Unraveling the Central Role of Sulfur-Oxidizing Acidiphilium multivorum LMS in Industrial Bioprocessing of Gold-Bearing Sulfide Concentrates
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation and Cultivation Conditions
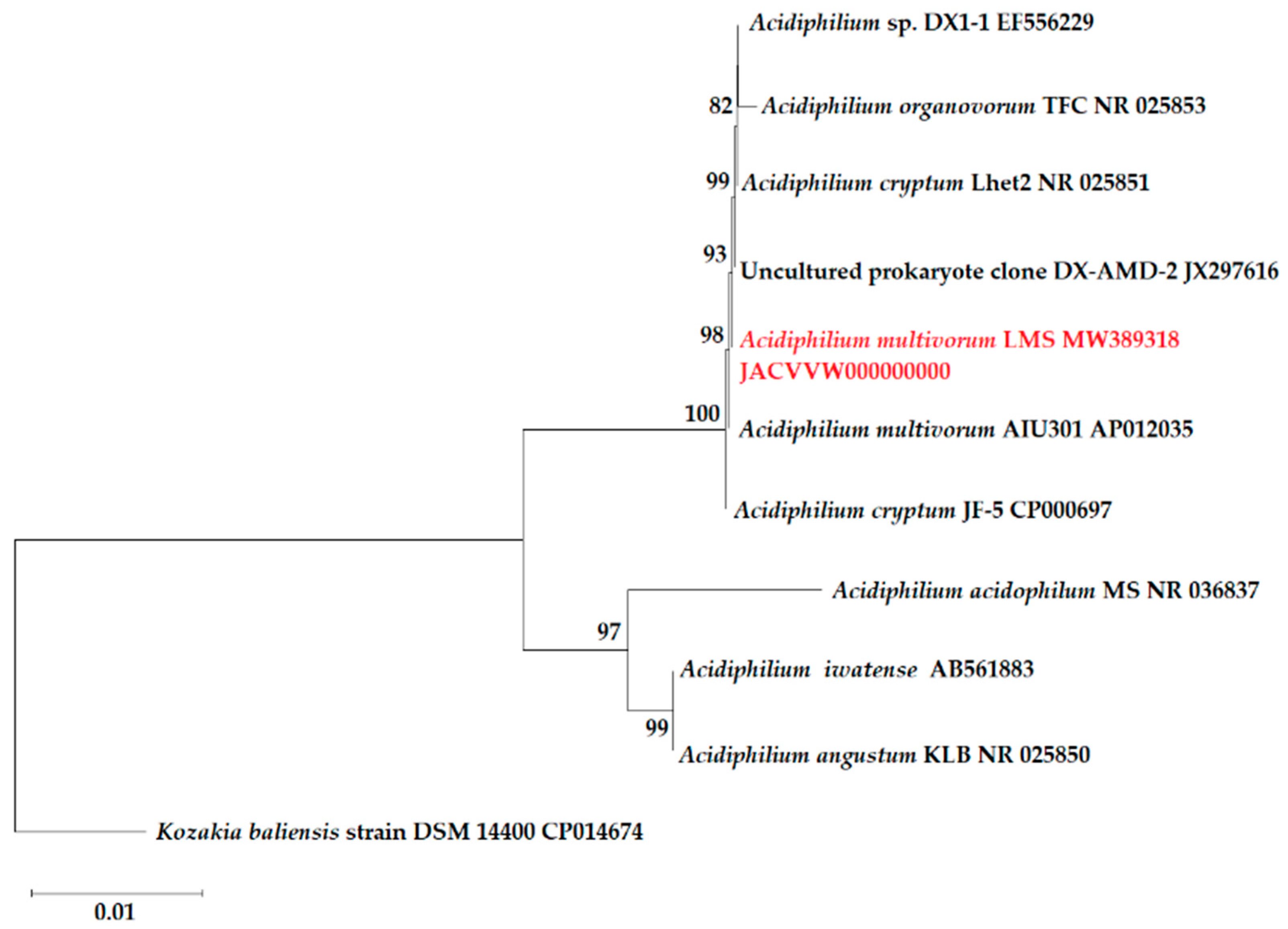
2.2. Taxonomic Research and Phylogenetic Analysis
2.3. Genome Sequencing and Assembly
2.4. Genome Annotation and Analysis
2.5. Analytical Techniques
3. Results and Discussion
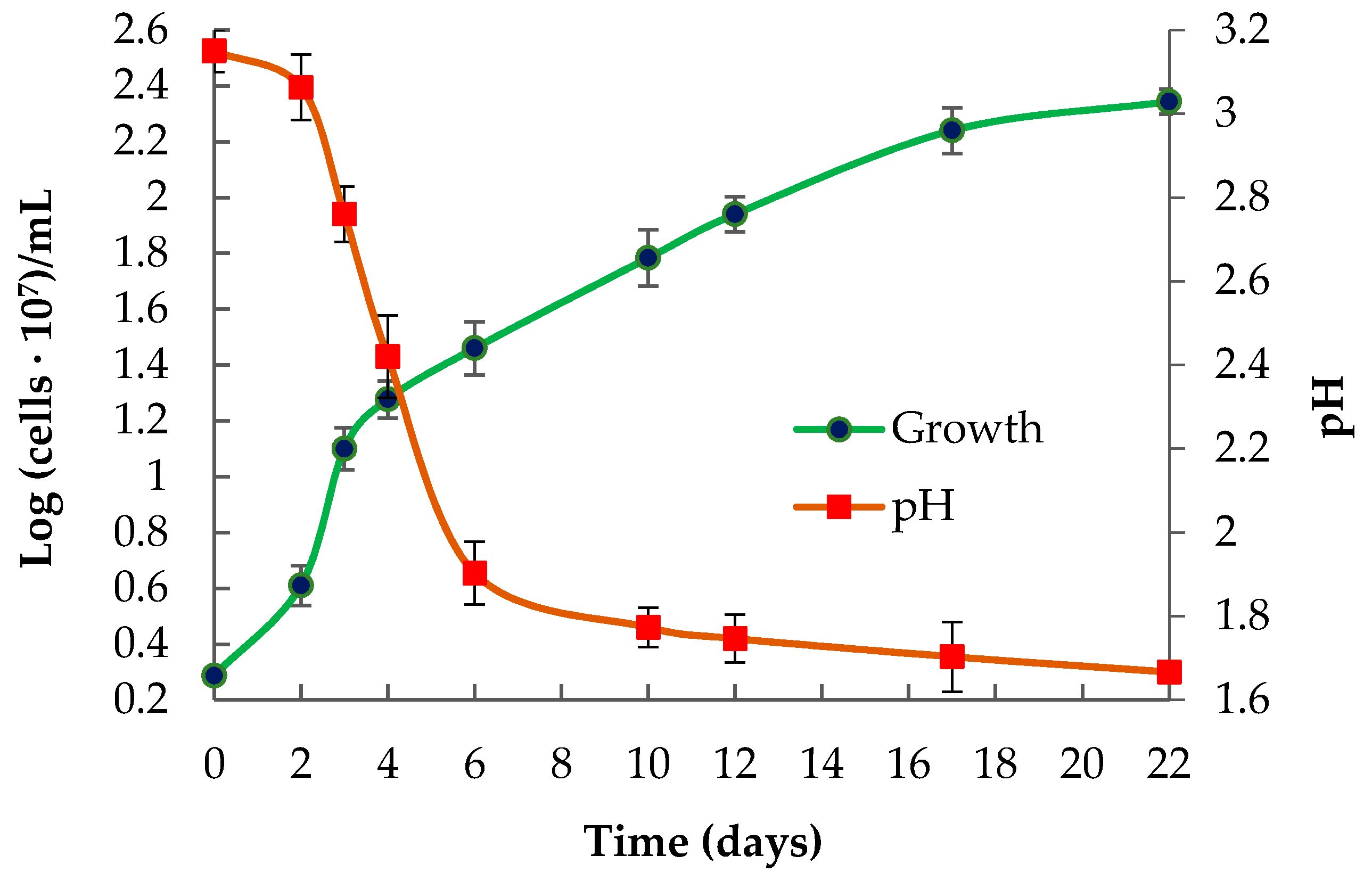
3.1. Isolation and Identification of the LMS Strain
3.2. Phenotypic Features of the Strain Ac. multivorum LMS
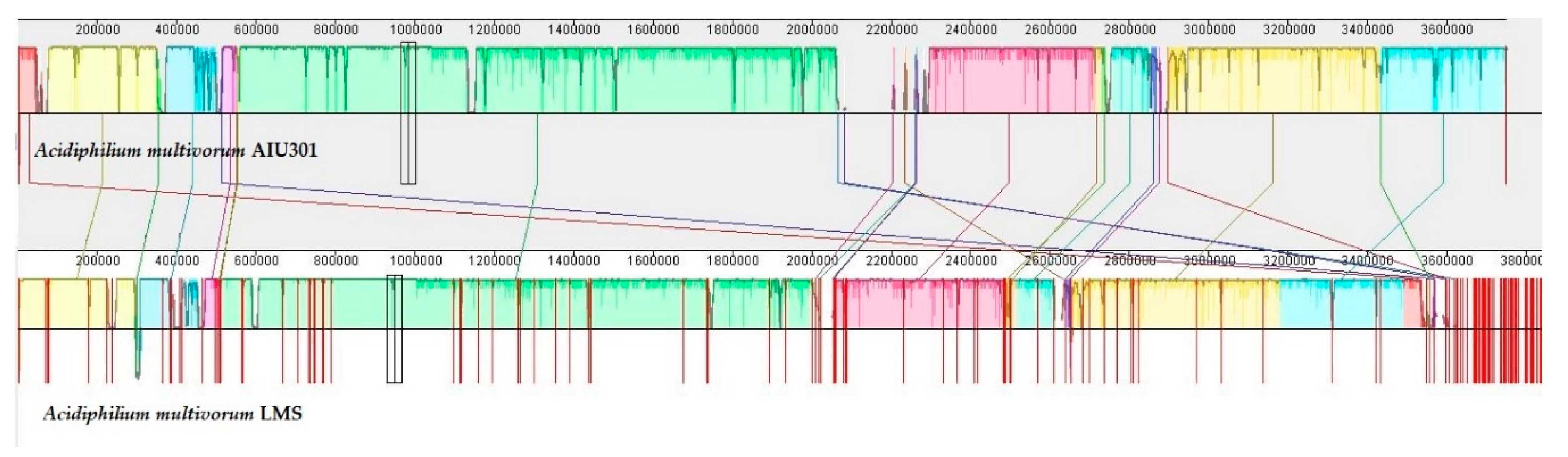
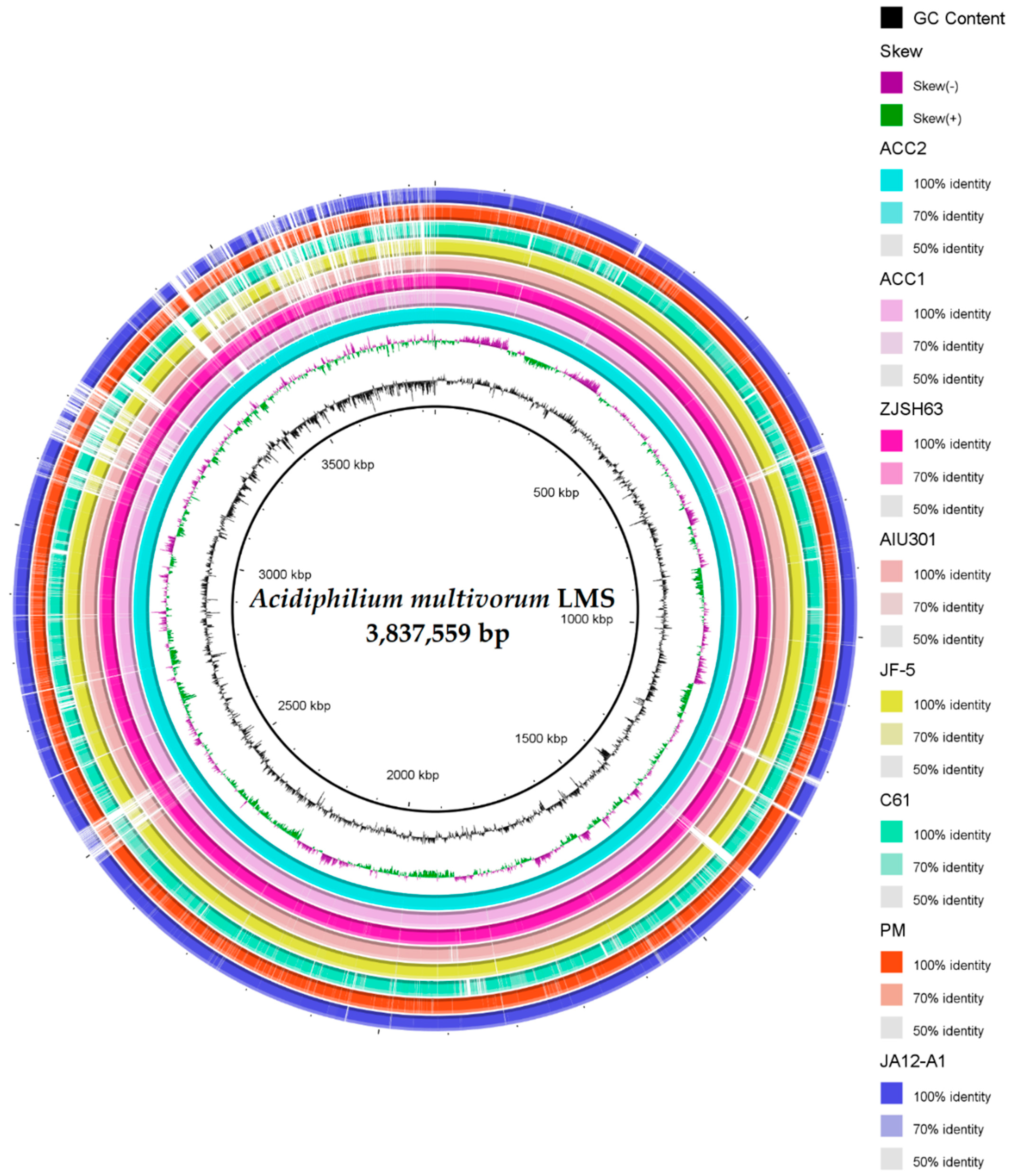
3.3. General Description and Specific Features of Ac. multivorum LMS Genome
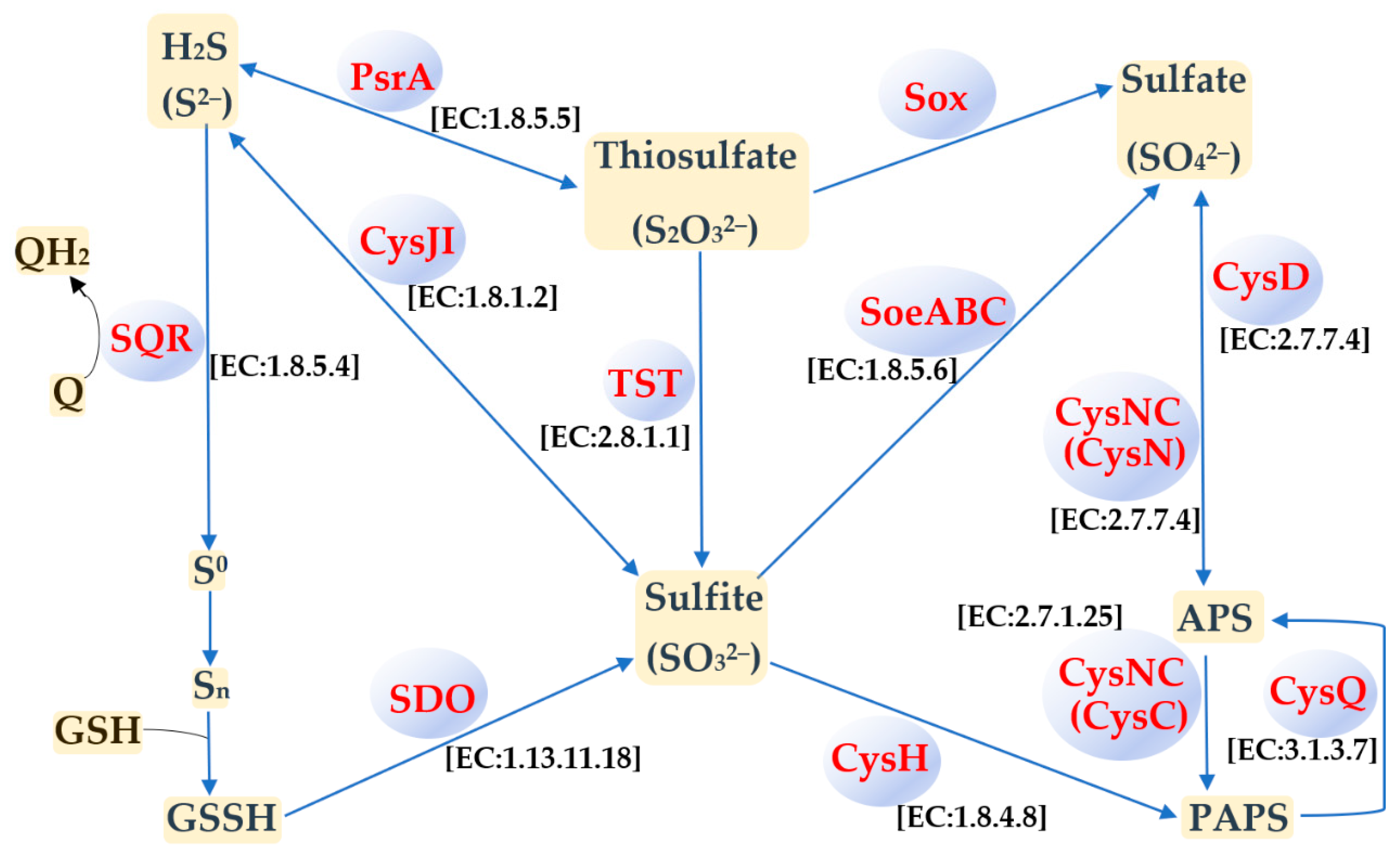
3.4. Sulfur Metabolism
4. Conclusions
- Ac. multivorum LMS proved to be one of the predominant sulfur oxidizers in the microbial populations during industrial processing of the gold-bearing pyrite-arsenopyrite ore concentrates;
- Phenotypic features of LMS strain included wide pH and temperature ranges for growth, as well as the ability to grow organotrophically, lithotrophically, or mixotrophically;
- The first draft genome of the sulfur-oxidizing Ac. multivorum was sequenced, annotated, and compared to other Acidiphilium genomes available in the databases;
- Based on the functional analysis of the genome and sulfur-oxidizing activity of the strain, a biochemical model for sulfur oxidation in Ac. multivorum LMS was proposed;
- Strain-specific genes of Ac. multivorum LMS may confer additional resistance to arsenic and heavy metals, such as copper, cadmium, and zinc;
- The commercial importance of Ac. multivorum LMS was proved by the efficient sulfur oxidation in the presence or absence of organic compounds, a complicated system of sulfur oxidation pathways, as well as additional metal resistance determinants specific for the LMS strain within the genus Acidiphilium.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Harrison, A.P., Jr. Acidiphilium cryptum gen. nov., sp. nov., heterotrophic bacterium from acidic mineral environments. Int. J. Syst. Evol. Microbiol. 1981, 31, 327–332. [Google Scholar] [CrossRef]
- Wakao, N.; Nagasawa, N.; Matsuura, T.; Matsukura, H.; Matsumoto, T.; Hiraishi, A.; Matsumoto, T.; Hiraishi, A.; Sakurai, Y.; Shiota, H. An acidophilic chemoorganotrophic bacterium from pyritic acid mine drainage. J. Gen. Appl. Microbiol. 1994, 40, 143–159. [Google Scholar] [CrossRef]
- Hallberg, K.B.; Johnson, D.B. Biodiversity of acidophilic prokaryotes. Adv. Appl. Microbiol. 2001, 49, 37–84. [Google Scholar] [CrossRef]
- Rawlings, D.E.; Johnson, D.B. The microbiology of biomining: Development and optimization of mineral-oxidizing microbial consortia. Microbiology 2007, 153, 315–324. [Google Scholar] [CrossRef]
- Hiraishi, A.; Nagashima, K.V.P.; Matsuura, K.; Shimada, K.; Takaichi, S.; Wakao, N.; Katayama, Y. Phylogeny and photosynthetic features of Thiobacillus acidophilus and related acidophilic bacteria: Its transfer to the genus Acidiphilium as Acidiphilium acidophilum comb. nov. Int. J. Syst. Bacteriol. 1998, 48, 1389–1398. [Google Scholar] [CrossRef]
- Xu, A.-I.; Xia, J.-I.; Song, Z.-W.; Jiang, P.; Xia, Y.; Wan, M.-X.; Zhang, R.-Y.; Yang, Y.; Liu, K.-K. The effect of energy substrates on PHB accumulation of Acidiphilium cryptum DX1-1. Curr. Microbiol. 2013, 67, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Bridge, T.A.M. Reduction of ferric iron by acidophilic heterotrophic bacteria: Evidence for constitutive and inducible enzyme systems in Acidiphilium spp. J. Appl. Microbiol. 2002, 92, 315–321. [Google Scholar] [CrossRef]
- Mahapatra, N.R.; Benerjee, P.C. Extreme tolerance to cadmium and high resistance to copper, nickel and zinc in different Acidiphilium strains. Lett. Appl. Microbiol. 1996, 23, 393–397. [Google Scholar] [CrossRef]
- Li, L.; Liu, Z.; Zhang, M.; Meng, D.; Liu, X.; Wang, P.; Li, X.; Jiang, Z.; Zhong, S.; Jiang, C.; et al. Insights into the metabolism and evolution of the genus Acidiphilium, a typical acidophile in acid mine drainage. mSystems 2020, 5, e00867-20. [Google Scholar] [CrossRef]
- San Martin-Uriz, P.; Gomez, M.J.; Arcas, A.; Bargiela, R.; Amils, R. Draft genome sequence of the electricigen Acidiphilium sp. strain PM (DSM 24941). J. Bacteriol. 2011, 193, 5585–5586. [Google Scholar] [CrossRef] [PubMed][Green Version]
- San Martin-Uriz, P.; Mirete, S.; Alcolea, P.J.; Gomez, M.J.; Amils, R.; Gonzalez-Pastor, J.E. Nickel-resistance determinants in Acidiphilium sp. PM identified by genome-wide functional screening. PLoS ONE 2014, 9, e95041. [Google Scholar] [CrossRef] [PubMed]
- Ullrich, S.R.; Poehlein, A.; Voget, S.; Hoppert, M.; Daniel, R.; Leimbach, A.; Tischler, J.S.; Schlömann, M.; Mühling, M. Permanent draft genome sequence of Acidiphilium sp. JA12-A1. Stand. Genom. Sci. 2015, 10, 56. [Google Scholar] [CrossRef]
- Li, Q.; Cooper, R.; Wegner, C.-E.; Küsel, K. Molecular mechanisms underpinning aggregation in Acidiphilium sp. C61 isolated from iron-rich pelagic aggregates. Microorganisms 2020, 8, 314. [Google Scholar] [CrossRef] [PubMed]
- Micciche, A.C.; Barabote, R.D.; Dittoe, D.K.; Ricke, S.C. In silico genome analysis of an acid mine drainage species, Acidiphilium multivorum, for potential commercial acetic acid production and biomining. J. Environ. Sci. Health B 2020, 55, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Bulaev, A.; Belyi, A.; Panyushkina, A.; Solopova, N.; Pivovarova, T. Microbial population of industrial biooxidation reactors. Solid State Phenom. 2017, 262, 48–52. [Google Scholar] [CrossRef]
- Kaksonen, A.H.; Boxall, N.J.; Gumulya, Y.; Khaleque, H.N.; Morris, C.; Bohu, T.; Cheng, K.Y.; Usher, K.M.; Lakaniemi, A.-M. Recent progress in biohydrometallurgy and microbial characterisation. Hydrometallurgy 2018, 180, 7–25. [Google Scholar] [CrossRef]
- Tuffin, I.M.; Hector, S.B.; Deane, S.M.; Rawlings, D.E. Resistance determinants of a highly arsenic-resistant strain of Leptospirillum ferriphilum isolated from a commercial biooxidation tank. Appl. Environ. Microbiol. 2006, 72, 2247–2253. [Google Scholar] [CrossRef] [PubMed]
- Tsaplina, I.A.; Panyushkina, A.E.; Melamud, V.S.; Grigor’eva, N.V.; Kondrat’eva, T.F. Leaching of pyrite–arsenopyrite concentrate in bioreactors during continuous cultivation of a thermoacidophilic microbial community. Microbiology 2014, 83, 568–576. [Google Scholar] [CrossRef]
- Panyushkina, A.; Matyushkina, D.; Pobeguts, O. Understanding stress response to high-arsenic gold-bearing sulfide concentrate in extremely metal-resistant acidophile Sulfobacillus thermotolerans. Microorganisms 2020, 8, 1076. [Google Scholar] [CrossRef]
- Belyi, A.V.; Chernov, D.V.; Solopova, N.V. Development of BIONORD® technology on Olimpiada deposit refractory arsenic-gold ores treatment in conditions of Extreme North. Hydrometallurgy 2018, 179, 188–191. [Google Scholar] [CrossRef]
- Panyushkina, A.E.; Tsaplina, I.A.; Kondrat’eva, T.F.; Belyi, A.V.; Bulaev, A.G. Physiological and morphological characteristics of acidophilic bacteria Leptospirillum ferriphilum and Acidithiobacillus thiooxidans, members of a chemolithotrophic microbial consortium. Microbiology 2018, 87, 326–338. [Google Scholar] [CrossRef]
- Panyushkina, A.E.; Babenko, V.V.; Nikitina, A.S.; Selezneva, O.V.; Tsaplina, I.A.; Letarova, M.A.; Kostryukova, E.S.; Letarov, A.V. Sulfobacillus thermotolerans: New insights into resistance and metabolic capacities of acidophilic chemolithotrophs. Sci. Rep. 2019, 9, 15069. [Google Scholar] [CrossRef] [PubMed]
- Bulaev, A.; Belyi, A.; Panyushkina, A.; Solopova, N. Molecular–biological monitoring of microbial population of industrial bioleach bioreactors. In Proceedings of the 23rd International Biohydromatallurgy Symposium (IBS), Fukuoka, Japan, 20–23 October 2019; p. 13. [Google Scholar]
- Silverman, M.P.; Lundgren, D.G. Studies on the chemoautotrophic iron bacteria Ferrobacillus ferrooxidans. An improved medium and harvesting procedure for securing high yields. J. Bacteriol. 1959, 77, 642–647. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Tamura, K.; Nei, M.; Kumar, S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc. Natl. Acad. Sci. USA 2004, 101, 11030–11035. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. ProgressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Laing, C.; Buchanan, C.; Taboada, E.N.; Zhang, Y.; Kropinski, A.; Villegas, A.; Thomas, J.E.; Gannon, V.P.J. Pan-genome sequence analysis using Panseq: An online tool for the rapid analysis of core and accessory genomic regions. BMC Bioinform. 2010, 11, 461. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Holmes, D. Metal resistance in acidophilic microorganisms and its significance for biotechnologies. Appl. Microbiol. Biotechnol. 2014, 98, 8133–8144. [Google Scholar] [CrossRef]
- Suzuki, K.; Wakao, N.; Kimura, T.; Sakka, K.; Ohmiya, K. Expression and regulation of the arsenic resistance operon of Acidiphilium multivorum AIU 301 plasmid pKW301 in Escherichia coli. Appl. Environ. Microbiol. 1998, 64, 411–418. [Google Scholar] [CrossRef]
- Mu, Y.; Zhou, X.; Liu, L.; Zhou, X.-K.; Zeng, X.-C.; Li, W.-J. Pseudaminobacter arsenicus sp. nov., an arsenic-resistant bacterium isolated from arsenic-rich aquifers. Int. J. Syst. Evol. Microbiol. 2019, 69, 791–797. [Google Scholar] [CrossRef]
- Jiang, H.; Liang, Y.; Yin, H.; Xiao, Y.; Guo, X.; Xu, Y.; Hu, Q.; Liu, H.; Liu, X. Effects of arsenite resistance on the growth and functional gene expression of Leptospirillum ferriphilum and Acidithiobacillus thiooxidans in pure culture and coculture. BioMed Res. Int. 2015, 2015, 203197. [Google Scholar] [CrossRef]
- Kishimoto, N.; Kosako, Y.; Wakao, N.; Tano, T.; Hiraishi, A. Transfer of Acidiphilium facilis and Acidiphilium aminolytica to the genus Acidocella gen. nov, and emendation of the genus Acidiphilium. Syst. Appl. Microbiol. 1995, 18, 85–91. [Google Scholar] [CrossRef]
- Dutta, S.J.; Liu, J.; Stemmler, A.J.; Mitra, B. Conservative and nonconservative mutations of the transmembrane CPC motif in ZntA: Effect on metal selectivity and activity. Biochemistry 2007, 46, 3692–3703. [Google Scholar] [CrossRef]
- Wang, R.; Lin, J.-Q.; Liu, X.-M.; Pang, X.; Zhang, C.-J.; Yang, C.-L.; Gao, X.-Y.; Lin, C.-M.; Li, Y.-Q.; Li, Y.; et al. Sulfur oxidation in the acidophilic autotrophic Acidithiobacillus spp. Front. Microbiol. 2019, 9, 3290. [Google Scholar] [CrossRef] [PubMed]
- Sugio, T.; Mizunashi, W.; Inagaki, K.; Tano, T. Purification and some properties of sulfur: Ferric ion oxidoreductase from Thiobacillus ferrooxidans. J. Bacteriol. 1987, 169, 4916–4922. [Google Scholar] [CrossRef] [PubMed]
- Rohwerder, T.; Sand, W. The sulfane sulfur of persulfides is the actual substrate of the sulfur-oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 2003, 149, 1699–1710. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, S.; Liu, X.; Li, X.; Wen, Q.; Lin, J. Identification and characterization of an ETHE1-like sulfur dioxygenase in extremely acidophilic Acidithiobacillus spp. Appl. Microbiol. Biotechnol. 2014, 98, 7511–7522. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Pang, X.; Lin, J.; Liu, X.; Wang, R.; Lin, J.; Chen, L. Discovery of a new subgroup of sulfur dioxygenases and characterization of sulfur dioxygenases in the sulfur metabolic network of Acidithiobacillus caldus. PLoS ONE 2017, 12, e0183668. [Google Scholar] [CrossRef] [PubMed]

| Parameter | LMS 1 | AIU301 2 |
|---|---|---|
| Source of isolation | Industrial bioreactor | AMD 3 |
| Morphology | Rods, single cells, and aggregates | Rods, single cells, and chains |
| Cell size, µm | 0.3–0.5 × 0.8–1.0 | 0.5–1.2 × 0.9–3.8 |
| Motility | + | + |
| pH range | 1.6–5.5 | 1.9–5.6 |
| pHopt 4 | 2.5–4.3 | 3.2–4.0 |
| T5 range, °C | <20–47 | 17–42 |
| Topt, °C | 30–40 | 27–35 |
| µmax6, h−1 | 0.02 (S0 + YE7) | 0.004–0.062 (organic substrates) |
| Growth with: | ||
| YE 7 | + | + |
| Glucose | + | + |
| YE + S0 | + | – |
| S0 | + | – |
| YE + Na2S2O3 | + | – |
| Na2S2O3 | + | – |
| Fe2+ | – | – |
| Reference | (present study) | [2] |
| Attribute | Value |
|---|---|
| Genome size (bp) | 3,837,559 |
| Number of contigs | 139 |
| Number of contigs (≥1000 bp) | 135 |
| Number of contigs (≥5000 bp) | 84 |
| Number of contigs (≥10,000 bp) | 62 |
| Number of contigs (≥25,000 bp) | 43 |
| Number of contigs (≥50,000 bp) | 23 |
| Largest contig | 308,818 |
| DNA, G + C (%) | 67.3 |
| Genes (total) | 3676 |
| CDSs (total) | 3623 |
| Genes (coding) | 3492 |
| CDSs (with protein) | 3492 |
| Genes (RNA) | 53 |
| Complete rRNAs | 1, 1, 1 (5S, 16S, 23S) |
| tRNAs | 46 |
| ncRNAs | 4 |
| Pseudogenes (total) | 131 |
| CRISPR arrays | 1 |
| Functional Category | Number of Proteins 1 |
|---|---|
| Carbohydrate metabolism | 242 |
| Protein families: Genetic information processing | 235 |
| Environmental information processing | 226 |
| Protein families: Signaling and cellular processes | 213 |
| Genetic information processing | 164 |
| Amino acid metabolism | 157 |
| Unclassified: Metabolism | 134 |
| Energy metabolism | 117 |
| Metabolism of cofactors and vitamins | 104 |
| Unclassified | 103 |
| Cellular processes | 100 |
| Nucleotide metabolism | 63 |
| Unclassified: Genetic information processing | 55 |
| Protein families: Metabolism | 53 |
| Lipid metabolism | 46 |
| Glycan biosynthesis and metabolism | 39 |
| Unclassified: Signaling and cellular processes | 39 |
| Xenobiotics biodegradation and metabolism | 36 |
| Metabolism of other amino acids | 28 |
| Metabolism of terpenoids and polyketides | 14 |
| Acidiphilium Strain | ANI Value (SD 1), % |
|---|---|
| Acidiphilium sp. AccII | 99.29 (1.43) |
| Acidiphilium sp. JA12-A1 | 99.23 (0.83) |
| Ac. multivorum AIU301T | 99.12 (1.92) |
| Ac. cryptum JF-5 | 99.18 (1.02) |
| Acidiphilium sp. ZJSH63 | 99.04 (1.52) |
| Acidiphilium sp. AccI | 98.97 (1.84) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Panyushkina, A.; Bulaev, A.; Belyi, A.V. Unraveling the Central Role of Sulfur-Oxidizing Acidiphilium multivorum LMS in Industrial Bioprocessing of Gold-Bearing Sulfide Concentrates. Microorganisms 2021, 9, 984. https://doi.org/10.3390/microorganisms9050984
Panyushkina A, Bulaev A, Belyi AV. Unraveling the Central Role of Sulfur-Oxidizing Acidiphilium multivorum LMS in Industrial Bioprocessing of Gold-Bearing Sulfide Concentrates. Microorganisms. 2021; 9(5):984. https://doi.org/10.3390/microorganisms9050984
Chicago/Turabian StylePanyushkina, Anna, Aleksandr Bulaev, and Aleksandr V. Belyi. 2021. "Unraveling the Central Role of Sulfur-Oxidizing Acidiphilium multivorum LMS in Industrial Bioprocessing of Gold-Bearing Sulfide Concentrates" Microorganisms 9, no. 5: 984. https://doi.org/10.3390/microorganisms9050984
APA StylePanyushkina, A., Bulaev, A., & Belyi, A. V. (2021). Unraveling the Central Role of Sulfur-Oxidizing Acidiphilium multivorum LMS in Industrial Bioprocessing of Gold-Bearing Sulfide Concentrates. Microorganisms, 9(5), 984. https://doi.org/10.3390/microorganisms9050984







