Revealing the Complexity of Sweepovirus-Deltasatellite–Plant Host Interactions: Expanded Natural and Experimental Helper Virus Range and Effect Dependence on Virus-Host Combination
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Samples
2.2. DNA Extraction and Cloning
2.3. Sequence Analysis
2.4. Plant Agroinoculation
2.5. Virus and Deltasatellite Detection and Quantification
3. Results
3.1. Widespread Presence of Sweepoviruses and Associated Deltasatellites Infecting Ipomoea indica in Spain
3.2. Detection of a Sweepovirus-Deltasatellite Chimera
3.3. Transreplication of Sweet Potato Leaf Curl Deltasatellite 1 by Old World and New World Begomoviruses as Well as by a Curtovirus in Nicotiana benthamiana Plants
3.4. Transreplication of Sweet Potato Leaf Curl Deltasatellite 1 in the Natural Plant Hosts of Helper Geminivirids
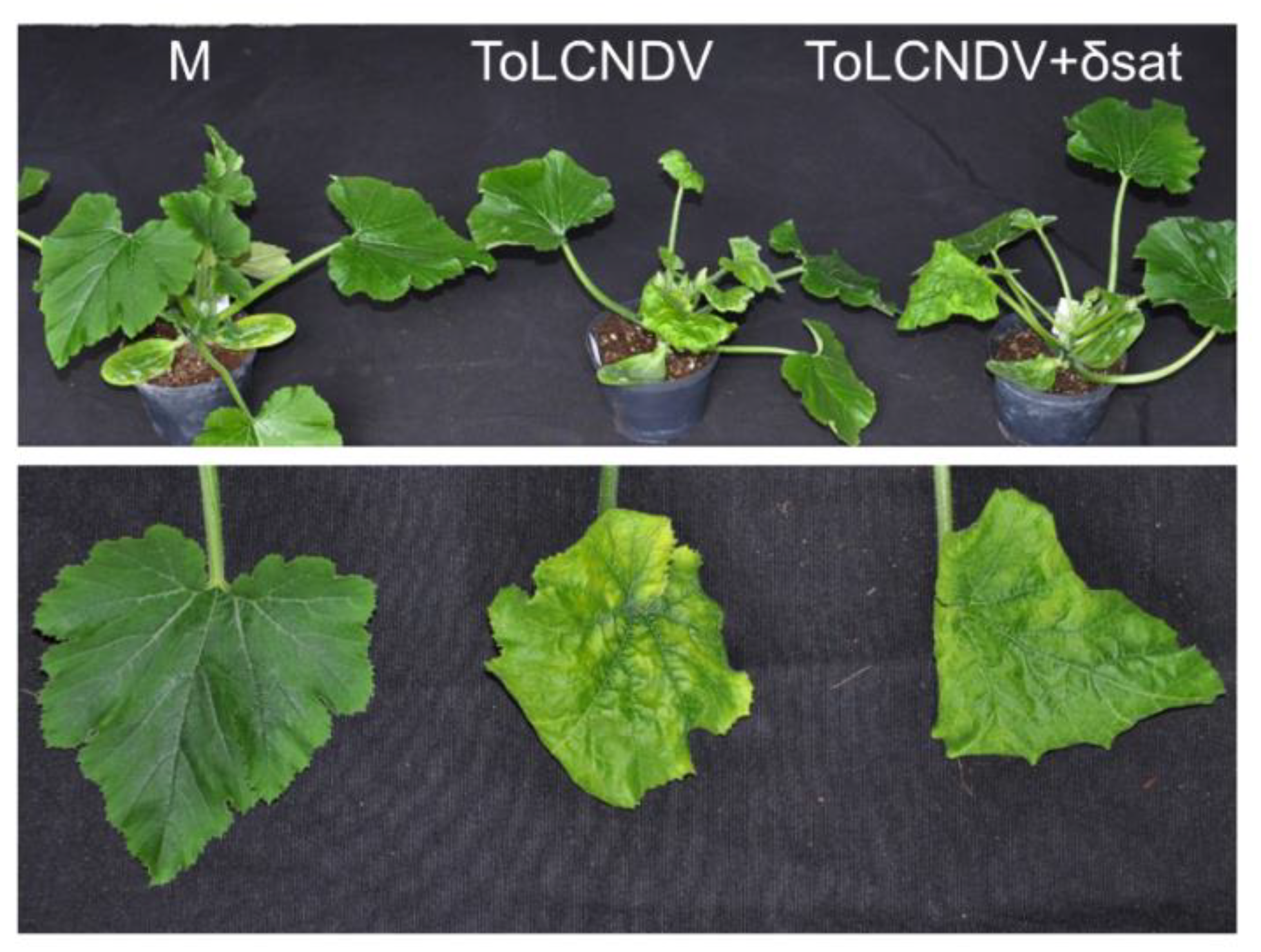
3.5. Effect of Sweet Potato Leaf Curl Deltasatellite 1 on Accumulation of Helper Geminivirids: Dependence on the Virus-Host Combination
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Navas-Castillo, J.; Fiallo-Olivé, E. Geminiviruses (Geminiviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; Volume 3, pp. 411–419. [Google Scholar]
- Navas-Castillo, J.; Fiallo-Olivé, E.; Sánchez-Campos, S. Emerging virus diseases transmitted by whiteflies. Annu. Rev. Phytopathol. 2011, 49, 219–248. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Pan, L.L.; Liu, S.S.; Navas-Castillo, J. Transmission of begomoviruses and other whitefly-borne viruses: Dependence on the vector species. Phytopathology 2020, 110, 10–17. [Google Scholar] [CrossRef]
- Fauquet, C.M.; Stanley, J. Geminivirus classification and nomenclature: Progress and problems. Ann. Appl. Biol. 2005, 142, 165–189. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Valverde, R.A.; Navas-Castillo, J. Novel begomovirus species of recombinant nature in sweet potato (Ipomoea batatas) and Ipomoea indica: Taxonomic and phylogenetic implications. J. Gen. Virol. 2009, 90, 2550–2562. [Google Scholar] [CrossRef]
- Albuquerque, L.C.; Inoue-Nagata, A.K.; Pinheiro, B.; Ribeiro, S.D.; Resende, R.O.; Moriones, E.; Navas-Castillo, J. A novel monopartite begomovirus infecting sweet potato in Brazil. Arch. Virol. 2011, 156, 1291–1294. [Google Scholar] [CrossRef]
- Banks, G.; Bedford, I.; Beitia, F.; Rodriguez-Cerezo, E.; Markham, P. A novel geminivirus of Ipomoea indica (Convolvulaceae) from Southern Spain. Plant Dis. 1999, 83, 486. [Google Scholar] [CrossRef]
- Esterhuizen, L.L.; van Heerden, S.W.; Rey, M.E.C.; van Heerden, H. Genetic identification of two sweet-potato-infecting begomoviruses in South Africa. Arch. Virol. 2012, 157, 2241–2245. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Katis, N.I.; Navas-Castillo, J. First report of Sweet potato leaf curl virus on blue morning glory in Greece. Plant Dis. 2014, 98, 700. [Google Scholar] [CrossRef]
- Lotrakul, P.; Valverde, R.; Clark, C.; Sim, J.; De La Torre, R. Detection of a geminivirus infecting sweet potato in the United States. Plant Dis. 1998, 82, 1253–1257. [Google Scholar] [CrossRef] [PubMed]
- Luan, Y.S.; Zhang, J.; Liu, D.M.; Li, W.L. Molecular characterization of sweet potato leaf curl virus isolate from China (SPLCV-CN) and its phylogenetic relationship with other members of the Geminiviridae. Virus Genes 2007, 35, 379–385. [Google Scholar] [CrossRef]
- Mohammed, H.S.; El Hussein, A.A.; El Siddig, M.A.; Ibrahim, F.A.; Navas-Castillo, J.; Fiallo-Olivé, E. First report of Sweet potato leaf curl virus infecting sweet potato in Sudan. Plant Dis. 2017, 101, 849. [Google Scholar] [CrossRef]
- Paprotka, T.; Boiteux, L.S.; Fonseca, M.E.N.; Resende, R.O.; Jeske, H.; Faria, J.C.; Ribeiro, S.G. Genomic diversity of sweet potato geminiviruses in a Brazilian germplasm bank. Virus Res. 2010, 149, 224–233. [Google Scholar] [CrossRef]
- Prasanth, G.; Hegde, V. Occurrence of Sweet potato feathery mottle virus and Sweet potato leaf curl Georgia virus on sweet potato in India. Plant Dis. 2008, 92, 311. [Google Scholar] [CrossRef]
- Wasswa, P.; Otto, B.; Maruthi, M.; Mukasa, S.; Monger, W.; Gibson, R. First identification of a sweet potato begomovirus (sweepovirus) in Uganda: Characterization, detection and distribution. Plant Pathol. 2011, 60, 1030–1039. [Google Scholar] [CrossRef]
- Rojas, M.R.; Hagen, C.; Lucas, W.J.; Gilbertson, R.L. Exploiting chinks in the plant’s armor: Evolution and emergence of geminiviruses. Annu. Rev. Phytopathol. 2005, 43, 361–394. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Idris, A.M.; Mansoor, S.; Bedford, I.A.; Dhawan, P.; Rishi, N.; Siwatch, S.S.; Abdel-Salam, A.M. Diversity of DNA β, a satellite molecule associated with some monopartite begomoviruses. Virology 2003, 312, 106–121. [Google Scholar] [CrossRef]
- Briddon, R.W.; Bull, S.E.; Amin, I.; Mansoor, S.; Bedford, I.D.; Rishi, N.; Siwatch, S.S.; Zafar, Y.; Abdel-Salam, A.M.; Markham, P.G. Diversity of DNA 1: A satellite-like molecule associated with monopartite begomovirus-DNA β complexes. Virology 2004, 324, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Lozano, G.; Trenado, H.P.; Fiallo-Olivé, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of non-coding DNA satellites associated with sweepoviruses (genus Begomovirus, Geminiviridae)-definition of a distinct class of begomovirus-associated satellites. Front. Microbiol. 2016, 7, 162. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Martínez-Zubiaur, Y.; Moriones, E.; Navas-Castillo, J. A novel class of DNA satellites associated with New World begomoviruses. Virology 2012, 426, 1–6. [Google Scholar] [CrossRef]
- Adams, M.J.; Kefkowitz, E.J.; King, A.M.Q.; Harrach, B.; Harrison, R.L.; Knowles, N.J.; Kropinski, A.M.; Krupovic, M.; Kuhn, J.H.; Mushegian, A.R.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2017). Arch. Virol. 2017, 162, 2505–2538. [Google Scholar] [CrossRef]
- Briddon, R.W.; Navas-Castillo, J.; Fiallo-Olivé, E. ICTV Taxonomic Proposal 2016.021a-kP.A.v2.Tolecusatellitidae. Create the Tolecusatellitidae, a New Family of Single-Stranded DNA Satellites with Two Genera. 2016. Available online: http://www.ictv.global/proposals-16/2016.021a-kP.A.v2.Tolecusatellitidae.pdf (accessed on 30 March 2021).
- Fiallo-Olivé, E.; Navas-Castillo, J. Molecular and biological characterization of a New World mono-/bipartite begomovirus/deltasatellite complex infecting Corchorus siliquosus. Front. Microbiol. 2020, 11, 1755. [Google Scholar] [CrossRef]
- Rosario, K.; Marr, C.; Varsani, A.; Kraberger, S.; Stainton, D.; Moriones, E.; Polston, J.E.; Breitbart, M. Begomovirus-associated satellite DNA diversity captured through Vector-Enabled Metagenomic (VEM) surveys using whiteflies (Aleyrodidae). Viruses 2016, 8, 36. [Google Scholar] [CrossRef] [PubMed]
- Fiallo-Olivé, E.; Lapeira, D.; Louro, D.; Navas-Castillo, J. First report of Sweet potato leaf curl virus and Sweet potato leaf curl deltasatellite 1 infecting blue morning glory in Portugal. Plant Dis. 2018, 102, 1043. [Google Scholar] [CrossRef]
- Hassan, I.; Orílio, A.F.; Fiallo-Olivé, E.; Briddon, R.W.; Navas-Castillo, J. Infectivity, effects on helper viruses and whitefly transmission of the deltasatellites associated with sweepoviruses (genus Begomovirus, family Geminiviridae). Sci. Rep. 2016, 6, 30204. [Google Scholar] [CrossRef] [PubMed]
- Haible, D.; Kober, S.; Jeske, H. Rolling circle amplification revolutionizes diagnosis and genomics of geminiviruses. J. Virol. Methods 2006, 135, 9–16. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Muhire, B.M.; Varsani, A.; Martin, D.P. SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 2014, 9, e108277. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Martin, D.P.; Murrell, B.; Golden, M.; Khoosal, A.; Muhire, B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015, 1, vev003. [Google Scholar] [CrossRef]
- Fortes, I.M.; Sánchez-Campos, S.; Fiallo-Olivé, E.; Díaz-Pendón, J.A.; Navas-Castillo, J.; Moriones, E. A novel strain of tomato leaf curl New Delhi virus has spread to the Mediterranean basin. Viruses 2016, 8, 307. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Tovar, R.; Navas-Castillo, J. Deciphering the biology of deltasatellites from the New World: Maintenance by New World begomoviruses and whitefly transmission. New Phytol. 2016, 212, 680–692. [Google Scholar] [CrossRef]
- Sánchez-Campos, S.; Martínez-Ayala, A.; Márquez-Martín, B.; Aragón-Caballero, L.; Navas-Castillo, J.; Moriones, E. Fulfilling Koch’s postulates confirms the monopartite nature of tomato leaf deformation virus: A begomovirus native to the New World. Virus Res. 2013, 173, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Briddon, R.W.; Watts, J.; Markham, P.G.; Stanley, J. The coat protein of beet curly top virus is essential for infectivity. Virology 1989, 172, 628–633. [Google Scholar] [CrossRef]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Shi, L.; Han, C.; Yu, J.; Li, D.; Zhang, Y. Validation of reference genes for gene expression studies in virus-infected Nicotiana benthamiana using quantitative real-time PCR. PLoS ONE 2012, 7, e46451. [Google Scholar] [CrossRef]
- Obrero, A.; Die, J.V.; Román, B.; Gómez, P.; Nadal, S.; González-Verdejo, C.I. Selection of reference genes for gene expression studies in zucchini (Cucurbita pepo) using qPCR. J. Agric. Food Chem. 2011, 59, 5402–5411. [Google Scholar] [CrossRef]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.; Fiallo-Olivé, E.; Briddon, R.W.; Hernandez-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef]
- Dry, I.B.; Krake, L.R.; Rigden, J.E.; Rezaian, M.A. A novel subviral agent associated with a geminivirus: The first report of a DNA satellite. Proc. Natl. Acad. Sci. USA 1997, 94, 7088–7093. [Google Scholar] [CrossRef]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Martin, D.P.; Hoareau, M.; Naze, F.; Delatte, H.; Thierry, M.; Varsani, A.; Becker, N.; Reynaud, B.; Lett, J.M. Begomovirus ‘melting pot’ in the south-west Indian Ocean islands: Molecular diversity and evolution through recombination. J. Gen. Virol. 2007, 88, 3458–3468. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Trenado, H.P.; Louro, D.; Navas-Castillo, J. Recurrent speciation of a tomato yellow leaf curl geminivirus in Portugal by recombination. Sci. Rep. 2019, 9, 1332. [Google Scholar] [CrossRef] [PubMed]
- Albuquerque, L.C.; Inoue-Nagata, A.K.; Pinheiro, B.; Resende, R.O.; Moriones, E.; Navas-Castillo, J. Genetic diversity and recombination analysis of sweepoviruses from Brazil. Virol. J. 2012, 9, 241. [Google Scholar] [CrossRef]
- Lefeuvre, P.; Lett, J.M.; Varsani, A.; Martin, D.P. Widely conserved recombination patterns among single-stranded DNA viruses. J. Virol. 2009, 83, 2697–2707. [Google Scholar] [CrossRef]
- Monjane, A.L.; van der Walt, E.; Varsani, A.; Rybicki, E.P.; Martin, D.P. Recombination hotspots and host susceptibility modulate the adaptive value of recombination during maize streak virus evolution. BMC Evol. Biol. 2011, 11, 350. [Google Scholar] [CrossRef]
- Valverde, R.A.; Clark, C.A.; Valkonen, J.P. Viruses and virus disease complexes of sweetpotato. Plant Viruses 2007, 1, 116–126. [Google Scholar]
- Berrie, L.C.; Rybicki, E.P.; Rey, M.E.C. Complete nucleotide sequence and host range of South African cassava mosaic virus: Further evidence for recombination amongst begomoviruses. J. Gen. Virol. 2001, 82, 53–58. [Google Scholar] [CrossRef]
- Martin, D.P.; Lefeuvre, P.; Varsani, A.; Hoareau, M.; Semegni, J.Y.; Dijoux, B.; Vincent, C.; Reynaud, B.; Lett, J.M. Complex recombination patterns arising during geminivirus coinfections preserve and demarcate biologically important intra-genome interaction networks. PLoS Pathog. 2011, 7, e1002203. [Google Scholar] [CrossRef]
- Zaidi, S.S.E.A.; Martin, D.P.; Amin, I.; Farooq, M.; Mansoor, S. Tomato leaf curl New Delhi virus: A widespread bipartite begomovirus in the territory of monopartite begomoviruses. Mol. Plant. Pathol. 2017, 18, 901–911. [Google Scholar] [CrossRef]
- Juárez, M.; Tovar, R.; Fiallo-Olivé, E.; Aranda, M.A.; Gosálvez, B.; Castillo, P.; Moriones, E.; Navas-Castillo, J. First detection of Tomato leaf curl New Delhi virus infecting zucchini squash in Spain. Plant Dis. 2014, 98. [Google Scholar] [CrossRef]
- Mar, T.B.; Mendes, I.R.; Lau, D.; Fiallo-Olivé, E.; Navas-Castillo, J.; Alves, M.S.; Zerbini, F.M. Interaction between the New World begomovirus Euphorbia yellow mosaic virus and its associated alphasatellite: Effects on infection and transmission by the whitefly Bemisia tabaci. J. Gen. Virol. 2017, 98, 1552–1562. [Google Scholar] [CrossRef] [PubMed]
- Saunders, K.; Briddon, R.W.; Stanley, J. Replication promiscuity of DNA-β satellites associated with monopartite begomoviruses; deletion mutagenesis of the Ageratum yellow vein virus DNA-β satellite localizes sequences involved in replication. J. Gen. Virol. 2008, 89, 3165–3172. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, Z.; Shafiq, M.; Ali, I.; Mansoor, S.; Briddon, R.W. Maintenance of cotton leaf curl Multan betasatellite by Tomato leaf curl New Delhi virus–analysis by mutation. Front. Plant. Sci. 2017, 8, 2208. [Google Scholar] [CrossRef] [PubMed]
- Nawaz-ul-Rehman, M.S.; Mansoor, S.; Briddon, R.W.; Fauquet, C.M. Maintenance of an Old World betasatellite by a New World helper begomovirus and possible rapid adaptation of the betasatellite. J. Virol. 2009, 83, 9347–9355. [Google Scholar] [CrossRef]
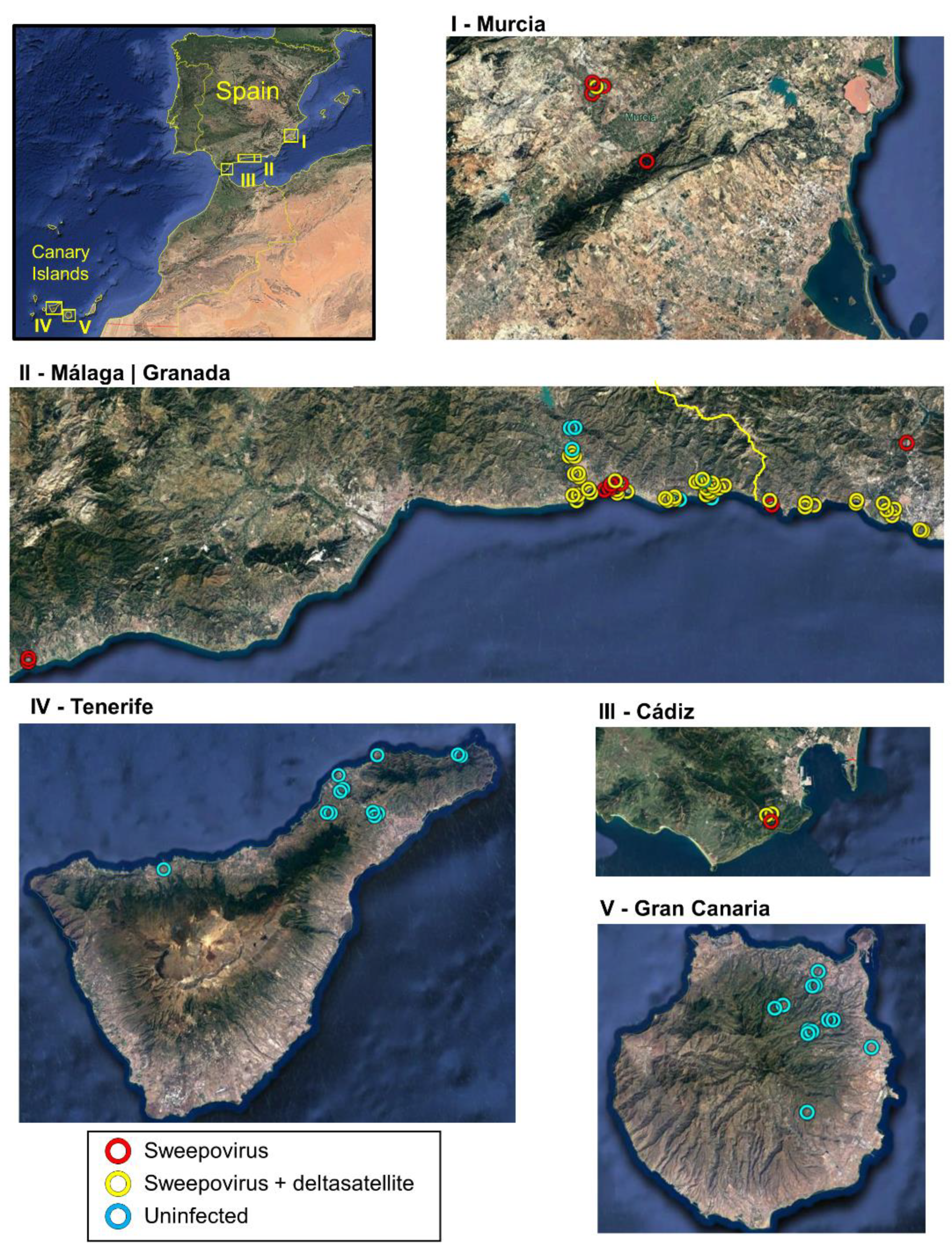





| Province/ Island | Number of Samples | Number of Symptomatic Samples | Number of Infected Samples | ||
|---|---|---|---|---|---|
| Yellow Veins (%) | Leaf Curling (%) | Sweepoviruses (%) | Deltasatellites (%) | ||
| Murcia | 5 | 1 (20.0) | 1 (20.0) | 5 (100.0) | 1 (20.0) |
| Granada | 5 | 0 (0.0) | 0 (0.0) | 5 (100.0) | 5 (100.0) |
| Málaga | 52 | 5 (9.6) | 5 (9.6) | 46 (88.5) | 38 (73.1) |
| Cádiz | 3 | 0 (0.0) | 0 (0.0) | 3 (100.0) | 2 (66.7) |
| Tenerife | 12 | 3 (25.0) | 3 (25.0) | 0 (0.0) | 0 (0.0) |
| Gran Canaria | 12 | 8 (66.7) | 7 (58.3) | 0 (0.0) | 0 (0.0) |
| Continental Spain 1 | 65 | 6 (9.2) | 6 (9.2) | 59 (90.8) | 46 (70.8) |
| Canary Islands 2 | 24 | 11 (45.8) | 10 (41.7) | 0 (0.0) | 0 (0.0) |
| Total | 89 | 17 (19.1) | 16 (18.0) | 59 (66.3) | 46 (51.7) |
| Virus (+Deltasatellite) | Number of Infected Plants/Number of Agroinoculated Plants | Total (%) | ||||
|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | |||||
| Virus | SPLCD1 | Virus | SPLCD1 | Virus | SPLCD1 | |
| ToLCNDV | 14/15 | 0/15 | 15/15 | 0/15 | 96.7 | 0.0 |
| ToLCNDV + SPLCD1 | 15/15 | 15/15 | 15/15 | 15/15 | 100.0 | 100.0 |
| SiGYVV | 15/15 | 0/15 | 15/15 | 0/15 | 100.0 | 0.0 |
| SiGYVV + SPLCD1 | 15/15 | 15/15 | 15/15 | 15/15 | 100.0 | 100.0 |
| ToLDeV | 15/15 | 0/15 | 15/15 | 0/15 | 100.0 | 0.0 |
| ToLDeV + SPLCD1 | 15/15 | 15/15 | 15/15 | 15/15 | 100.0 | 100.0 |
| BCTV | 15/15 | 0/15 | 15/15 | 0/15 | 100.0 | 0.0 |
| BCTV + SPLCD1 | 15/15 | 7/15 | 15/15 | 8/15 | 100.0 | 50.0 |
| SPLCV (positive control) | 12/12 | 0/12 | 12/12 | 0/12 | 100.0 | 0.0 |
| SPLCV + SPLCD1 (positive control) | 12/12 | 12/12 | 12/15 | 12/12 | 100.0 | 100.0 |
| Mock (negative control) | 0/15 | 0/15 | 0/15 | 0/15 | 0.0 | 0.0 |
| Plant Host | Virus (+Deltasatellite) | Number of Infected Plants/Number of Agroinoculated Plants | Total (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Experiment 1 | Experiment 2 | Experiment 3 | |||||||
| Virus | SPLCD1 | Virus | SPLCD1 | Virus | SPLCD1 | Virus | SPLCD1 | ||
| Zucchini | ToLCNDV | 15/15 | 0/15 | 15/15 | 0/15 | – | – | 100.0 | 0.0 |
| ToLCNDV + SPLCD1 | 15/15 | 5/15 | 14/15 | 9/15 | – | – | 96.6 | 46.6 | |
| Mock (negative control) | 0/15 | 0/15 | 0/15 | 0/15 | – | – | 0.0 | 0.0 | |
| M. c. | SiGYVV | 17/32 | 0/32 | 16/32 | 0/32 | – | – | 51.5 | 0.0 |
| SiGYVV + SPLCD1 | 16/32 | 0/32 | 15/32 | 0/32 | – | – | 48.4 | 0.0 | |
| Mock (negative control) | 0/15 | 0/15 | 0/15 | 0/15 | – | – | 0.0 | 0.0 | |
| Tomato | ToLDeV | 15/15 | 0/15 | 60/60 | 0/60 | 48/48 | 0/48 | 100.0 | 0.0 |
| ToLDeV + SPLCD1 | 14/15 | 0/15 | 55/60 | 0/60 | 103/108 | 0/108 | 93.9 | 0.0 | |
| BCTV | 15/15 | 0/15 | 66/66 | 0/66 | – | – | 100.0 | 0.0 | |
| BCTV + SPLCD1 | 15/15 | 0/15 | 57/66 | 0/66 | – | – | 88.8 | 0.0 | |
| Mock (negative control) | 0/15 | 0/15 | 0/15 | 0/15 | 0/15 | 0/15 | 0.0 | 0.0 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferro, C.G.; Zerbini, F.M.; Navas-Castillo, J.; Fiallo-Olivé, E. Revealing the Complexity of Sweepovirus-Deltasatellite–Plant Host Interactions: Expanded Natural and Experimental Helper Virus Range and Effect Dependence on Virus-Host Combination. Microorganisms 2021, 9, 1018. https://doi.org/10.3390/microorganisms9051018
Ferro CG, Zerbini FM, Navas-Castillo J, Fiallo-Olivé E. Revealing the Complexity of Sweepovirus-Deltasatellite–Plant Host Interactions: Expanded Natural and Experimental Helper Virus Range and Effect Dependence on Virus-Host Combination. Microorganisms. 2021; 9(5):1018. https://doi.org/10.3390/microorganisms9051018
Chicago/Turabian StyleFerro, Camila G., F. Murilo Zerbini, Jesús Navas-Castillo, and Elvira Fiallo-Olivé. 2021. "Revealing the Complexity of Sweepovirus-Deltasatellite–Plant Host Interactions: Expanded Natural and Experimental Helper Virus Range and Effect Dependence on Virus-Host Combination" Microorganisms 9, no. 5: 1018. https://doi.org/10.3390/microorganisms9051018
APA StyleFerro, C. G., Zerbini, F. M., Navas-Castillo, J., & Fiallo-Olivé, E. (2021). Revealing the Complexity of Sweepovirus-Deltasatellite–Plant Host Interactions: Expanded Natural and Experimental Helper Virus Range and Effect Dependence on Virus-Host Combination. Microorganisms, 9(5), 1018. https://doi.org/10.3390/microorganisms9051018








