A Tentative Study of the Effects of Heat-Inactivation of the Probiotic Strain Shewanella putrefaciens Pdp11 on Senegalese Sole (Solea senegalensis) Intestinal Microbiota and Immune Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Approval
2.2. Probiotic Microorganism
2.3. Experimental Diets
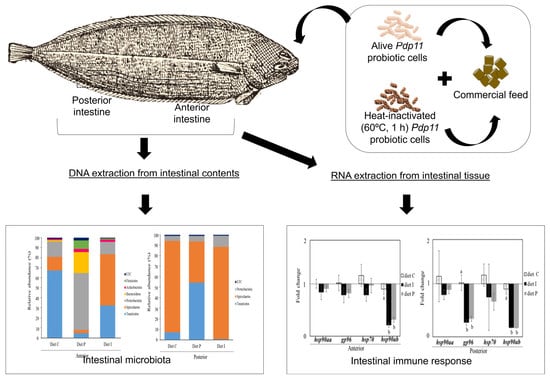
2.4. Experimental Design and Sample Collection
2.5. RNA Isolation and Gene Expression Analysis
2.6. DNA Extraction and Microbiota Characterization
2.7. Growth Performance Calculations
2.8. Statistical Analysis
3. Results
3.1. Growth Performance
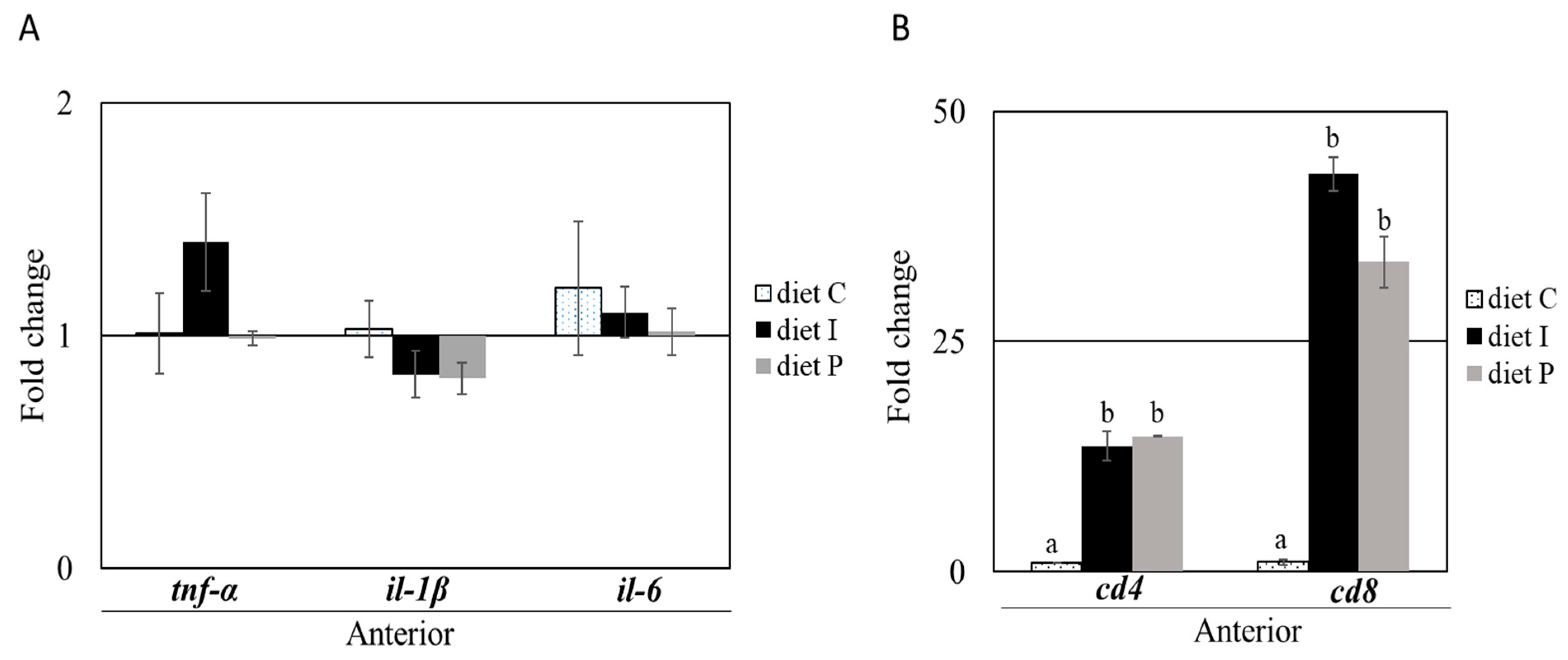
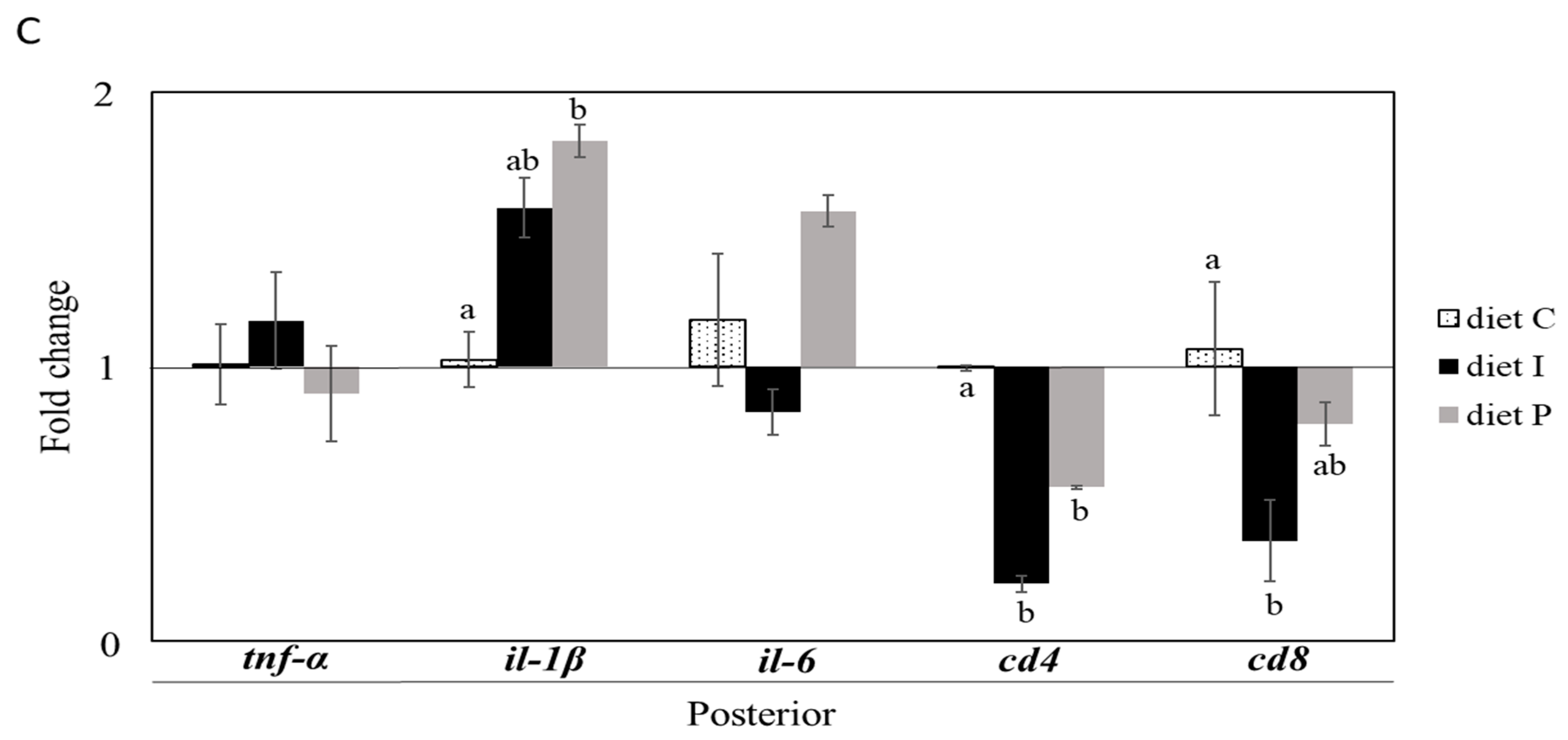
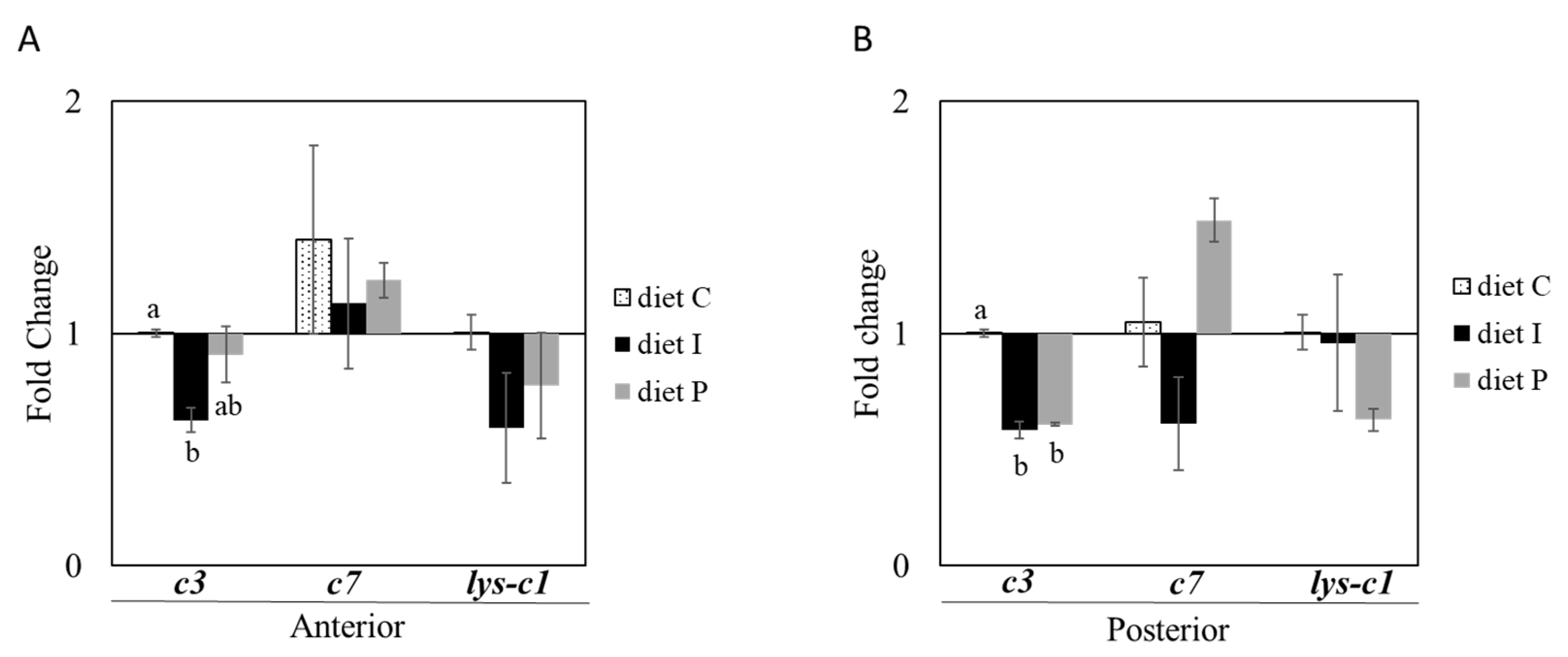
3.2. Gene Expression Analysis
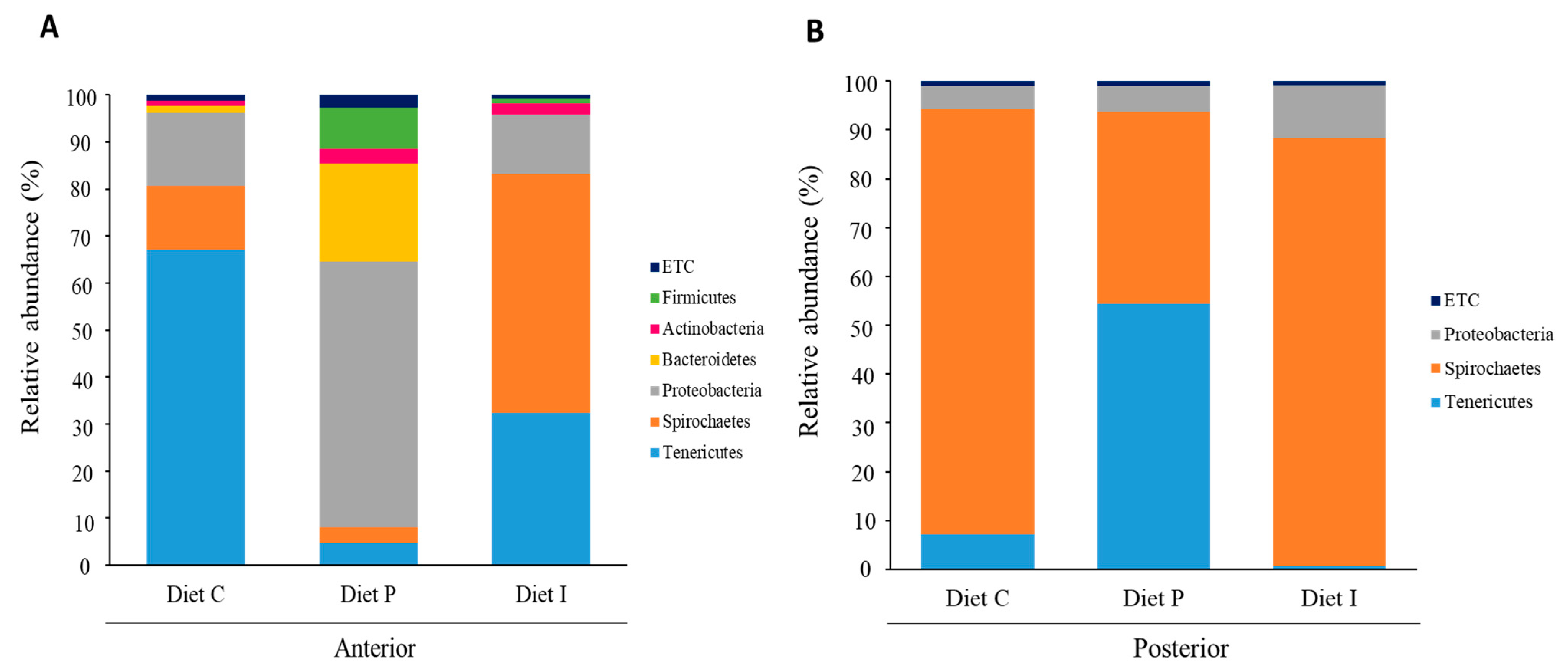
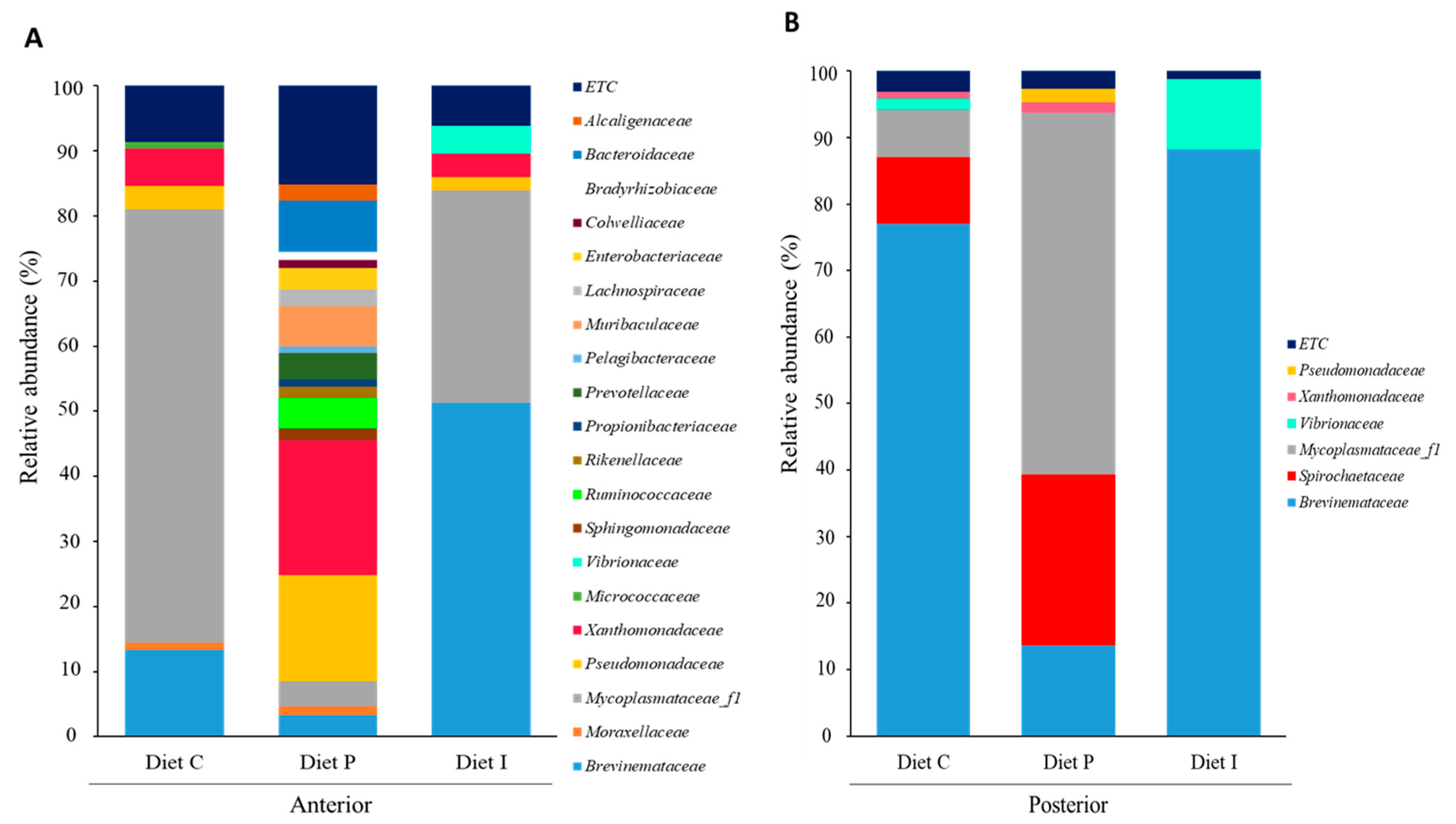
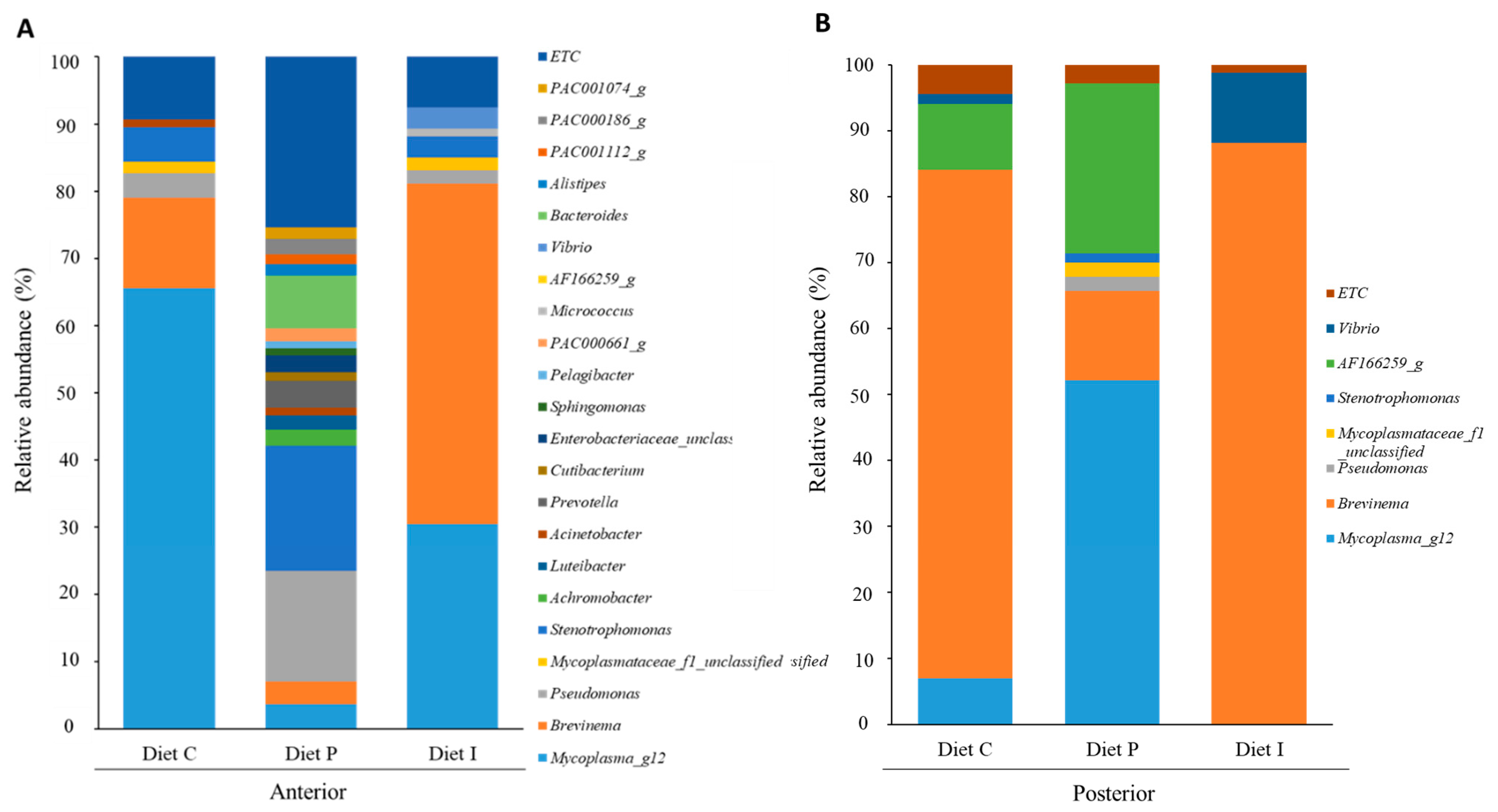
3.3. Analysis of the Intestinal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). World Aquaculture 2015: A Brief Overview; FAO: Rome, Italy, 2017. [Google Scholar]
- Wedemeyer, G. Physiology of Fish in Intensive Culture Systems; Springer Science & Business Media (SSBM): Berlin, Germany, 1996. [Google Scholar]
- Bowden, T.J. Modulation of the immune system of fish by their environment. Fish Shellfish Immunol. 2008, 25, 373–383. [Google Scholar] [CrossRef]
- Plumb, J.A. Health Maintenance of Cultured Fishes: Principal Microbial Diseases; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Ringø, E.; Hoseinifar, S.H.; Ghosh, K.; Doan, H.V.; Beck, B.R.; Song, S.K. Lactic acid bacteria in finfish—An update. Front. Microbiol. 2018, 9, 1818. [Google Scholar] [CrossRef] [PubMed]
- Lazado, C.C.; Caipang, C.M.A. Mucosal immunity and probiotics in fish. Fish Shellfish Immunol. 2014, 39, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Salinas, I.; Zhang, Y.A.; Sunyer, J.O. Mucosal immunoglobulins and B cells of teleost fish. Dev. Comp. Immunol. 2011, 35, 1346–1365. [Google Scholar] [CrossRef]
- Rombout, J.H.; Abelli, L.; Picchietti, S.; Scapigliati, G.; Kiron, V. Teleost intestinal immunology. Fish Shellfish Immunol. 2011, 31, 616–626. [Google Scholar] [CrossRef] [PubMed]
- Llewellyn, M.; Boutin, S.; Hoseinifar, S.H.; Derome, N. Teleost microbiomes: Progress towards their characterization, manipulation and applications in aquaculture and fisheries. Front. Microbiol. 2014, 5, 207. [Google Scholar] [CrossRef]
- Standen, B.T.; Rodiles, A.; Peggs, D.L.; Davies, S.J.; Santos, G.A.; Merrifield, D.L. Modulation of the intestinal microbiota and morphology of tilapia, Oreochromis niloticus, following the application of a multi-species probiotic. Appl. Microbiol. Biotechnol. 2015, 99, 8403–8417. [Google Scholar] [CrossRef]
- Caipang, C.M.A.; Lazado, C.C. Nutritional impacts on fish mucosa: Immunostimulants, pre- and probiotics. Mucosal Health Aquac. 2015, 211–272. [Google Scholar] [CrossRef]
- Sayes, C.; Leyton, Y.; Riquelme, C. Probiotic Bacteria as an Healthy Alternative for Fish Aquaculture. Antibiotics Use in Animals; Savic, S., Ed.; InTech Publishers: Rijeka, Croatia, 2018; pp. 115–132. [Google Scholar] [CrossRef]
- Merrifield, D.L.; Dimitroglou, A.; Foey, A.; Davies, S.J.; Baker, R.T.M.; Bøgwald, J.; Castex, M.; Ringø, E. The current status and future focus of probiotic and prebiotic applications for salmonids. Aquaculture 2010, 302, 1–18. [Google Scholar] [CrossRef]
- Nayak, S.K. Probiotics and immunity: A fish perspective. Fish Shellfish Immunol. 2010, 29, 2–14. [Google Scholar] [CrossRef]
- Pérez-Sánchez, T.; Balcázar, J.L.; Merrifield, D.L.; Carnevali, O.; Gioacchini, G.; de Blas, I.; Ruiz-Zarzuela, I. Expression of immune-related genes in rainbow trout (Oncorhynchus mykiss) induced by probiotic bacteria during Lactococcus garvieae infection. Fish Shellfish Immunol. 2011, 31, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Muñoz-Atienza, E.; Araújo, C.; Lluch, N.; Hernández, P.E.; Herranz, C.; Cintas, L.M.; Magadán, S. Different impact of heat-inactivated and viable lactic acid bacteria of aquatic origin on turbot (Scophthalmus maximus L.) head-kidney leucocytes. Fish Shellfish Immunol. 2015, 44, 214–223. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Rosales, P.; Arijo, S.; Chabrillón, M.; Alarcón, F.J.; Tapia-Paniagua, S.T.; Martínez-Manzanares, E.; Balebona, M.C.; Moriñigo, M.A. Effects of two closely related probiotics on respiratory burst activity of Senegalese sole (Solea senegalensis, Kaup) phagocytes, and protection against Photobacterium damselae subsp. Piscicida. Aquaculture 2009, 293, 16–21. [Google Scholar] [CrossRef]
- Sáenz de Rodrigáñez, M.A.; Díaz-Rosales, P.; Chabrillón, M.; Smidt, H.; Arijo, S.; León-Rubio, J.M.; Alarcón, F.J.; Balebona, M.C.; Moriñigo, M.A.; Cara, J.B.; et al. Effect of dietary administration of probiotics on growth and intestine functionality of juvenile Senegalese sole (Solea senegalensis, Kaup 1858). Aquac. Nutr. 2009, 15, 177–185. [Google Scholar] [CrossRef]
- De La Banda, I.G.; Lobo, C.; León-Rubio, J.M.; Tapia-Paniagua, S.T.; Balebona, M.C.; Moriñigo, M.A.; Moreno-Ventas, X.; Lucas, M.L.; Linares, F.; Arce, F.; et al. Influence of two closely related probiotics on juvenile Senegalese sole (Solea senegalensis, Kaup 1858) performance and protection against Photobacterium damselae subsp. piscicida. Aquaculture 2010, 306, 281–288. [Google Scholar] [CrossRef]
- Lobo, C.; Tapia-Paniagua, S.T.; Moreno-Ventas, X.; Alarcón, F.J.; Rodríguez, C.; Balebona, M.C.; Moriñigo, M.A.; García de la Banda, I. Benefits of probiotic administration on growth and performance along metamorphosis and weaning of Senegalese sole (Solea senegalensis). Aquaculture 2014, 433, 183–195. [Google Scholar] [CrossRef]
- Cordero, H.; Guardiola, F.A.; Tapia-Paniagua, S.T.; Cuesta, A.; Meseguer, J.; Balebona, M.C.; Moriñigo, M.A.; Esteban, M.A. Modulation of immunity and gut microbiota after dietary administration of alginate encapsulated Shewanella putrefaciens Pdp11 to gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol. 2015, 45, 608–618. [Google Scholar] [CrossRef]
- Chabrillón, M.; Rico, R.M.; Balebona, M.C.; Moriñigo, M.A. Adhesion to sole, Solea senegalensis Kaup, mucus of microorganisms isolated from farmed fish, and their interaction with Photobacterium damselae subsp. piscicida. J. Fish Dis. 2005, 28, 229–237. [Google Scholar] [CrossRef]
- Tapia-Paniagua, S.T.; Vidal, S.; Lobo, C.; Prieto-Alamo, M.J.; Jurado, J.; Cordero, H.; Cerezuela, R.; de la Banda, I.G.; Esteban, M.A.; Balebona, M.C.; et al. The treatment with the probiotic Shewanella putrefaciens Pdp11 of specimens of Solea senegalensis exposed to high stocking densities to enhance their resistance to disease. Fish Shellfish Immunol. 2014, 41, 209–221. [Google Scholar] [CrossRef]
- Tapia-Paniagua, S.T.; Chabrillón, M.; Díaz-Rosales, P.; de la Banda, I.G.; Lobo, C.; Balebona, M.C.; Morinigo, M.A. Intestinal microbiota diversity of the flat fish Solea senegalensis (Kaup, 1858) following probiotic administration. Microb. Ecol. 2010, 60, 310–319. [Google Scholar] [CrossRef]
- Tapia-Paniagua, S.T.; Lobo, C.; Moreno-Ventas, X.; de la Banda, I.G.; Moriñigo, M.A.; Balebona, M.C. Probiotic supplementation influences the diversity of the intestinal microbiota during early stages of farmed Senegalese sole (Solea senegalensis, Kaup 1858). Mar. Biotechnol. 2014, 16, 716–728. [Google Scholar] [CrossRef]
- Tapia-Paniagua, S.T.; Díaz-Rosales, P.; García de la Banda, I.; Lobo, C.; Clavijo, E.; Balebona, M.C.; Moriñigo, M.A. Modulation of certain liver fatty acids in Solea senegalensis is influenced by the dietary administration of probiotic microorganisms. Aquaculture 2014, 424, 234–238. [Google Scholar] [CrossRef]
- Tapia-Paniagua, S.T.; Vidal, S.; Lobo, C.; de la Banda, I.G.; Esteban, M.A.; Balebona, M.C.; Moriñigo, M.A. Dietary administration of the probiotic SpPdp11: Effects on the intestinal microbiota and immune-related gene expression of farmed Solea senegalensis treated with oxytetracycline. Fish Shellfish Immunol. 2015, 46, 449–458. [Google Scholar] [CrossRef]
- Choudhury, T.G.; Kamilya, D. Paraprobiotics: An aquaculture perspective. Rev. Aquac. 2019, 11, 1258–1270. [Google Scholar] [CrossRef]
- Panigrahi, A.; Kiron, V.; Satoh, S.; Watanabe, T. Probiotic bacteria Lactobacillus rhamnosus influences the blood profile in rainbow trout, Oncorhynchus mykiss (Walbaum). Fish Physiol. Biochem. 2010, 36, 969–977. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.O.; Koshio, S.; Ishikawa, M.; Yokoyama, S. Immune responses and stress resistance in red sea bream, Pagrus major, after oral administration of heat-killed Lactobacillus plantarum and vitamin C. Fish Shellfish Immunol. 2016, 54, 266–275. [Google Scholar] [CrossRef]
- Sawada, D.; Sugawara, T.; Ishida, Y.; Aihara, K.; Aoki, Y.; Takehara, I.; Takano, K.; Fujiwara, S. Effect of continuous ingestion of a beverage prepared with Lactobacillus gasseri CP2305 inactivated by heat treatment on the regulation of intestinal function. Food Res. Int. 2016, 79, 33–39. [Google Scholar] [CrossRef]
- Tavernity, V.; Guglielmetti, S. The immunomodulatory properties of probiotic microorganisms beyond their viability (ghost probiotics: Proposal of paraprobiotic concept). Genes Nutr. 2011, 6, 261–274. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Rosales, P.; Salinas, I.; Rodríguez, A.; Cuesta, A.; Chabrillón, M.; Balebona, M.C.; Moriñigo, M.A.; Esteban, M.A.; Meseguer, J. Gilthead seabream (Sparus aurata L.) innate immune response after dietary administration of heat-inactivated potential probiotics. Fish Shellfish Immunol. 2006, 20, 482–492. [Google Scholar] [CrossRef] [PubMed]
- Salinas, I.; Díaz-Rosales, P.; Cuesta, A.; Meseguer, J.; Chabrillón, M.; Moriñigo, M.A.; Esteban, M.A. Effect of heat-inactivated fish and non-fish derived probiotics on the innate immune parameters of a teleost fish (Sparus aurata L.). Vet. Immunol. Immunopathol 2006, 111, 279–286. [Google Scholar] [CrossRef] [PubMed]
- De Almada, C.N.; Almada, C.N.; Martinez, R.C.; Sant’Ana, A.S. Paraprobiotics: Evidences on their ability to modify biological responses, inactivation methods and perspectives on their application in foods. Trends Food Sci. Technol. 2016, 58, 96–114. [Google Scholar] [CrossRef]
- Andresen, V.; Gschossmann, J.; Layer, P. Heat-inactivated Bifidobacterium bifidum MIMBb75 (SYN-HI-001) in the treatment of irritable bowel syndrome: A multicentre, randomised, double-blind, placebo-controlled clinical trial. Lancet Gastroenterol. Hepatol. 2020, 5, 658–666. [Google Scholar] [CrossRef]
- Chuang, L.; Wu, K.G.; Pai, C.; Hsieh, P.S.; Tsai, J.J.; Yen, J.H.; Lin, M.Y. Heat-killed cells of lactobacilli skew the immune response toward T helper 1 polarization in mouse splenocytes and dendritic cell-treated T cells. J. Agric. Food Chem. 2007, 55, 11080–11086. [Google Scholar] [CrossRef]
- Ishikawa, H.; Kutsukake, E.; Fukui, T.; Sato, I.; Shirai, T.; Kurihara, T.; Maeda, Y. Oral administration of heat-killed Lactobacillus plantarum strain b240 protected mice against Salmonella enterica Serovar Typhimurium. Biosci. Biotechnol. Biochem. 2010, 74, 1338–1342. [Google Scholar] [CrossRef]
- Chabrillón, M.; Arijo, S.; Díaz-Rosales, P.; Balebona, M.C.; Moriñigo, M.A. Interference of Listonella anguillarum with potential probiotic microorganisms isolated from farmed gilthead seabream (Sparus aurata, L.). Aquac. Res. 2006, 37, 78–86. [Google Scholar] [CrossRef]
- Infante, C.; Matsuoka, M.P.; Asensio, E.; Cañavate, J.P.; Reith, M.; Manchado, M. Selection of housekeeping genes for gene expression studies in larvae from flatfish using real-time PCR. BMC Mol. Biol. 2008, 9, 28. [Google Scholar] [CrossRef] [PubMed]
- Montero, D.; Benitez-Dorta, V.; Caballero, M.J.; Ponce, M.; Torrecillas, S.; Izquierdo, M.; Zamorano, M.J.; Manchado, M. Dietary vegetable oils: Effects on the expression of immune-related genes in Senegalese sole (Solea senegalensis) intestine. Fish Shellfish Immunol. 2015, 44, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Prieto-Álamo, M.J.; Abril, N.; Osuna-Jiménez, I.; Pueyo, C. Solea senegalensis genes responding to lipopolysaccharide and copper sulphate challenges: Large-scale identification by suppression subtractive hybridization and absolute quantification of transcriptional profiles by real-time RT-PCR. Aquat. Toxicol. 2009, 91, 312–319. [Google Scholar] [CrossRef] [PubMed]
- Osuna-Jiménez, I.; Williams, T.D.; Prieto-Álamo, M.J.; Abril, N.; Chipman, J.K.; Pueyo, C. Immune-and stress-related transcriptomic responses of Solea senegalensis stimulated with lipopolysaccharide and copper sulphate using heterologous cDNA microarrays. Fish Shellfish Immunol. 2009, 26, 699–706. [Google Scholar] [CrossRef] [PubMed]
- Manchado, M.; Salas-Leiton, E.; Infante, C.; Ponce, M.; Asensio, E.; Crespo, A.; Zuasti, E.; Cañavate, J.P. Molecular characterization, gene expression and transcriptional regulation of cytosolic HSP90 genes in the flatfish Senegalese sole (Solea senegalensis Kaup). Gene 2008, 416, 77–84. [Google Scholar] [CrossRef]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C T method. Nat. Protoc. 2008, 3, 1101. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Gao, T.; Zheng, Y.; Wang, W.; Cheng, Y.; Wang, G. Microbial diversity of intestinal contents and mucus in yellow catfish (Pelteobagrus fulvidraco). Aquaculture 2010, 303, 1–7. [Google Scholar] [CrossRef]
- Hao, K.; Wu, Z.Q.; Li, D.L.; Yu, X.B.; Wang, G.X.; Ling, F. Effects of dietary administration of Shewanella xiamenensis A-1, Aeromonas veronii A-7, and Bacillus subtilis, single or combined, on the grass carp (Ctenopharyngodon idella) intestinal microbiota. Probiotics Antimicrob. Proteins 2017, 9, 386–396. [Google Scholar] [CrossRef]
- Tapia-Paniagua, S.T.; Balebona, M.D.C.; Firmino, J.P.; Rodríguez, C.; Polo, J.; Moriñigo, M.A.; Gisbert, E. The effect of spray-dried porcine plasma on gilthead seabream (Sparus aurata) intestinal microbiota. Aquac. Nutr. 2020, 26, 801–811. [Google Scholar] [CrossRef]
- Martínez, G.; Shaw, E.M.; Carrillo, M.; Zanuy, S. Protein salting-out method applied to genomic DNA isolation from fish whole blood. BioTechniques 1998, 24, 238–239. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Van Nguyen, N.; Onoda, S.; Van Khanh, T.; Hai, P.D.; Trung, N.T.; Hoang, L.; Koshio, S. Evaluation of dietary Heat-killed Lactobacillus plantarum strain L-137 supplementation on growth performance, immunity and stress resistance of Nile tilapia (Oreochromis niloticus). Aquaculture 2019, 498, 371–379. [Google Scholar] [CrossRef]
- Hasan, M.T.; Jang, W.J.; Lee, B.J.; Kim, K.W.; Hur, S.W.; Lim, S.G.; Bai, S.C.; Kong, I.S. Heat-killed Bacillus sp. SJ-10 probiotic acts as a growth and humoral innate immunity response enhancer in olive flounder (Paralichthys olivaceus). Fish Shellfish Immunol. 2019, 88, 424–431. [Google Scholar] [CrossRef]
- Jurado, J.; Villasanta-González, A.; Tapia-Paniagua, S.T.; Balebona, M.C.; de la Banda, I.G.; Moríñigo, M.Á.; Prieto-Álamo, M.J. Dietary administration of the probiotic Shewanella putrefaciens Pdp11 promotes transcriptional changes of genes involved in growth and immunity in Solea senegalensis larvae. Fish Shellfish Immunol. 2018, 77, 350–363. [Google Scholar] [CrossRef]
- Chen, Z.; Ceballos-Francisco, D.; Guardiola, F.A.; Esteban, M.Á. Influence of skin wounds on the intestinal inflammatory response and barrier function: Protective role of dietary Shewanella putrefaciens SpPdp11 administration to gilthead seabream (Sparus aurata L.). Fish Shellfish Immunol. 2020, 99, 414–423. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.Y.; Chen, S.W.; Hu, S.Y. Improvements in the growth performance, immunity, disease resistance, and gut microbiota by the probiotic Rummeliibacillus stabekisii in Nile tilapia (Oreochromis niloticus). Fish Shellfish Immunol. 2019, 92, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Bano, N.; DeRae Smith, A.; Bennett, W.; Vasquez, L.; Hollibaugh, J.T. Dominance of Mycoplasma in the guts of the Long-Jawed Mudsucker, Gillichthys mirabilis, from five California salt marshes. Environ. Microb. 2007, 9, 2636–2641. [Google Scholar] [CrossRef] [PubMed]
- Yúfera, M.; Darías, M.J. Changes in the gastrointestinal pH from larvae to adult in Senegal sole (Solea senegalensis). Aquaculture 2007, 267, 94–99. [Google Scholar] [CrossRef]
- Cicala, F.; Lago-Lestón, A.; Gomez-Gil, B.; Gollas-Galván, T.; Chong-Robles, J.; Cortés-Jacinto, E.; Martínez-Porchas, M. Gut microbiota shifts in the giant tiger shrimp, Penaeus monodon, during the postlarvae, juvenile, and adult stages. Aquac. Int. 2020, 1–13. [Google Scholar] [CrossRef]
- Fidopiastis, M.; Bezdek, D.J.; Horn, M.H.; Kandel, J.S. Characterizing the resident, fermentative microbial consortium in the hindgut of the temperate-zone herbivorous fish, Hermosilla azurea (Teleostei: Kyphosidae). Mar. Biol. 2006, 148, 631–642. [Google Scholar] [CrossRef]
- Tran, N.T.; Xiong, F.; Hao, Y.T.; Zhang, J.; Wu, S.G.; Wang, G.T. Two biomass preparation methods provide insights into studying microbial communities of intestinal mucosa in grass carp (Ctenopharyngodon idellus). Aquac. Res. 2017, 48, 4272–4283. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Y.; Zhang, X.; Liu, J.; Zhang, Q.; Zhao, Y.; Peng, J.; Feng, Q.; Dai, J.; Sun, S.; et al. Gut Microbial Dysbiosis Is Associated with Altered Hepatic Functions and Serum Metabolites in Chronic Hepatitis B Patients. Front. Microbiol. 2017, 8, 2222. [Google Scholar] [CrossRef]
- Zhang, J.; Xiong, F.; Tang Wang, G.T.; Li, W.X.; Li, M.; Zou, H.; Wu, S.G. The influence of diet on the grass carp intestinal microbiota and bile acids. Aquac. Res. 2017, 48, 4934–4944. [Google Scholar] [CrossRef]
- Watanabe, M.; Houten, S.M.; Wang, L.; Moschetta, A.; Mangelsdorf, D.J.; Heyman, R.A.; Moore, D.D.; Auwerx, J. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J. Clin. Investig. 2004, 113, 1408–1418. [Google Scholar] [CrossRef]
- Givens, C.E.; Ransom, B.; Bano, N.; Hollibaugh, J.T. Comparison of the gut microbiomes of 12 bony fish and 3 shark species. Mar. Ecol. Prog. Ser. 2015, 518, 209–223. [Google Scholar] [CrossRef]
- Corrigan, A.; de Leeuw, M.; Penaud-Frézet, S.; Dimova, D.; Murphy, R.A. Phylogenetic and functional alterations in bacterial community compositions in broiler ceca as a result of mannan oligosaccharide supplementation. Appl. Environ. Microbiol. 2015, 81, 3460–3470. [Google Scholar] [CrossRef] [PubMed]
- Polakof, S.; Panserat, S.; Soengas, J.L.; Moon, T.W. Glucose metabolism in fish: A review. J. Comp. Physiol. B 2012, 182, 1015–1045. [Google Scholar] [CrossRef] [PubMed]
- Conde-Sieira, M.; Salas-Leiton, E.; Duarte, M.; Pelusio, N.; Soengas, J.; Valente, L. Short- and long-term metabolic responses to diets with different protein:carbohydrate ratios in Senegalese sole (Solea senegalensis, Kaup 1858). Br. J. Nut. 2016, 115, 1896–1910. [Google Scholar] [CrossRef] [PubMed]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The gut microbiota of marine fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- Xu, G.; Xing, W.; Li, T.; Xue, M.; Ma, Z.; Jiang, N.; Luo, L. Comparative study on the effects of different feeding habits and diets on intestinal microbiota in Acipenser baeri Brandt and Huso huso. BMC Microbiol. 2019, 19, 297. [Google Scholar] [CrossRef]
- Wang, A.R.; Ran, C.; Ringø, E.; Zhou, Z.G. Progress in fish gastrointestinal microbiota research. Rev. Aquac. 2018, 10, 626–640. [Google Scholar] [CrossRef]
- Butt, R.L.; Volkoff, H. Gut microbiota and energy homeostasis in fish. Front. Endocrinol. 2019, 10, 9. [Google Scholar] [CrossRef]
- Basu, N.; Todgham, A.E.; Ackerman, P.A.; Bibeau, M.R.; Nakano, K.; Schulte, P.M.; Iwama, G.K. Heat shock protein genes and their functional significance in fish. Gene 2002, 295, 173–183. [Google Scholar] [CrossRef]
- Huo, D.; Sun, L.; Zhang, L.; Yang, H.; Liu, S.; Sun, J.; Su, F. Time course analysis of immunity-related gene expression in the sea cucumber Apostichopus japonicus during exposure to thermal and hypoxic stress. Fish Shellfish Immunol. 2019, 95, 383–390. [Google Scholar] [CrossRef]
- Chen, Y.M.; Kuo, C.E.; Wang, T.Y.; Shie, P.S.; Wang, W.C.; Huang, S.L.; Chen, T.Y. Cloning of an orange-spotted grouper Epinephelus coioides heat shock protein 90AB (HSP90AB) and characterization of its expression in response to nodavirus. Fish Shellfish Immunol. 2010, 28, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Schild, H.; Rammensee, H.G. gp96-the immune system’s Swiss army knife. Nat. Immunol. 2000, 1, 100–101. [Google Scholar] [CrossRef]
- Núñez-Díaz, J.A.; Fumanal, M.; Mancera, J.M.; Moriñigo, M.A.; Balebona, M.C. Two routes of infection with Photobacterium damselae subsp. piscicida are effective in the modulation of the transcription of immune related genes in Solea senegalensis. Vet. Immunol. Immunopathol. 2016, 179, 8–17. [Google Scholar] [CrossRef]
- Trusch, F.; Loebach, L.; Wawra, S.; Durward, E.; Wuensch, A.; Iberahim, N.A.; de Bruijn, I.; MacKenzie, K.; Willems, A.; Toloczko, A.; et al. Cell entry of a host-targeting protein of oomycetes requires gp96. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Cara, J.B.; Aluru, N.; Moyano, F.J.; Vijayan, M.M. Food-deprivation induces HSP70 and HSP90 protein expression in larval gilthead sea bream and rainbow trout. Comp. Biochem. Physiol. B. Biochem. Mol. Biol. 2005, 142, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Li, B.; Wu, L.; Yin, X.; Zhong, X.; Li, Y.; Ye, J. Interleukin-6 gets involved in response to bacterial infection and promotes antibody production in Nile tilapia (Oreochromis niloticus). Dev. Comp. Immunol. 2018, 89, 141–151. [Google Scholar] [CrossRef]
- Bird, S.; Zou, J.; Savan, R.; Kono, T.; Sakai, M.; Woo, J.; Secombes, C. Characterisation and expression analysis of an interleukin 6 homologue in the Japanese pufferfish, Fugu rubripes. Dev. Comp. Immunol. 2005, 29, 775–789. [Google Scholar] [CrossRef]
- Panigrahi, A.; Kiron, V.; Satoh, S.; Hirono, I.; Kobayashi, T.; Sugita, H.; Puangkaew, J.; Aoki, T. Immune modulation and expression of cytokine genes in rainbow trout Oncorhynchus mykiss upon probiotic feeding. Dev. Comp. Immunol. 2007, 31, 372–382. [Google Scholar] [CrossRef] [PubMed]
- Caipang, C.M.A.; Hynes, N.; Puangkaew, J.; Brinchmann, M.F.; Kiron, V. Intraperitoneal vaccination of Atlantic cod, Gadus morhua with heat-killed Listonella anguillarum enhances serum antibacterial activity and expression of immune response genes. Fish Shellfish Immunol. 2008, 24, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.Y.; Xia, H.Q.; Yang, H.L.; Hoseinifar, S.H.; Sun, Y.Z. Effects of dietary live or heat-inactivated autochthonous Bacillus pumilus SE 5 on growth performance, immune responses and immune gene expression in grouper Epinephelus coioides. Aquac. Nutr. 2016, 22, 698–707. [Google Scholar] [CrossRef]
- Sun, Z.; Tan, X.; Ye, H.; Zou, C.; Ye, C.; Wang, A. Effects of dietary Panax notoginseng extract on growth performance, fish composition, immune responses, intestinal histology and immune related genes expression of hybrid grouper (Epinephelus lanceolatus × Epinephelus fuscoguttatus) fed high lipid diets. Fish Shellfish Immunol. 2018, 73, 234–244. [Google Scholar] [CrossRef]
- Holland, M.C.H.; Lambris, J.D. The complement system in teleosts. Fish Shellfish Immunol. 2002, 12, 399–420. [Google Scholar] [CrossRef]
- Li, H.; Zhou, Y.; Ling, H.; Luo, L.; Qi, D.; Feng, L. The effect of dietary supplementation with Clostridium butyricum on the growth performance, immunity, intestinal microbiota and disease resistance of tilapia (Oreochromis niloticus). PLoS ONE 2019, 14, e0223428. [Google Scholar] [CrossRef] [PubMed]
- Rebl, A.; Goldammer, T. Under control: The innate immunity of fish from the inhibitors’ perspective. Fish Shellfish Immunol. 2018, 77, 328–349. [Google Scholar] [CrossRef] [PubMed]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef] [PubMed]
- Dawood, M.A.; Koshio, S.; Abdel-Daim, M.M.; Van Doan, H. Probiotic application for sustainable aquaculture. Rev. Aquac. 2019, 11, 907–924. [Google Scholar] [CrossRef]
- Liu, B.; Xu, L.; Ge, X.; Xie, J.; Xu, P.; Shou, Q.; Pan, L.; Zhang, Y. Effects of mannan oligosaccharide on the physiological responses, HSP70 gene expression and disease resistance of Allogynogenetic crucian carp (Carassius auratus gibelio) under Aeromonas hydrophila infection. Fish Shellfish Immunol. 2013, 34, 1395–1403. [Google Scholar] [CrossRef] [PubMed]
- Haase, M.; Fitze, G. HSP90AB1: Helping the good and the bad. Gene 2016, 575, 171–186. [Google Scholar] [CrossRef]
- Sung, Y.Y.; MacRae, T.H. Heat Shock Proteins and disease control in aquatic organisms. J. Aquac. Res. Dev. 2011, 3, 1–10. [Google Scholar] [CrossRef]
- Zeng, F.; Tee, C.; Liu, M.; Sherry, V.; Dixon, B.; Duncker, B.P.; Bols, N.C. The p53/ HSP70 inhibitor, 2-phenylethynesulfonamide, causes oxidative stress, unfolded protein response and apoptosis in rainbow trout cells. Aquat. Toxicol. 2014, 146, 45–51. [Google Scholar] [CrossRef]
- Núñez-Díaz, J.A.; de la Banda, I.G.; Lobo, C.; Moriñigo, M.A.; Balebona, M.C. Transcription of immune related genes in Solea senegalensis vaccinated against Photobacterium damselae subsp. piscicida. Identification of surrogates of protection. Fish Shellfish Immunol. 2017, 66, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Shi, D.; Zhou, X.Q.; Yin, L.; Feng, L.; Jiang, W.D.; Liu, Y.; Tang, L.; Wu, P.; Zhao, Y. Vitamin D inhibits lipopolysaccharide-induced inflammatory response potentially through the Toll-like receptor 4 signaling pathway in the intestine and enterocytes of juvenile jian carp (Cyprinus carpio var. jian). Br. J. Nutr. 2015, 114, 1560–1568. [Google Scholar] [CrossRef]
- Bo, Y.X.; Song, X.H.; Wu, K.; Hy, B.; Sun, B.Y.; Liu, Z.J.; Fu, J.G. Characterization of interleukin-1beta as a proinflammatory cytokine in grass carp (Ctenopharyngodon idella). Fish Shellfish Immunol. 2015, 46, 584–595. [Google Scholar] [CrossRef]
- Burrello, C.; Garavaglia, F.; Cribiù, F.M.; Ercoli, G.; Bosari, S.I.; Caprioli, F.; Facciotti, F. Short-term oral antibiotics treatment promotes inflammatory activation of colonic invariant natural killer T and conventional CD4+ T cells. Front. Med. 2018, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.K.; Nakanishi, T. Direct antibacterial activity of CD8+/CD4+ T-cells in ginbuna crucian carp, Carassius auratus langsdorfii. Fish Shellfish Immunol. 2013, 34, 136–141. [Google Scholar] [CrossRef] [PubMed]

| Gene | Code | Sequence | Reference |
|---|---|---|---|
| Beta actin 2 | actb2 | AATCGTGACCTCTGCTTCCCCCTGT (F) TCTGGCACCCCATGTTACCCCATC (R) | [40] |
| Ribosomal protein S4 | rps4 | GTGAAGAAGCTCCTTGTCGGCACCA (F) AGGGGGTCGGGGTAGCGGATG (R) | [40] |
| Interleukin 1 beta | il-1β | CGCAGAAAGTGACATGTTGAGATTT (F) GGAAGCGGGCAGACATGA (R) | [41] |
| Interleukin 6 | il-6 | ACAATTTCCTGCAGAGATAAAAGTAAGCT (F) CAAGCCCTCAGGCCTACAATATTAA (R) | [41] |
| Complement C3 | c3 | ACCTTAGACTGCCCTACTCTGCTGTCCGTG (F) GCACTGCACACATCATCCGTCTCAGAC (R) | [42] |
| Complement C7 | c7 | GGCACACACTATCTGTCGCAGGGCTC (F) GGCGAACGCCTGATGGTTTAACTCCAG (R) | [42] |
| C1 type lysozyme | lys-c1 | CAGATCAACAGCCGCTATTGG (F) GCTGATTCCACATGCATTTGAAGTG (R) | [41] |
| Cluster of differentiation 4 | cd4 | GACCTCAGGCTGCAATGGT (F) TGAGCAGAGTGATGGACAGACT (R) | [41] |
| Cluster of differentiation 8 alpha | cd8α | GTGCCAGCATTAAAAGCAACGA (F) GCAGTCACAACTTCCGCTCTTT (R) | [41] |
| Tumor necrosis factor alpha | tnfα | CTGGGACTGCTGGCACTTGGATTTG (F) CAGTTCTCCACGCTGACGTACTGTCGAAC (R) | [43] |
| Heat shock protein GP96 | gp96 | GAGTCTTCTCCCTTTGTTGAGCGGCTG (F) TGATGCCTTCCTTTGCCACGTTCTG (R) | [43] |
| Heat shock protein 90AA | hsp90aa | GACCAAGCCTATCTGGACCCGCAAC (F) TTGACAGCCAGGTGGTCCTCCCAGT (R) | [44] |
| Heat shock protein 90AB | hsp90ab | TCAGTTTGGTGTGGGTTTCTACTCGGCTTA (F) GCCAAGGGGCTCACCTGTGTCG (R) | [44] |
| Heat shock protein 70 | hsp70 | GCTATACCAGGGAGGGATGGAAGGAGGG (F) CGACCTCCTCAATATTTGGGCCAGCA (R) | [43] |
| Diet C | Diet P | Diet I | |
|---|---|---|---|
| Initial body weight (g) | 28.52 ± 1.42 a | 30.18 ± 1.37 a | 30.87 ± 1.37 a |
| Final body weight (g) | 34.86 ± 2.44 a | 46.40 ± 2.18 b | 37.10 ± 2.18 a |
| SR (%) | 100.00 ± 0.00 a | 100.00 ± 0.00 a | 100.00 ± 0.00 a |
| WGR (%) | 22.23 ± 3.35 a | 48.81 ± 4.12 b | 20.18 ± 3.01 a |
| SGR (%/d) | 0.45 ± 0.02 a | 0.88 ± 0.05 b | 0.41 ± 0.02 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Domínguez-Maqueda, M.; Cerezo, I.M.; Tapia-Paniagua, S.T.; De La Banda, I.G.; Moreno-Ventas, X.; Moriñigo, M.Á.; Balebona, M.C. A Tentative Study of the Effects of Heat-Inactivation of the Probiotic Strain Shewanella putrefaciens Pdp11 on Senegalese Sole (Solea senegalensis) Intestinal Microbiota and Immune Response. Microorganisms 2021, 9, 808. https://doi.org/10.3390/microorganisms9040808
Domínguez-Maqueda M, Cerezo IM, Tapia-Paniagua ST, De La Banda IG, Moreno-Ventas X, Moriñigo MÁ, Balebona MC. A Tentative Study of the Effects of Heat-Inactivation of the Probiotic Strain Shewanella putrefaciens Pdp11 on Senegalese Sole (Solea senegalensis) Intestinal Microbiota and Immune Response. Microorganisms. 2021; 9(4):808. https://doi.org/10.3390/microorganisms9040808
Chicago/Turabian StyleDomínguez-Maqueda, Marta, Isabel M. Cerezo, Silvana Teresa Tapia-Paniagua, Inés García De La Banda, Xabier Moreno-Ventas, Miguel Ángel Moriñigo, and Maria Carmen Balebona. 2021. "A Tentative Study of the Effects of Heat-Inactivation of the Probiotic Strain Shewanella putrefaciens Pdp11 on Senegalese Sole (Solea senegalensis) Intestinal Microbiota and Immune Response" Microorganisms 9, no. 4: 808. https://doi.org/10.3390/microorganisms9040808
APA StyleDomínguez-Maqueda, M., Cerezo, I. M., Tapia-Paniagua, S. T., De La Banda, I. G., Moreno-Ventas, X., Moriñigo, M. Á., & Balebona, M. C. (2021). A Tentative Study of the Effects of Heat-Inactivation of the Probiotic Strain Shewanella putrefaciens Pdp11 on Senegalese Sole (Solea senegalensis) Intestinal Microbiota and Immune Response. Microorganisms, 9(4), 808. https://doi.org/10.3390/microorganisms9040808