Prophage Genomics and Ecology in the Family Rhodobacteraceae
Abstract
1. Introduction
2. Methods
2.1. Prophage Identification
2.2. Prophage Taxonomy and Phylogenomics
2.3. Auxiliary Metabolic Genes and Gene Transfer Agents
2.4. Presence of Prophages in an Algae Bloom
2.5. Global Distribution of Roseobacter Prophages
3. Results
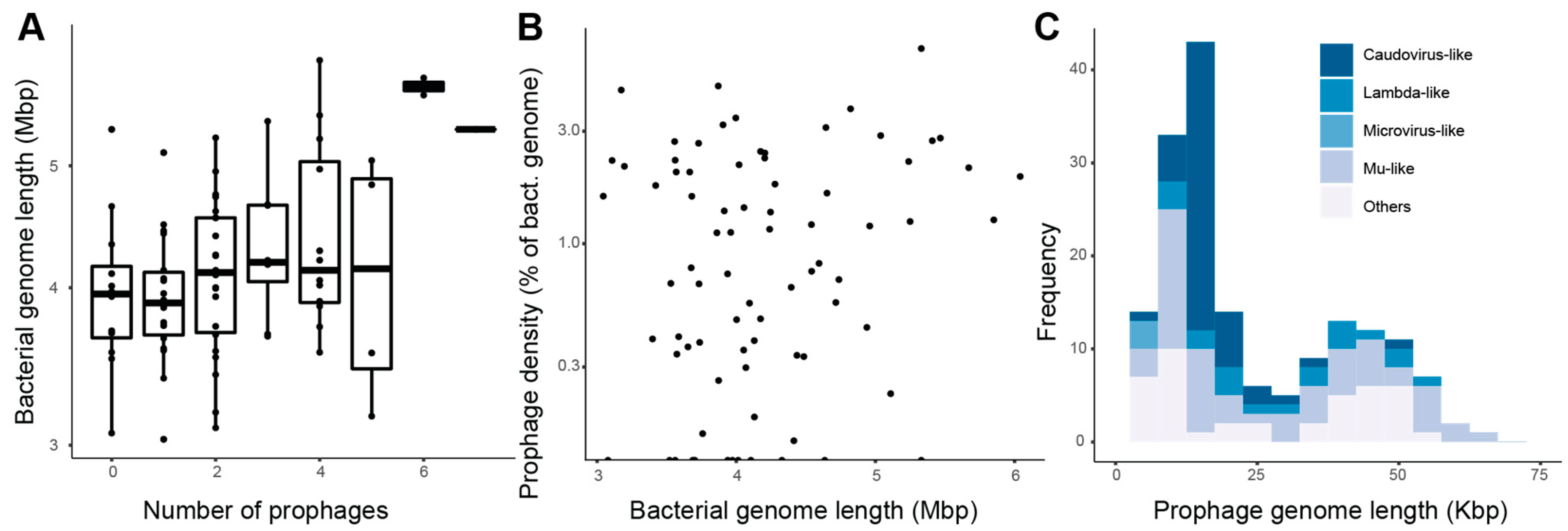
3.1. Putative Prophages in Roseobacter Genomes
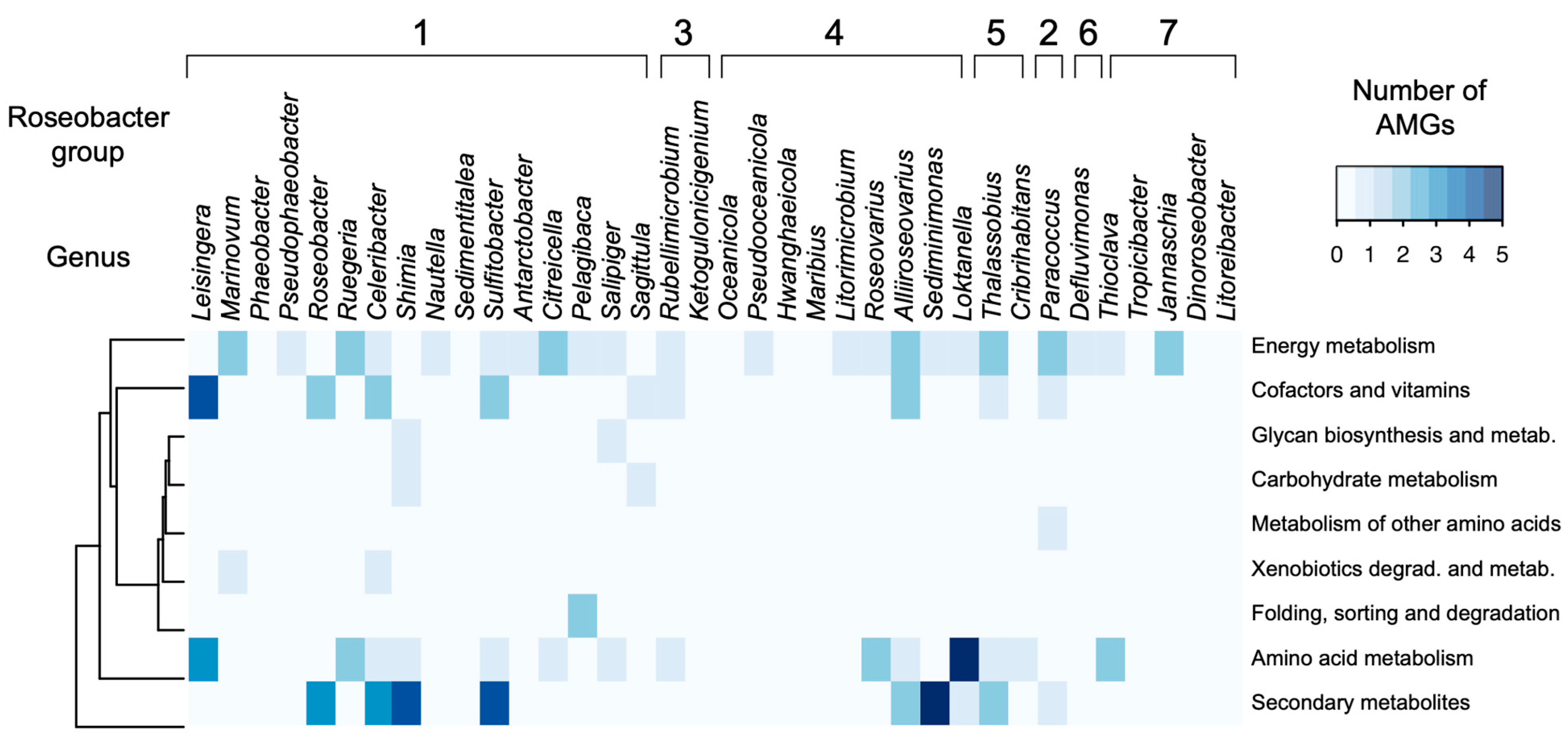
3.2. Auxiliary Metabolic Genes
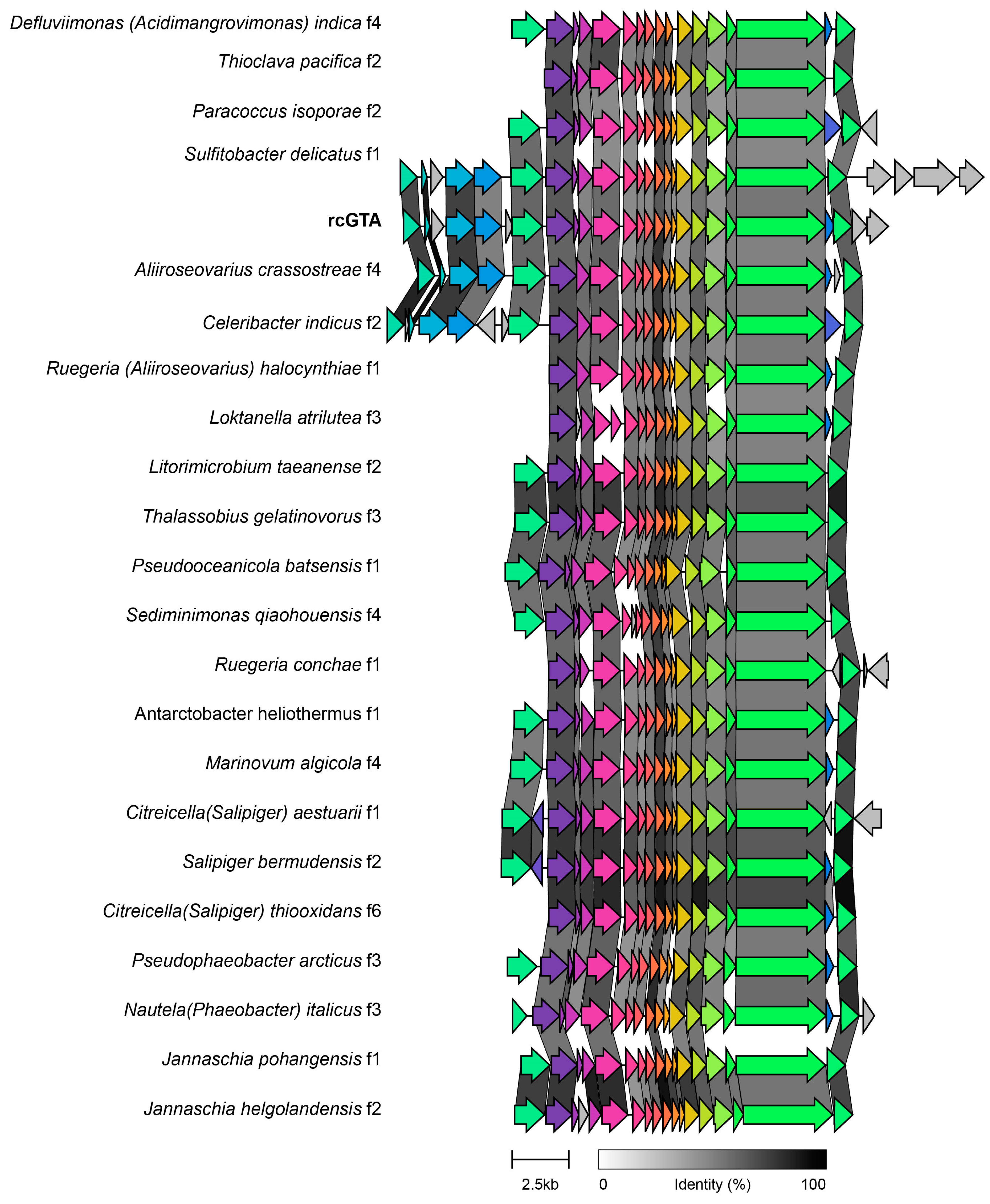
3.3. Gene Transfer Agents
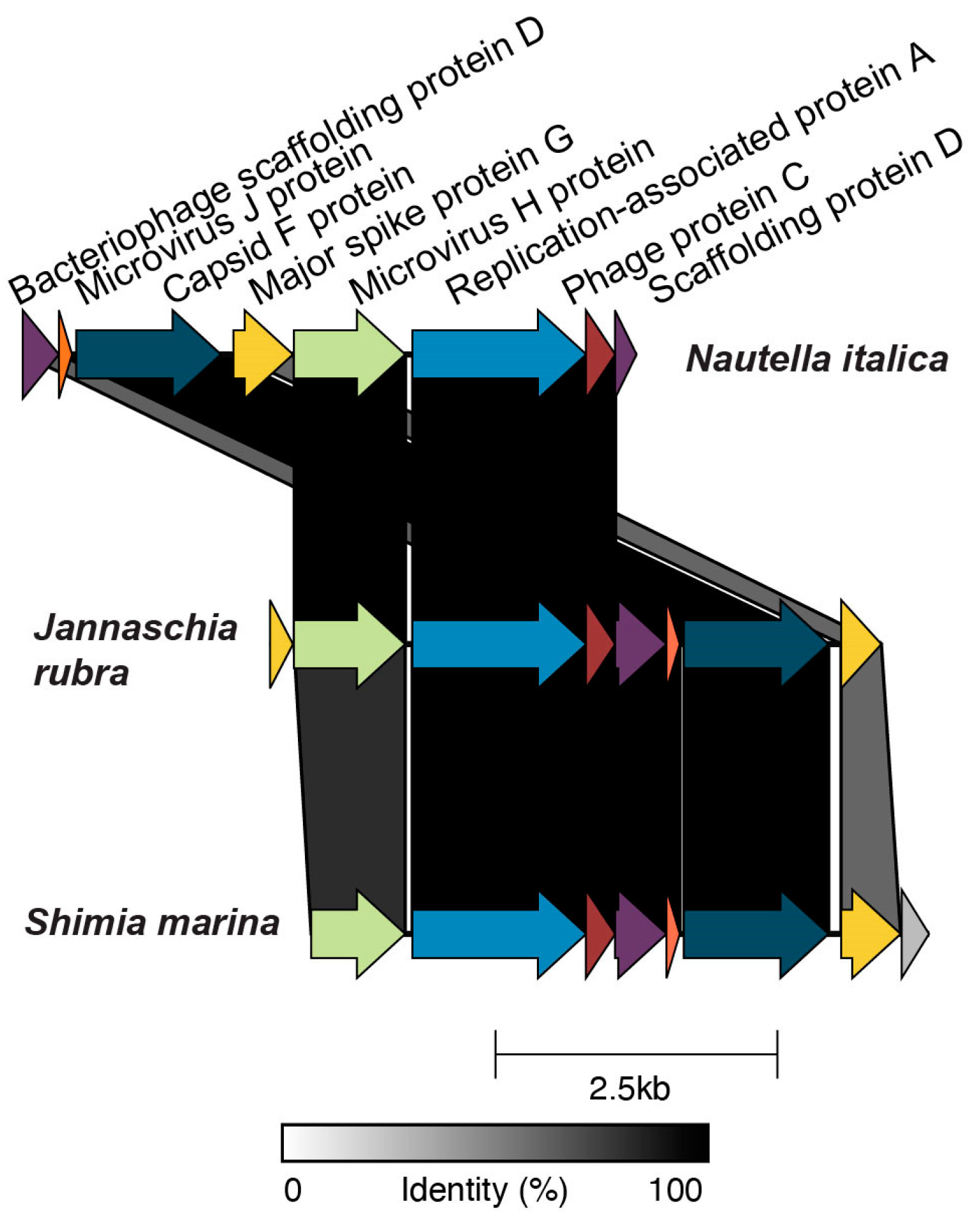
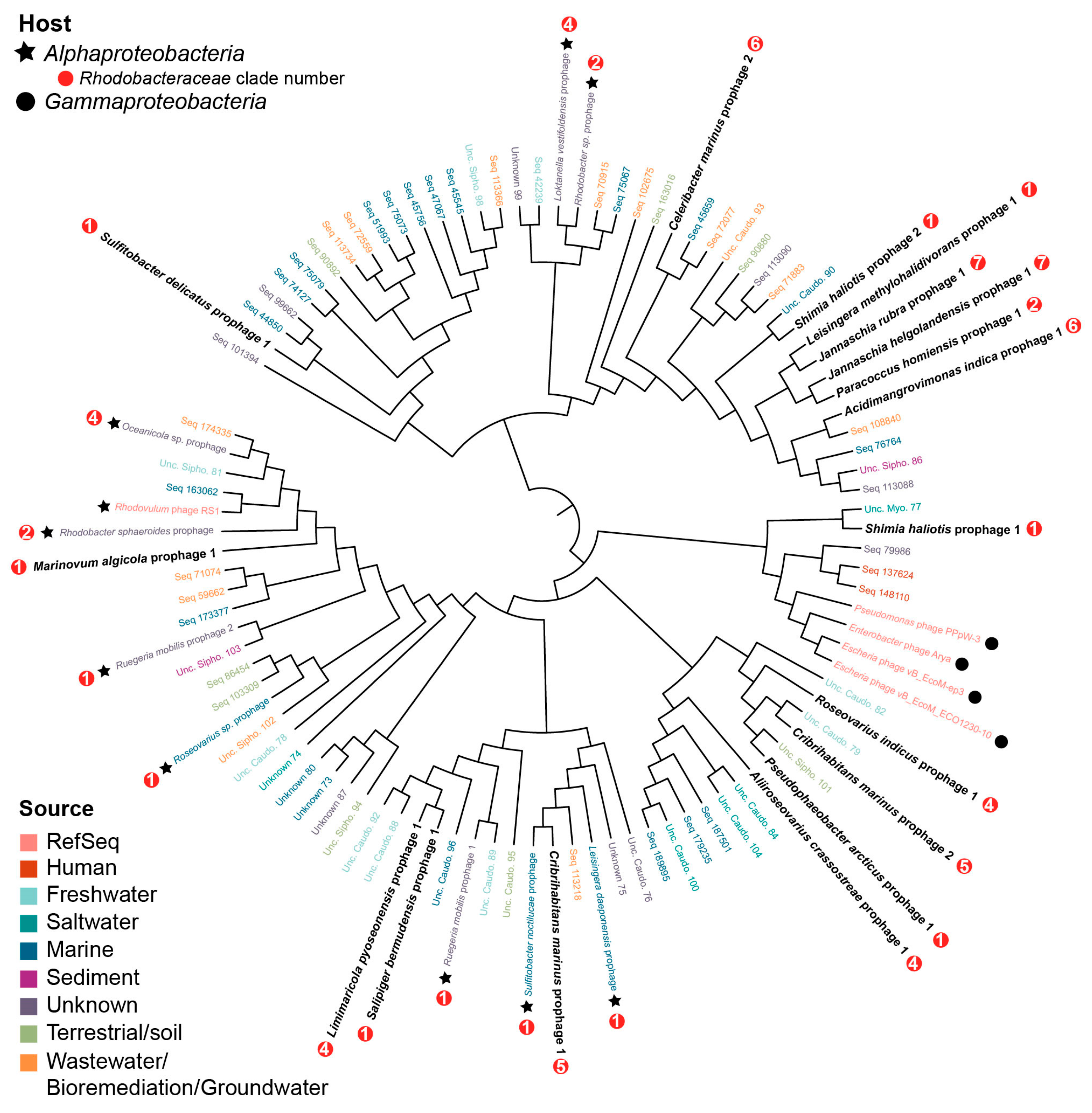
3.4. Prophage Phylogenomics
3.5. Roseobacter Prophages in a Phytoplankton Bloom
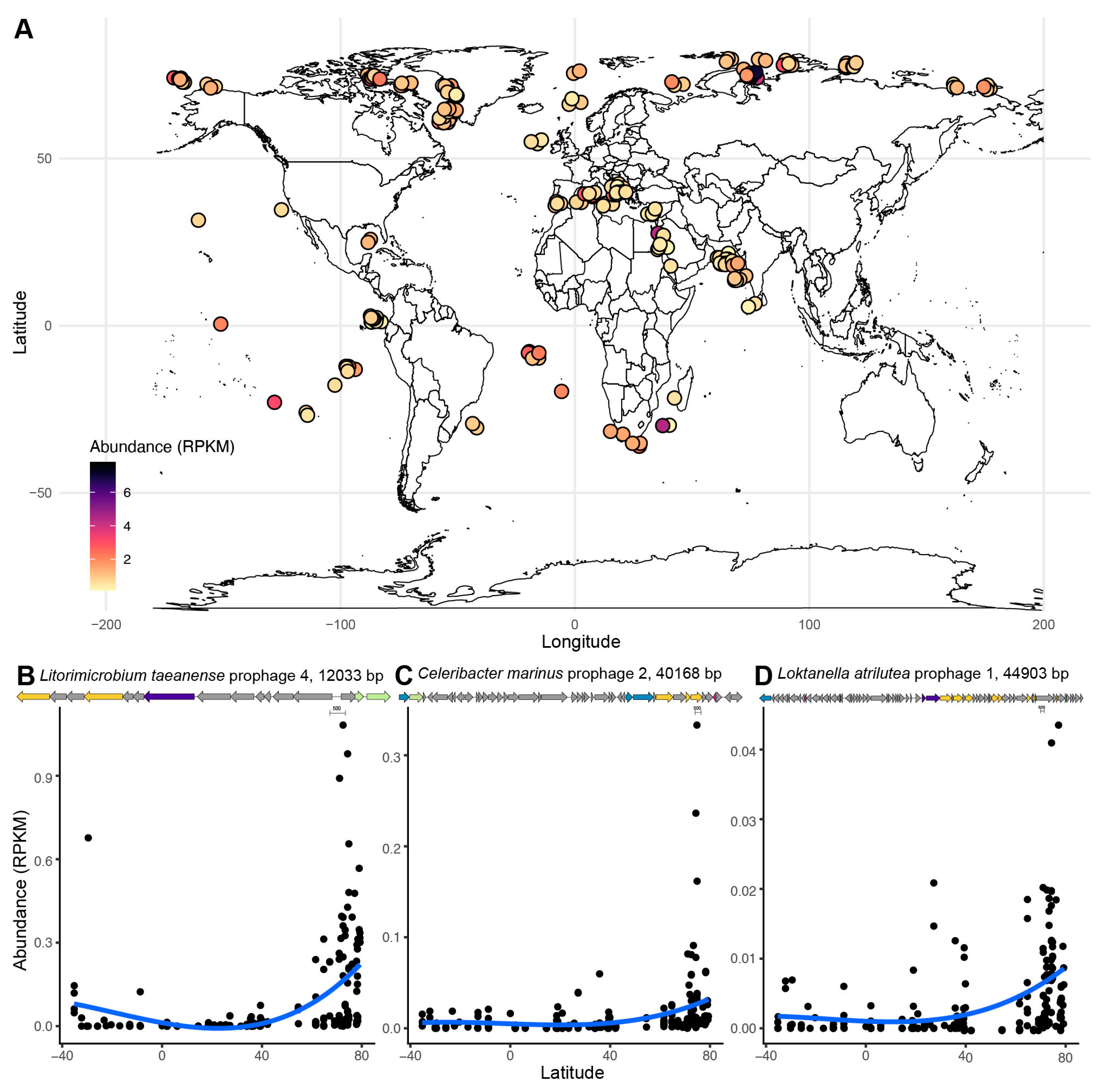
3.6. Global Distribution of Roseobacter Prophages
4. Discussion
4.1. Prophages Contribute to the Evolution of Roseobacter Genomes
4.2. Roseobacter Prophages Have Narrow Host Range
4.3. Auxiliary Metabolic Genes and Gene Transfer Agents
4.4. Roseobacter Prophages Represent Novel Viral Groups
4.5. Roseobacter Prophages Are Abundant in a Phytoplankton Bloom
4.6. The Global Distribution of Roseobacter Prophages Is Associated with Latitude
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bobay, L.-M.; Touchon, M.; Rocha, E.P.C. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 12127–12132. [Google Scholar] [CrossRef]
- Bobay, L.-M.; Rocha, E.P.C.; Touchon, M. The adaptation of temperate bacteriophages to their host genomes. Mol. Biol. Evol. 2013, 30, 737–751. [Google Scholar] [CrossRef] [PubMed]
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, A.E.; Howard-Varona, C.; Needham, D.M.; John, S.G.; Worden, A.Z.; Sullivan, M.B.; Waldbauer, J.R.; Coleman, M.L. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat. Rev. Microbiol. 2020, 18, 21–34. [Google Scholar] [CrossRef]
- Matos, R.C.; Lapaque, N.; Rigottier-Gois, L.; Debarbieux, L.; Meylheuc, T.; Gonzalez-Zorn, B.; Repoila, F.; de Fatima Lopes, M.; Serror, P. Enterococcus faecalis Prophage Dynamics and Contributions to Pathogenic Traits. PLoS Genet. 2013, 9, e1003539. [Google Scholar] [CrossRef]
- Touchon, M.; Bernheim, A.; Rocha, E.P.C. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016, 10, 2744–2754. [Google Scholar] [CrossRef]
- Brüssow, H.; Canchaya, C.; Hardt, W.; Bru, H. Phages and the evolution of bacterial pathogens: From genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 2004, 68, 560–602. [Google Scholar] [CrossRef]
- Touchon, M.; De Sousa, J.A.M.; Rocha, E.P.C. Embracing the enemy: The diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 2017, 38, 66–73. [Google Scholar] [CrossRef]
- Luo, H.; Moran, M.A. Evolutionary ecology of the marine Roseobacter clade. Microbiol. Mol. Biol. Rev. 2014, 78, 573–587. [Google Scholar] [CrossRef]
- Buchan, A.; González, J.M.; Moran, M.A. Overview of the Marine Roseobacter Lineage. Appl. Environ. Microbiol. 2005, 71, 5665–5677. [Google Scholar] [CrossRef]
- Giebel, H.-A.; Brinkhoff, T.; Zwisler, W.; Selje, N.; Simon, M. Distribution of Roseobacter RCA and SAR11 lineages and distinct bacterial communities from the subtropics to the Southern Ocean. Environ. Microbiol. 2009, 11, 2164–2178. [Google Scholar] [CrossRef] [PubMed]
- Rappé, M.S.; Vergin, K.; Giovannoni, S.J. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol. Ecol. 2000, 33, 219–232. [Google Scholar] [CrossRef]
- Geng, H.; Belas, R. Molecular mechanisms underlying roseobacter–phytoplankton symbioses. Curr. Opin. Biotechnol. 2010, 21, 332–338. [Google Scholar] [CrossRef]
- Brinkhoff, T.; Bach, G.; Heidorn, T.; Liang, L.; Schlingloff, A.; Simon, M. Antibiotic Production by a Roseobacter Clade-Affiliated Species from the German Wadden Sea and Its Antagonistic Effects on Indigenous Isolates. Appl. Environ. Microbiol. 2004, 70, 2560–2565. [Google Scholar] [CrossRef] [PubMed]
- Lenk, S.; Moraru, C.; Hahnke, S.; Arnds, J.; Richter, M.; Kube, M.; Reinhardt, R.; Brinkhoff, T.; Harder, J.; Amann, R.; et al. Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J. 2012, 6, 2178–2187. [Google Scholar] [CrossRef]
- Sañudo-Wilhelmy, S.A.; Gómez-Consarnau, L.; Suffridge, C.; Webb, E.A. The role of B vitamins in marine biogeochemistry. Ann. Rev. Mar. Sci. 2014, 6, 339–367. [Google Scholar] [CrossRef]
- Helliwell, K.E.; Wheeler, G.L.; Leptos, K.C.; Goldstein, R.E.; Smith, A.G. Insights into the evolution of vitamin B12 auxotrophy from sequenced algal genomes. Mol. Biol. Evol. 2011, 28, 2921–2933. [Google Scholar] [CrossRef]
- Bertrand, E.M.; Allen, A.E. Influence of vitamin B auxotrophy on nitrogen metabolism in eukaryotic phytoplankton. Front. Microbiol. 2012, 3, 375. [Google Scholar] [CrossRef]
- Pujalte, M.J.; Lucena, T.; Ruvira, M.A.; Arahal, D.R.; Macián, M.C. The Family Rhodobacteraceae. In The Prokaryotes: Alphaproteobacteria and Betaproteobacteria; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2014; pp. 439–512. ISBN 978-3-642-30197-1. [Google Scholar]
- Harrison, P.W.; Lower, R.P.J.; Kim, N.K.D.; Young, J.P.W. Introducing the bacterial ‘chromid’: Not a chromosome, not a plasmid. Trends Microbiol. 2010, 18, 141–148. [Google Scholar] [CrossRef]
- Petersen, J.; Frank, O.; Göker, M.; Pradella, S. Extrachromosomal, extraordinary and essential—The plasmids of the Roseobacter clade. Appl. Microbiol. Biotechnol. 2013, 97, 2805–2815. [Google Scholar] [CrossRef]
- Frank, O.; Michael, V.; Päuker, O.; Boedeker, C.; Jogler, C.; Rohde, M.; Petersen, J. Plasmid curing and the loss of grip—The 65-kb replicon of Phaeobacter inhibens DSM 17395 is required for biofilm formation, motility and the colonization of marine algae. Syst. Appl. Microbiol. 2015, 38, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Chen, F.; Chu, X.; Zhang, H.; Luo, H.; Qin, F.; Zhai, Z.; Yang, M.; Sun, J.; Zhao, Y. Diverse, Abundant, and Novel Viruses Infecting the Marine Roseobacter RCA Lineage. mSystems 2019, 4, e00494-19. [Google Scholar] [CrossRef] [PubMed]
- Breitbart, M.; Bonnain, C.; Malki, K.; Sawaya, N.A. Phage puppet masters of the marine microbial realm. Nat. Microbiol. 2018, 3, 754–766. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Brum, J.R.; Dutilh, B.E.; Sunagawa, S.; Duhaime, M.B.; Loy, A.; Poulos, B.T.; Solonenko, N.; Lara, E.; Poulain, J.; et al. Ecogenomics and potential biogeochemical impacts of globally abundant ocean viruses. Nature 2016, 537, 689–693. [Google Scholar] [CrossRef]
- Kaneko, H.; Blanc-Mathieu, R.; Endo, H.; Chaffron, S.; Delmont, T.O.; Gaia, M.; Henry, N.; Hernández-Velázquez, R.; Nguyen, C.H.; Mamitsuka, H. Eukaryotic virus composition can predict the efficiency of carbon export in the global ocean. iScience 2021, 24, 102002. [Google Scholar] [CrossRef]
- Silveira, C.; Cavalcanti, G.; Walter, J.; Silva-Lima, A.; Dinsdale, E.; Bourne, D.; Thompson, C.; Thompson, F. Microbial processes driving coral reef organic carbon flow. FEMS Microbiol. Rev. 2017, 41, 575–595. [Google Scholar] [CrossRef]
- Knowles, B.; Silveira, C.B.; Bailey, B.A.; Barott, K.; Cantu, V.A.; Cobian-Guëmes, A.G.; Coutinho, F.H.; Dinsdale, E.A.; Felts, B.; Furby, K.A.; et al. Lytic to temperate switching of viral communities. Nature 2016, 531, 466–470. [Google Scholar] [CrossRef]
- Silveira, C.B.; Rohwer, F.L. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Microbiomes 2016, 2, 16010. [Google Scholar] [CrossRef]
- Faruque, S.M.; Rahman, M.M.; Asadulghani; Islam, K.M.N.; Mekalanos, J.J. Lysogenic Conversion of Environmental Vibrio mimicus Strains by CTXΦ. Infect. Immun. 1999, 67, 5723–5729. [Google Scholar] [CrossRef]
- Silveira, C.B.; Coutinho, F.H.; Cavalcanti, G.S.; Benler, S.; Doane, M.P.M.P.; Dinsdale, E.A.; Edwards, R.A.; Francini-Filho, R.B.; Thompson, C.C.; Luque, A.; et al. Genomic and ecological attributes of marine bacteriophages encoding bacterial virulence genes. BMC Genom. 2020, 21, 1–11. [Google Scholar] [CrossRef]
- Steinberg, K.M.; Levin, B.R. Grazing protozoa and the evolution of the Escherichia coli O157:H7 Shiga toxin-encoding prophage. Proc. R. Soc. B Biol. Sci. 2007, 274, 1921–1929. [Google Scholar] [CrossRef]
- Rabinovich, L.; Sigal, N.; Borovok, I.; Nir-Paz, R.; Herskovits, A.A. Prophage excision activates Listeria competence genes that promote phagosomal escape and virulence. Cell 2012, 150, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Brüssow, H. Bacteria between protists and phages: From antipredation strategies to the evolution of pathogenicity. Mol. Microbiol. 2007, 65, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Zeng, Q.; Kelly, L.; Huang, K.H.; Singer, A.U.; Stubbe, J.; Chisholm, S.W. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc. Natl. Acad. Sci. USA 2011, 108, E757–E764. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anderson, R.E.; Buchan, A.; Campbell, B.J.; Hallam, S.; Hess, M.; Sullivan, M.B.; Walsh, D.A.; Roux, S. Ecology of inorganic sulfur auxiliary metabolism in widespread bacteriophages. bioRxiv 2020. [Google Scholar] [CrossRef]
- Rohwer, F.; Segall, A.; Steward, G.; Seguritan, V.; Breitbart, M.; Wolven, F.; Azam, F.F. The complete genomic sequence of the marine phage Roseophage SIO1 shares homology with nonmarine phages. Limnol. Oceanogr. 2000, 45, 408–418. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, F.; Yang, J.; Jiao, N. Host responses of a marine bacterium, Roseobacter denitrificans OCh114, to phage infection. Arch. Microbiol. 2012, 194, 323–330. [Google Scholar] [CrossRef][Green Version]
- Zhan, Y.; Chen, F. Bacteriophages that infect marine roseobacters: Genomics and ecology. Environ. Microbiol. 2019, 21, 1885–1895. [Google Scholar] [CrossRef]
- Zhan, Y.; Huang, S.; Voget, S.; Simon, M.; Chen, F. A novel roseobacter phage possesses features of podoviruses, siphoviruses, prophages and gene transfer agents. Sci. Rep. 2016, 6, 30372. [Google Scholar] [CrossRef]
- Huang, S.; Zhang, Y.; Chen, F.; Jiao, N. Complete genome sequence of a marine roseophage provides evidence into the evolution of gene transfer agents in alphaproteobacteria. Virol. J. 2011, 8, 124. [Google Scholar] [CrossRef]
- Ankrah, N.Y.D.; Budinoff, C.R.; Wilson, W.H.; Wilhelm, S.W.; Buchan, A. Genome Sequence of the Sulfitobacter sp. Strain 2047-Infecting Lytic Phage ΦCB2047-B. Genome Announc. 2014, 2. [Google Scholar] [CrossRef]
- Bischoff, V.; Bunk, B.; Meier-Kolthoff, J.P.; Spröer, C.; Poehlein, A.; Dogs, M.; Nguyen, M.; Petersen, J.; Daniel, R.; Overmann, J.; et al. Cobaviruses—A new globally distributed phage group infecting Rhodobacteraceae in marine ecosystems. ISME J. 2019, 13, 1404–1421. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhang, S.; Long, L.; Huang, S. Characterization and Complete Genome Sequences of Three N4-Like Roseobacter Phages Isolated from the South China Sea. Curr. Microbiol. 2016, 73, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.Z.-M.; Millard, A.D.; Mann, N.H.; Schäfer, H. Comparative genomics defines the core genome of the growing N4-like phage genus and identifies N4-like Roseophage specific genes. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, K.; Jiao, N.; Chen, F. Genome sequences of two novel phages infecting marine roseobacters. Environ. Microbiol. 2009, 11, 2055–2064. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, J.; Turner, D.; Millard, A.D.; Mahadevan, P.; Kropinski, A.M.; Adriaenssens, E.M. From Orphan Phage to a Proposed New Family–the Diversity of N4-Like Viruses. Antibiotics 2020, 9, 663. [Google Scholar] [CrossRef]
- Ceyssens, P.-J.; Brabban, A.; Rogge, L.; Lewis, M.S.; Pickard, D.; Goulding, D.; Dougan, G.; Noben, J.-P.; Kropinski, A.; Kutter, E.; et al. Molecular and physiological analysis of three Pseudomonas aeruginosa phages belonging to the “N4-like viruses”. Virology 2010, 405, 26–30. [Google Scholar] [CrossRef]
- Angly, F.E.; Felts, B.; Breitbart, M.; Salamon, P.; Edwards, R.A.; Carlson, C.; Chan, A.M.; Haynes, M.; Kelley, S.; Liu, H.; et al. The marine viromes of four oceanic regions. PLoS Biol. 2006, 4, 2121–2131. [Google Scholar] [CrossRef]
- Sullivan, M.B.; Coleman, M.L.; Weigele, P.; Rohwer, F.; Chisholm, S.W. Three Prochlorococcus Cyanophage Genomes: Signature Features and Ecological Interpretations. PLoS Biol. 2005, 3. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, L.; Wang, Y.; Jiao, N.; Zhang, R. Microarray analysis of gene expression of Dinoroseobacter shibae DFL12T in response to phage R2C infection. Mar. Genom. 2018, 42, 53–57. [Google Scholar] [CrossRef]
- Ankrah, N.Y.D.; May, A.L.; Middleton, J.L.; Jones, D.R.; Hadden, M.K.; Gooding, J.R.; LeCleir, G.R.; Wilhelm, S.W.; Campagna, S.R.; Buchan, A. Phage infection of an environmentally relevant marine bacterium alters host metabolism and lysate composition. ISME J. 2014, 8, 1089–1100. [Google Scholar] [CrossRef] [PubMed]
- Ankrah, N.Y.D.; Budinoff, C.R.; Wilson, W.H.; Wilhelm, S.W.; Buchan, A. Genome Sequences of Two Temperate Phages, CB2047-A and CB2047-C, Infecting Sulfitobacter sp. Strain 2047. Genome Announc. 2014, 2, e00108-14. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Wang, K.; Ackermann, H.-W.; Halden, R.U.; Jiao, N.; Chen, F. Searching for a “Hidden” Prophage in a Marine Bacterium. Appl. Environ. Microbiol. 2010, 76, 589–595. [Google Scholar] [CrossRef]
- Tang, K.; Lin, D.; Zheng, Q.; Liu, K.; Yang, Y.; Han, Y.; Jiao, N. Genomic, proteomic and bioinformatic analysis of two temperate phages in Roseobacter clade bacteria isolated from the deep-sea water. BMC Genom. 2017, 18, 485. [Google Scholar] [CrossRef] [PubMed]
- Basso, J.T.R.; Ankrah, N.Y.D.; Tuttle, M.J.; Grossman, A.S.; Sandaa, R.-A.; Buchan, A. Genetically similar temperate phages form coalitions with their shared host that lead to niche-specific fitness effects. ISME J. 2020, 14, 1688–1700. [Google Scholar] [CrossRef] [PubMed]
- Wemheuer, B.; Wemheuer, F.; Hollensteiner, J.; Meyer, F.-D.; Voget, S.; Daniel, R. The green impact: Bacterioplankton response toward a phytoplankton spring bloom in the southern North Sea assessed by comparative metagenomic and metatranscriptomic approaches. Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef] [PubMed]
- Simon, M.; Scheuner, C.; Meier-Kolthoff, J.P.; Brinkhoff, T.; Wagner-Döbler, I.; Ulbrich, M.; Klenk, H.-P.; Schomburg, D.; Petersen, J.; Göker, M. Phylogenomics of Rhodobacteraceae reveals evolutionary adaptation to marine and non-marine habitats. ISME J. 2017, 11, 1483–1499. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Kieft, K.; Zhou, Z.; Anantharaman, K. VIBRANT: Automated recovery, annotation and curation of microbial viruses, and evaluation of viral community function from genomic sequences. Microbiome 2020, 8, 90. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Sonnhammer, E.L.L.; Eddy, S.R.; Durbin, R. Pfam: A comprehensive database of protein domain families based on seed alignments. Proteins Struct. Funct. Bioinform. 1997, 28, 405–420. [Google Scholar] [CrossRef]
- Grazziotin, A.L.; Koonin, E.V.; Kristensen, D.M. Prokaryotic Virus Orthologous Groups (pVOGs): A resource for comparative genomics and protein family annotation. Nucleic Acids Res. 2017, 45, D491–D498. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Páez-Espino, D.; Chen, I.-M.A.; Palaniappan, K.; Ratner, A.; Chu, K.; Reddy, T.B.K.; Nayfach, S.; Schulz, F.; Call, L.; et al. IMG/VR v3: An integrated ecological and evolutionary framework for interrogating genomes of uncultivated viruses. Nucleic Acids Res. 2021, 49, D764–D775. [Google Scholar] [CrossRef]
- Coutinho, F.H.; Edwards, R.A.; Rodríguez-Valera, F. Charting the diversity of uncultured viruses of Archaea and Bacteria. BMC Biol. 2019, 17, 109. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2007, 23, 127–128. [Google Scholar] [CrossRef]
- Schliep, K.; Potts, A.J.; Morrison, D.A.; Grimm, G.W. Intertwining phylogenetic trees and networks. Methods Ecol. Evol. 2017, 8, 1212–1220. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Gilchrist, C.L.M.; Chooi, Y.-H. clinker & clustermap.js: Automatic generation of gene cluster comparison figures. Bioinformatics 2021, btab007. [Google Scholar] [CrossRef]
- Michael, V.; Frank, O.; Bartling, P.; Scheuner, C.; Göker, M.; Brinkmann, H.; Petersen, J. Biofilm plasmids with a rhamnose operon are widely distributed determinants of the ‘swim-or-stick’ lifestyle in roseobacters. ISME J. 2016, 10, 2498–2513. [Google Scholar] [CrossRef]
- Miller, T.R.; Belas, R. Dimethylsulfoniopropionate Metabolism by Pfiesteria-Associated Roseobacter spp. Appl. Environ. Microbiol. 2004, 70, 3383–3391. [Google Scholar] [CrossRef]
- Howard, E.C.; Henriksen, J.R.; Buchan, A.; Reisch, C.R.; Bürgmann, H.; Welsh, R.; Ye, W.; González, J.M.; Mace, K.; Joye, S.B.; et al. Bacterial Taxa That Limit Sulfur Flux from the Ocean. Science 2006, 314, 649–652. [Google Scholar] [CrossRef] [PubMed]
- Du, X.-P.; Cai, Z.-H.; Zuo, P.; Meng, F.-X.; Zhu, J.-M.; Zhou, J. Temporal Variability of Virioplankton during a Gymnodinium catenatum Algal Bloom. Microorganisms 2020, 8, 107. [Google Scholar] [CrossRef]
- Huang, X.; Zhu, J.; Cai, Z.; Lao, Y.; Jin, H.; Yu, K.; Zhang, B.; Zhou, J. Profiles of quorum sensing (QS)-related sequences in phycospheric microorganisms during a marine dinoflagellate bloom, as determined by a metagenomic approach. Microbiol. Res. 2018, 217, 1–13. [Google Scholar] [CrossRef]
- Bushnell, B.; Rood, J.; Singer, E. BBMerge—Accurate paired shotgun read merging via overlap. PLoS ONE 2017, 12, e0185056. [Google Scholar] [CrossRef]
- Ponstingl, H.; Ning, Z. SMALT—A new mapper for DNA sequencing reads. F1000 Posters 2010, 1, 313. [Google Scholar]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef]
- Brum, J.R.; Ignacio-Espinoza, J.C.; Roux, S.; Doulcier, G.; Acinas, S.G.; Alberti, A.; Chaffron, S.; Cruaud, C.; de Vargas, C.; Gasol, J.M. Patterns and ecological drivers of ocean viral communities. Science 2015, 348, 1261498. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.C.; Zayed, A.A.; Conceição-Neto, N.; Temperton, B.; Bolduc, B.; Alberti, A.; Ardyna, M.; Arkhipova, K.; Carmichael, M.; Cruaud, C.; et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell 2019, 177, 1109–1123.e14. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Wilks, C.; Antonescu, V.; Charles, R. Scaling read aligners to hundreds of threads on general-purpose processors. Bioinformatics 2019, 35, 421–432. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Brownrigg, R. Mapdata: Extra Map Databases 2018. R Package Version 2.3.0. Available online: https://CRAN.R-project.org/package=mapdata (accessed on 17 May 2021).
- Liaw, A.; Wiener, M. Classification and Regression by RandomForest. R News 2002, 2, 18–22. [Google Scholar]
- Cherwa, J.E.; Fane, B.A. Microviridae: Microviruses and Gokushoviruses. In eLS; John Wiley & Sons, Ltd: Chichester, UK, 2011. [Google Scholar] [CrossRef]
- Roux, S.; Krupovic, M.; Poulet, A.; Debroas, D.; Enault, F. Evolution and Diversity of the Microviridae Viral Family through a Collection of 81 New Complete Genomes Assembled from Virome Reads. PLoS ONE 2012, 7, e40418. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-M.; Frank, M.W.; Virga, K.G.; Lee, R.E.; Rock, C.O.; Jackowski, S. Acyl Carrier Protein Is a Cellular Target for the Antibacterial Action of the Pantothenamide Class of Pantothenate Antimetabolites. J. Biol. Chem. 2004, 279, 50969–50975. [Google Scholar] [CrossRef]
- Edwards, P.; Nelsen, J.S.; Metz, J.G.; Dehesh, K. Cloning of the fabF gene in an expression vector and in vitro characterization of recombinant fabF and fabB encoded enzymes from Escherichia coli. FEBS Lett. 1997, 402, 62–66. [Google Scholar] [CrossRef]
- Lai, C.-Y.; Cronan, J.E. Isolation and Characterization of β-Ketoacyl-Acyl Carrier Protein Reductase (fabG) Mutants of Escherichia coli and Salmonella enterica Serovar Typhimurium. J. Bacteriol. 2004, 186, 1869–1878. [Google Scholar] [CrossRef]
- Shakya, M.; Soucy, S.M.; Zhaxybayeva, O. Insights into origin and evolution of α-proteobacterial gene transfer agents. Virus Evol. 2017, 3, vex036. [Google Scholar] [CrossRef] [PubMed]
- González, J.M.; Simó, R.; Massana, R.; Covert, J.S.; Casamayor, E.O.; Pedrós-Alió, C.; Moran, M.A. Bacterial Community Structure Associated with a Dimethylsulfoniopropionate-Producing North Atlantic Algal Bloom. Appl. Environ. Microbiol. 2000, 66, 4237–4246. [Google Scholar] [CrossRef]
- Luo, H.; Csűros, M.; Hughes, A.L.; Moran, M.A. Evolution of Divergent Life History Strategies in Marine Alphaproteobacteria. mBio 2013, 4. [Google Scholar] [CrossRef]
- Brueggemann, A.B.; Harrold, C.L.; Rezaei Javan, R.; van Tonder, A.J.; McDonnell, A.J.; Edwards, B.A. Pneumococcal prophages are diverse, but not without structure or history. Sci. Rep. 2017, 7, 42976. [Google Scholar] [CrossRef] [PubMed]
- Crispim, J.S.; Dias, R.S.; Vidigal, P.M.P.; de Sousa, M.P.; da Silva, C.C.; Santana, M.F.; de Paula, S.O. Screening and characterization of prophages in Desulfovibrio genomes. Sci. Rep. 2018, 8, 9273. [Google Scholar] [CrossRef]
- Khan, A.; Burmeister, A.R.; Wahl, L.M. Evolution along the parasitism-mutualism continuum determines the genetic repertoire of prophages. PLoS Comput. Biol. 2020, 16, e1008482. [Google Scholar] [CrossRef]
- Dekel-Bird, N.P.; Sabehi, G.; Mosevitzky, B.; Lindell, D. Host-dependent differences in abundance, composition and host range of cyanophages from the Red Sea. Environ. Microbiol. 2015, 17, 1286–1299. [Google Scholar] [CrossRef]
- Zheng, Q.; Chen, Q.; Xu, Y.; Suttle, C.A.; Jiao, N. A virus infecting marine photoheterotrophic Alphaproteobacteria (Citromicrobium spp.) Defines a new lineage of ssDNA viruses. Front. Microbiol. 2018, 9, 1418. [Google Scholar] [CrossRef]
- Krupovic, M.; Forterre, P. Microviridae goes temperate: Microvirus-related proviruses reside in the genomes of Bacteroidetes. PLoS ONE 2011, 6, e19893. [Google Scholar] [CrossRef]
- McKenna, R.; Ilag, L.L.; Rossmann, M.G. Analysis of the single-stranded DNA bacteriophage phi X174, refined at a resolution of 3.0 A. J. Mol. Biol. 1994, 237, 517–543. [Google Scholar] [CrossRef]
- Bono, L.M.; Gensel, C.L.; Pfennig, D.W.; Burch, C.L. Competition and the origins of novelty: Experimental evolution of niche-width expansion in a virus. Biol. Lett. 2013, 9, 20120616. [Google Scholar] [CrossRef] [PubMed]
- Bertozzi Silva, J.; Storms, Z.; Sauvageau, D. Host receptors for bacteriophage adsorption. FEMS Microbiol. Lett. 2016, 363. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Jiao, N.; Zhang, R. The Genomic Content and Context of Auxiliary Metabolic Genes in Roseophages. Environ. Microbiol. 2021. [Google Scholar] [CrossRef]
- Shen, Z.; Byers, D.M. Isolation of Vibrio harveyi acyl carrier protein and the fabG, acpP, and fabF genes involved in fatty acid biosynthesis. J. Bacteriol. 1996, 178, 571–573. [Google Scholar] [CrossRef][Green Version]
- Kutchma, A.J.; Hoang, T.T.; Schweizer, H.P. Characterization of a Pseudomonas aeruginosa Fatty Acid Biosynthetic Gene Cluster: Purification of Acyl Carrier Protein (ACP) and Malonyl-Coenzyme A:ACP Transacylase (FabD). J. Bacteriol. 1999, 181, 5498–5504. [Google Scholar] [CrossRef]
- Rawlings, M. The gene encoding Escherichia coli acyl carrier protein lies within a cluster of fatty acid biosynthetic genes. J. Biol. Chem. 1991, 267, 5751–5754. [Google Scholar] [CrossRef]
- Roitman, S.; Hornung, E.; Flores-Uribe, J.; Sharon, I.; Feussner, I.; Béjà, O. Cyanophage-encoded lipid desaturases: Oceanic distribution, diversity and function. ISME J. 2018, 12, 343–355. [Google Scholar] [CrossRef]
- Lang, A.S.; Beatty, J.T. Importance of widespread gene transfer agent genes in α-proteobacteria. Trends Microbiol. 2007, 15, 54–62. [Google Scholar] [CrossRef]
- Kawato, Y.; Yasuike, M.; Nakamura, Y.; Shigenobu, Y.; Fujiwara, A.; Sano, M.; Nakai, T. Complete Genome Sequence Analysis of Two Pseudomonas plecoglossicida Phages, Potential Therapeutic Agents. Appl. Environ. Microbiol. 2015, 81, 874–881. [Google Scholar] [CrossRef]
- Tikhe, C.V.; Gissendanner, C.R.; Husseneder, C. Whole-Genome Sequence of the Novel Enterobacter Bacteriophage Arya with an Integrase Pseudogene, Isolated from the Gut of the Formosan Subterranean Termite. Genome Announc. 2018, 6. [Google Scholar] [CrossRef]
- Fuentes, J.L.; Garbayo, I.; Cuaresma, M.; Montero, Z.; González-del-Valle, M.; Vílchez, C. Impact of Microalgae-Bacteria Interactions on the Production of Algal Biomass and Associated Compounds. Mar. Drugs 2016, 14, 100. [Google Scholar] [CrossRef]
- Seyedsayamdost, M.R.; Carr, G.; Kolter, R.; Clardy, J. Roseobacticides: Small Molecule Modulators of an Algal-Bacterial Symbiosis. J. Am. Chem. Soc. 2011, 133, 18343–18349. [Google Scholar] [CrossRef]
- Wong, H.C.; Liu, G.; Zhang, Y.-M.; Rock, C.O.; Zheng, J. The Solution Structure of Acyl Carrier Protein from Mycobacterium tuberculosis. J. Biol. Chem. 2002, 277, 15874–15880. [Google Scholar] [CrossRef]
- Boels, I.C.; Beerthuyzen, M.M.; Kosters, M.H.W.; Van Kaauwen, M.P.W.; Kleerebezem, M.; de Vos, W.M. Identification and Functional Characterization of the Lactococcus lactis rfb Operon, Required for dTDP-Rhamnose Biosynthesis. J. Bacteriol. 2004, 186, 1239–1248. [Google Scholar] [CrossRef]
- Giraud, M.-F.; Naismith, J.H. The rhamnose pathway. Curr. Opin. Struct. Biol. 2000, 10, 687–696. [Google Scholar] [CrossRef]
- Marolda, C.L.; Valvano, M.A. Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: Identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 1995, 177, 5539–5546. [Google Scholar] [CrossRef]
- Liu, D.; Reeves, P.R. Escherichia coli K12 regains its O antigen. Microbiology 1994, 140 Pt 1, 49–57. [Google Scholar] [CrossRef]
- Broeker, N.K.; Barbirz, S. Not a barrier but a key: How bacteriophages exploit host’s O-antigen as an essential receptor to initiate infection. Mol. Microbiol. 2017, 105, 353–357. [Google Scholar] [CrossRef]
- Hwang, J.; Park, S.Y.; Park, M.; Lee, S.; Lee, T.-K. Seasonal Dynamics and Metagenomic Characterization of Marine Viruses in Goseong Bay, Korea. PLoS ONE 2017, 12, e0169841. [Google Scholar] [CrossRef]
- Cai, L.; Zhang, R.; He, Y.; Feng, X.; Jiao, N. Metagenomic Analysis of Virioplankton of the Subtropical Jiulong River Estuary, China. Viruses 2016, 8, 35. [Google Scholar] [CrossRef]
- Liang, Y.; Wang, L.; Wang, Z.; Zhao, J.; Yang, Q.; Wang, M.; Yang, K.; Zhang, L.; Jiao, N.; Zhang, Y. Metagenomic Analysis of the Diversity of DNA Viruses in the Surface and Deep Sea of the South China Sea. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Ghai, R.; Hernandez, C.M.; Picazo, A.; Mizuno, C.M.; Ininbergs, K.; Díez, B.; Valas, R.; DuPont, C.L.; McMahon, K.D.; Camacho, A.; et al. Metagenomes of Mediterranean Coastal Lagoons. Sci. Rep. 2012, 2, 490. [Google Scholar] [CrossRef] [PubMed]
- Selje, N.; Simon, M.; Brinkhoff, T. A newly discovered Roseobacter cluster in temperate and polar oceans. Nature 2004, 427, 445–448. [Google Scholar] [CrossRef] [PubMed]
- Labonté, J.M.; Swan, B.K.; Poulos, B.; Luo, H.; Koren, S.; Hallam, S.J.; Sullivan, M.B.; Woyke, T.; Wommack, K.E.; Stepanauskas, R. Single-cell genomics-based analysis of virus–host interactions in marine surface bacterioplankton. ISME J. 2015, 9, 2386–2399. [Google Scholar] [CrossRef]
- Luo, E.; Eppley, J.M.; Romano, A.E.; Mende, D.R.; DeLong, E.F. Double-stranded DNA virioplankton dynamics and reproductive strategies in the oligotrophic open ocean water column. ISME J. 2020, 1–12. [Google Scholar] [CrossRef]
- Srinivas, T.N.R.; Nageswara Rao, S.S.S.; Vishnu Vardhan Reddy, P.; Pratibha, M.S.; Sailaja, B.; Kavya, B.; Hara Kishore, K.; Begum, Z.; Singh, S.M.; Shivaji, S. Bacterial Diversity and Bioprospecting for Cold-Active Lipases, Amylases and Proteases, from Culturable Bacteria of Kongsfjorden and Ny-Ålesund, Svalbard, Arctic. Curr. Microbiol. 2009, 59, 537–547. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Forcone, K.; Coutinho, F.H.; Cavalcanti, G.S.; Silveira, C.B. Prophage Genomics and Ecology in the Family Rhodobacteraceae. Microorganisms 2021, 9, 1115. https://doi.org/10.3390/microorganisms9061115
Forcone K, Coutinho FH, Cavalcanti GS, Silveira CB. Prophage Genomics and Ecology in the Family Rhodobacteraceae. Microorganisms. 2021; 9(6):1115. https://doi.org/10.3390/microorganisms9061115
Chicago/Turabian StyleForcone, Kathryn, Felipe H. Coutinho, Giselle S. Cavalcanti, and Cynthia B. Silveira. 2021. "Prophage Genomics and Ecology in the Family Rhodobacteraceae" Microorganisms 9, no. 6: 1115. https://doi.org/10.3390/microorganisms9061115
APA StyleForcone, K., Coutinho, F. H., Cavalcanti, G. S., & Silveira, C. B. (2021). Prophage Genomics and Ecology in the Family Rhodobacteraceae. Microorganisms, 9(6), 1115. https://doi.org/10.3390/microorganisms9061115




