17G2R1 (2) | - | |
| BE4 | 9/4 | 10/1 | 4 | 1 | - | - | - | - | - | - | - | - | IIaA15G2R1 (3) | IIaA15G2R1 (1) | |
| BE5 | 10/4 | 10/1 | 2 | 1 | 1 | - | - | - | 1 | - | - | - | IIaA15G2R1 (2) | IIaA15G2R1 (1) | |
| BE6 | 10/1 | 10/1 | 1 | 1 | - | - | - | - | - | - | - | - | IIaA15G2R1 (1) | - | |
| BE7 | 10/4 | 10/0 | 2 | - | 1 | - | - | - | 1 | - | - | - | - | - | |
| BE8 | 10/6 | 10/1 | 2 | - | 4 | 1 | - | - | - | - | - | - | IIaA15G2R1 (2) | - | |
| BE9 | 10/1 | 10/0 | 1 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (1) | - | |
| BE10 | 10/1 | 8/0 | - | - | 1 | - | - | - | - | - | - | - | - | - | |
| BE11 | 10/6 | 10/2 | 6 | 2 | - | - | - | - | - | - | - | - | IIaA13G2R1 (6) | IIaA13G2R1 (2) | |
| BE12 | 7/3 | 1/0 | - | - | 3 | - | - | - | - | - | - | - | - | - | |
| BE13 | 10/4 | 10/0 | 4 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (4) | - | |
| BE14 | 10/3 | 10/0 | 3 | - | - | - | - | - | - | - | - | - | IIaA16G3R1 (3) | - | |
| BE15 | 10/0 | 9/0 | - | - | - | - | - | - | - | - | - | - | - | - | |
| BE16 | 10/2 | 10/1 | - | 1 | 2 | - | - | - | - | - | - | - | - | - | |
| BE17 | 8/2 | 8/0 | 2 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (2) | - | |
| France | FR18 | 9/0 | 9/1 | - | 1 | - | - | - | - | - | - | - | - | - | IIaA16G2R1 (1) |
| FR19 | 11/3 | 11/0 | 3 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (3) | - | |
| FR20 | 10/5 | 10/0 | 5 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (5) | - | |
| FR21 | 10/6 | 10/0 | 6 | - | - | - | - | - | - | - | - | - | IIaA16G2R1 (6) | - | |
| FR22 | 10/4 | 10/1 | - | - | 3 | 1 | - | - | - | - | 1 | - | - | - | |
| FR23 | 10/7 | 10/1 | 7 | 1 | - | - | - | - | - | - | - | - | IIaA15G2R1 (7) | - | |
| FR24 | 10/4 | 10/3 | - | - | 4 | 2 | - | 1 | - | - | - | - | - | - | |
| FR25 | 10/4 | 10/0 | 3 | - | 1 | - | - | - | - | - | - | - | IIaA15G2R1 (3) | - | |
| FR26 | 10/7 | 10/0 | 5 | - | 1 | - | - | - | 1 | - | - | - | IIaA16G2R1 (5) | - | |
| FR27 | 9/5 | 9/0 | - | - | 4 | - | - | - | 1 | - | - | - | - | - | |
| FR28 | 9/1 | 10/1 | - | - | 1 | - | - | - | - | 1 | - | - | - | - | |
| FR29 | 5/2 | 5/0 | 2 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (2) | - | |
| FR30 | 10/4 | 10/0 | 1 | - | 3 | - | - | - | - | - | - | - | IIaA16G1R1 (1) | - | |
| FR31 | 3/3 | 3/0 | 3 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (3) | - | |
| FR32 | 4/1 | 4/0 | 1 | - | - | - | - | - | - | - | - | - | - | - | |
| FR33 | 7/3 | 7/0 | 3 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (3) | - | |
| FR34 | 10/3 | 10/0 | 2 | - | 1 | - | - | - | - | - | - | - | IIaA15G2R1 (2) | - | |
| FR35 | 11/4 | 11/0 | 4 | - | - | - | - | - | - | - | - | - | IIaA18G2R1 (4) | - | |
| FR36 | 9/5 | 9/0 | 4 | - | - | - | - | 1 | - | - | - | IIaA15G2R1 (2) | - | ||
| FR37 | 8/1 | 7/0 | - | - | 1 | - | - | - | - | - | - | - | - | - | |
| The Netherlands | NL38 | 10/0 | 10/0 | - | - | - | - | - | - | - | - | - | - | - | - |
| NL39 | 10/0 | 10/0 | - | - | - | - | - | - | - | - | - | - | - | - | |
| NL40 | 10/2 | 9/0 | 2 | - | - | - | - | - | - | - | - | - | - | - | |
| NL41 | 10/2 | 10/1 | 1 | 1 | 1 | - | - | - | - | - | - | - | IIaA15G2R1 (1) | - | |
| NL42 | 10/3 | 10/0 | 3 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (2) | - | |
| NL43 | 10/0 | 10/0 | - | - | - | - | - | - | - | - | - | - | - | - | |
| NL44 | 10/4 | 10/0 | 3 | - | 1 | - | - | - | - | - | - | - | IIaA15G2R1 (3) | - | |
| NL45 | 10/3 | 10/0 | 3 | - | - | - | - | - | - | - | - | - | IIaA13G2R1 (3) | - | |
| NL46 | 10/5 | 10/1 | 3 | 1 | - | - | - | - | 2 | - | - | - | IIaA15G2R1 (2) | - | |
| NL47 | 10/4 | 10/1 | 4 | 1 | - | - | - | - | - | - | - | - | IIaA15G2R1 (4) | - | |
| NL48 | 10/5 | 10/0 | 5 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (5) | - | |
| NL49 | 10/5 | 10/0 | 2 | - | 2 | - | - | - | 1 | - | - | - | IIaA15G2R1 (1) | - | |
| NL50 | 10/6 | 10/0 | 6 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (6) | - | |
| NL51 | 10/1 | 10/0 | - | - | 1 | - | - | - | - | - | - | - | - | - | |
| NL52 | 10/3 | 10/1 | 3 | - | - | 1 | - | - | - | - | - | - | IIaA14G1R1 (2) | - | |
| NL53 | 10/2 | 10/0 | 1 | - | - | - | - | 1 | - | - | - | - | - | ||
| NL54 | 10/3 | 10/0 | 3 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (3) | - | |
| NL55 | 10/5 | 10/0 | 5 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (4) | - | |
| NL56 | 10/6 | 10/0 | 6 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (5) | - | |
| NL57 | 10/6 | 10/0 | 6 | - | - | - | - | - | - | - | - | - | IIaA15G2R1 (6) | - | |
| Country | Age Group | Number of Screened/Positive Animals | Genotyping and Number of Positive Samples (%) | Genotyping of C. parvum at the gp60 (n/%) | ||||
|---|---|---|---|---|---|---|---|---|
| C. parvum | C. bovis | C. andersoni | C. ryanae | C. xiaoi | ||||
| Belgium | Calf | 170/53 | 38 (73.1) | 12 (23.1) | - | 3 (5.8) | - | IIaA15G2R1 (23/67.7), IIaA17G2R1 (2/5.9), IIaA13G2R1 (6/17.6), IIaA16G3R1 (3/8.8) |
| Cow | 165/10 | 7 (70.0) | 2 (20.0) | - | 1 (10.0) | - | IIaA15G2R1 (3/60.0), IIaA13G2R1 (2/40.0), | |
| Overall | 335/63 | 45 (71.4) | 14 (22.2) | - | 4 (6.4) | - | IIaA15G2R1 (26/66.7), IIaA17G2R1 (2/5.1), IIaA13G2R1 (8/20.5), IIaA16G3R1 (3/7.7) | |
| France | Calf | 175/72 | 49 (68.1) | 19 (26.4) | - | 3 (4.2) | 1 (1.3) | IIaA15G2R1 (30/65.2), IIaA16G2R1 (11/23.9), IIaA16G1R1 (1/2.2), IIaA18G2R1 (4/8.7) |
| Cow | 175/7 | 2 (28.6) | 3 (42.8) | 1 (14.3) | 1 (14.3) | - | IIaA16G2R1 (1/100.0) | |
| Overall | 350/79 | 51 (64.6) | 22 (27.8) | 1 (1.3) | 4 (5.0) | 1 (1.3) | IIaA15G2R1 (30/63.8), IIaA16G2R1 (12/25.6), IIaA16G1R1 (1/2.1), IIaA18G2R1 (4/8.5) | |
| Netherlands | Calf | 200/65 | 56 (86.2) | 5 (7.7) | - | 4 (6.1) | - | IIaA15G2R1 (42/89.4), IIaA13G2R1 (3/6.4), IIaA14G1R1 (2/4.2) |
| Cow | 199/4 | 3 (75.0) | 1 (25.0) | - | - | - | - | |
| Overall | 399/69 | 59 (85.5) | 6 (8.7) | - | 4 (5.8) | - | IIaA15G2R1 (42/89.4), IIaA13G2R1 (3/6.4), IIaA14G1R1 (2/4.2) | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pinto, P.; Ribeiro, C.A.; Hoque, S.; Hammouma, O.; Leruste, H.; Détriché, S.; Canniere, E.; Daandels, Y.; Dellevoet, M.; Roemen, J.; et al. Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe. Microorganisms 2021, 9, 2394. https://doi.org/10.3390/microorganisms9112394
Pinto P, Ribeiro CA, Hoque S, Hammouma O, Leruste H, Détriché S, Canniere E, Daandels Y, Dellevoet M, Roemen J, et al. Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe. Microorganisms. 2021; 9(11):2394. https://doi.org/10.3390/microorganisms9112394
Chicago/Turabian StylePinto, Pedro, Cláudia A. Ribeiro, Sumaiya Hoque, Ourida Hammouma, Hélène Leruste, Sébastien Détriché, Evi Canniere, Yvonne Daandels, Martine Dellevoet, Janine Roemen, and et al. 2021. "Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe" Microorganisms 9, no. 11: 2394. https://doi.org/10.3390/microorganisms9112394
APA StylePinto, P., Ribeiro, C. A., Hoque, S., Hammouma, O., Leruste, H., Détriché, S., Canniere, E., Daandels, Y., Dellevoet, M., Roemen, J., Barbier Bourgeois, A., Kváč, M., Follet, J., & Tsaousis, A. D. (2021). Cross-Border Investigations on the Prevalence and Transmission Dynamics of Cryptosporidium Species in Dairy Cattle Farms in Western Mainland Europe. Microorganisms, 9(11), 2394. https://doi.org/10.3390/microorganisms9112394






