Plant Growth-Promoting Bacteria as an Emerging Tool to Manage Bacterial Rice Pathogens
Abstract
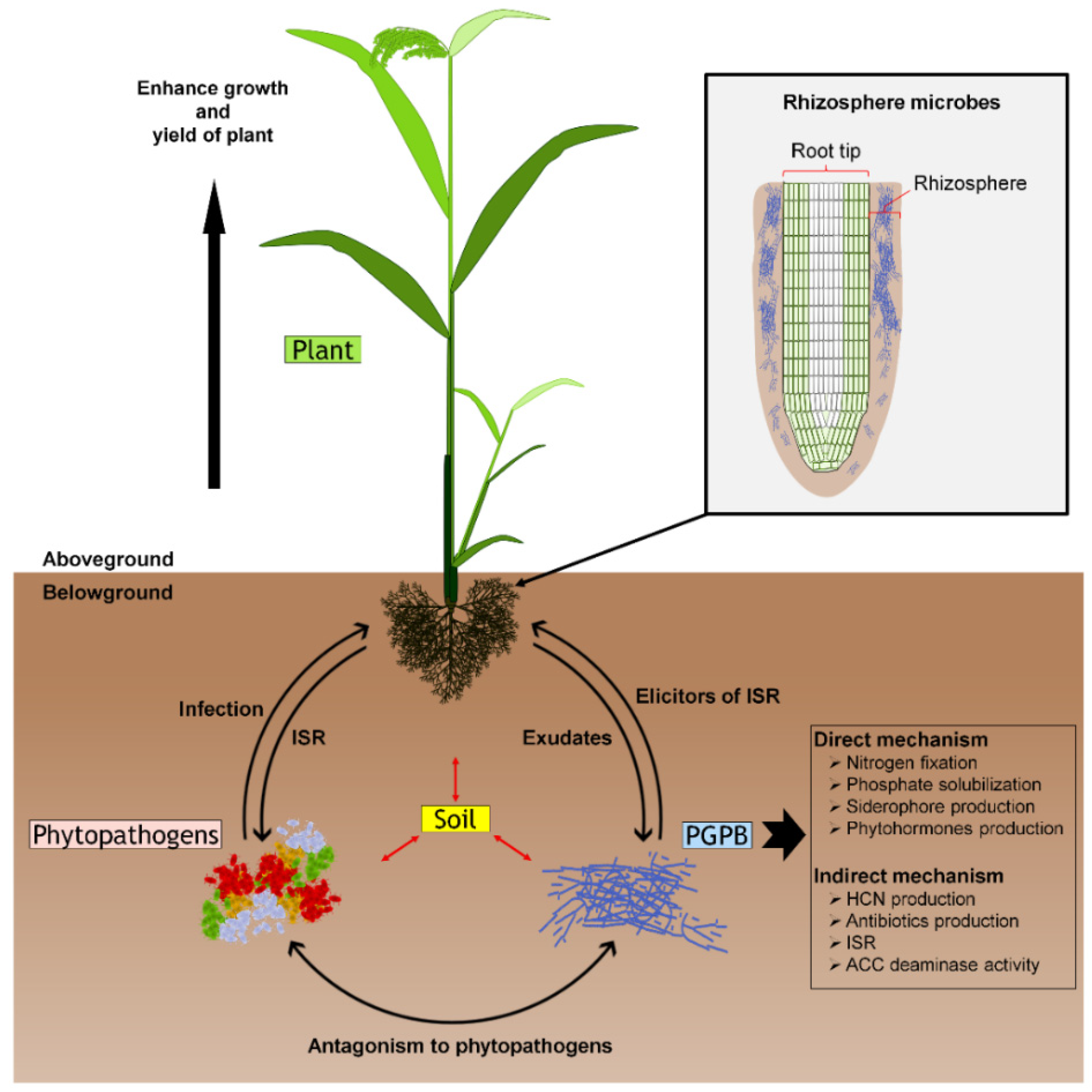
1. Introduction
2. Bacterial Rice Pathogens
| Diseases | Bacterial Pathogens | References | |
|---|---|---|---|
| Seedling | Seedling blight | Burkholderia plantarii | [50] |
| Bacterial Brown Stripe of Rice (BBSR) | Pseudomonas syringae pv. panici | [51] | |
| Acidovorax avenae subsp. avenae | [52] | ||
| Foliar | Bacterial Blight (BB) or Bacteria Leaf Blight (BLB) | Xanthomonas oryzae pv. oryzae | [53] |
| Pantoea ananatis | [54] | ||
| Pantoea stewartii subsp. indologenes | [55] | ||
| Pantoea stewartii | [54] | ||
| Pantoea agglomerans | [56] | ||
| Bacterial Leaf Streak (BLS) | Xanthomonas oryzae pv. oryzicola | [57] | |
| Halo blight | Pseudomonas syringae pv. oryzae | [58] | |
| Leaf sheath and grain rot | Sheath brown rot | Pseudomonas fuscovaginae | [59] |
| Sheath rot | Pseudomonas syringae pv. syringae | [60] | |
| Bacterial Panicle Blight (BPB) | Burkholderia glumae or Burkholderia gladioli | [29] | |
| Bacterial palea browning | Erwinia herbicola | [61] | |
| Pantoea ananatis | [62] | ||
| Enterobacter cloacae | [63] | ||
| Culm and root | Bacterial foot rot | Erwinia chrysanthemi | [64] |
| Dickeya zeae | [65] | ||
3. An Overview of In Vitro Characterizations of Promising PGPB
4. The PGPB as Biocontrol Agent
4.1. Bacillus spp.
4.2. Pseudomonas spp.
4.3. Enterobacter spp.
4.4. Streptomyces spp.
4.5. Other Bacterial Genus
5. Bioformulations of PGPB in Rice: Applications, Challenges, and Future Prospects
5.1. Applications on Bioformulations
5.2. Challenges and Future Prospects
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Firdaus, R.B.R.; Leong Tan, M.; Rahmat, S.R.; Senevi Gunaratne, M. Paddy, rice and food security in Malaysia: A review of climate change impacts. Cogent Soc. Sci. 2020, 6, 1818373. [Google Scholar] [CrossRef]
- Saha, S.; Garg, R.; Biswas, A.; Rai, A.B. Bacterial diseases of rice: An overview. J. Pure Appl. Microbiol. 2015, 9, 725–736. [Google Scholar]
- Jiang, N.; Yan, J.; Liang, Y.; Shi, Y.; He, Z.; Wu, Y.; Zeng, Q.; Liu, X.; Peng, J. Resistance genes and their interactions with Bacterial Blight/Leaf Streak pathogens (Xanthomonas oryzae) in rice (Oryza sativa L.)-An updated review. Rice 2020, 13, 3. [Google Scholar] [CrossRef]
- Azizi, M.M.F.; Ismail, S.I.; Ina-Salwany, M.Y.; Hata, E.M.; Zulperi, D. The emergence of Pantoea species as a future threat to global rice production. J. Plant. Prot. Res. 2020, 60, 1–9. [Google Scholar]
- Shew, A.M.; Durand-Morat, A.; Nalley, L.L.; Zhou, X.G.; Rojas, C.; Thoma, G. Warming increases Bacterial Panicle Blight (Burkholderia glumae) occurrences and impacts on USA rice production. PLoS ONE 2019, 14, e0219199. [Google Scholar] [CrossRef] [PubMed]
- Rani, U.; Kumar, V. Microbial bioformulations: Present and future aspects. In Nanobiotechnology in Bioformulations; Springer: Chem, Switzerland, 2019; pp. 243–258. [Google Scholar]
- Prasad, M.; Srinivasan, R.; Chaudhary, M.; Choudhary, M.; Jat, L.K. Plant growth promoting rhizobacteria (PGPR) for sustainable agriculture: Perspectives and challenges. In PGPR Amelioration in Sustainable Agriculture; Woodhead Publishing: Cambridge, UK, 2019; pp. 129–157. [Google Scholar]
- Sharma, T.; Kumar, N.; Rai, N. Isolation, screening and characterization of PGPR isolates from rhizosphere of rice plants in Kashipur region (Tarai region). Biotechnol. Int. 2012, 5, 69–84. [Google Scholar]
- Jeyanthi, V.; Kanimozhi, S. Plant growth promoting rhizobacteria (PGPR)-prospective and mechanisms: A review. J. Pure Appl. Microbiol. 2018, 12, 733–749. [Google Scholar] [CrossRef]
- Tiwari, S.; Prasad, V.; Chauhan, P.S.; Lata, C. Bacillus amyloliquefaciens confers tolerance to various abiotic stresses and modulates plant response to phytohormones through osmoprotection and gene expression regulation in rice. Front. Plant. Sci. 2017, 8, 1510. [Google Scholar] [CrossRef]
- Xiao, A.W.; Li, Z.; Li, W.C.; Ye, Z.H. The effect of plant growth-promoting rhizobacteria (PGPR) on arsenic accumulation and the growth of rice plants (Oryza sativa L.). Chemosphere 2020, 242, 125136. [Google Scholar]
- Ghaffari, H.; Gholizadeh, A.; Biabani, A.; Fallah, A.; Mohammadian, M. Plant growth promoting rhizobacteria (PGPR) application with different nitrogen fertilizer levels in rice (Oryza sativa L.). Pertanika J. Trop. Agric. Sci. 2018, 41, 715–728. [Google Scholar]
- Mitra, S.; Pramanik, K.; Sarkar, A.; Ghosh, P.K.; Soren, T.; Maiti, T.K. Bioaccumulation of cadmium by Enterobacter sp. and enhancement of rice seedling growth under cadmium stress. Ecotoxicol. Environ. Saf. 2018, 156, 183–196. [Google Scholar] [CrossRef]
- Gopalakrishnan, S.; Srinivas, V.; Vidya, M.S.; Rathore, A. Plant growth-promoting activities of Streptomyces spp. in sorghum and rice. Springerplus 2013, 2, 574. [Google Scholar] [CrossRef] [PubMed]
- Jambhulkar, P.P.; Sharma, P. Promotion of rice seedling growth characteristics by development and use of bioformulation of Pseudomonas fluorescens. Indian J. Agric. Sci. 2013, 83, 136–142. [Google Scholar]
- Jambhulkar, P.P.; Sharma, P. Development of bioformulation and delivery system of Pseudomonas fluorescens against Bacterial Leaf Blight of rice (Xanthomonas oryzae pv. oryzae). J. Environ. Biol. 2014, 35, 843–849. [Google Scholar] [PubMed]
- Suryadi, Y.; Susilowati, D.N.; Kadir, T.S.; Zaffan, Z.R.; Hikmawati, N.; Mubarik, N.S. Bioformulation of antagonistic bacterial consortium for controlling Blast, Sheath Blight and Bacterial Blight disease on rice. Asian J. Plant. Pathol. 2013, 7, 92–108. [Google Scholar] [CrossRef][Green Version]
- Sharma, A.; Shankhdhar, D.; Sharma, A.; Shankhdhar, S.C. Growth promotion of the rice genotypes by PGPRs isolated from rice rhizosphere. J. Soil Sci. Plant. Nutr. 2014, 14, 505–517. [Google Scholar] [CrossRef]
- Harikrishnan, H.; Shanmugaiah, V.; Balasubramanian, N. Optimization for production of indole acetic acid (IAA) by plant growth promoting Streptomyces sp. VSMGT1014 isolated from rice rhizosphere. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 158–171. [Google Scholar]
- Ashrafuzzaman, M.; Hossen, F.A.; Ismail, M.R.; Hoque, M.A.; Islam, M.Z.; Shahidullah, S.M.; Meon, S. Efficiency of plant growth-promoting rhizobacteria (PGPR) for the enhancement of rice growth. African J. Biotechnol. 2009, 8, 1247–1252. [Google Scholar]
- Peñaloza Atuesta, G.C.; Murillo Arango, W.; Eras, J.; Oliveros, D.F.; Méndez Arteaga, J.J. Rice-associated rhizobacteria as a source of secondary metabolites against Burkholderia glumae. Molecules 2020, 25, 2567. [Google Scholar] [CrossRef]
- Suryadi, Y.; Susilowati, D.N.; Fauziah, F. Management of plant diseases by PGPR-mediated induced resistance with special reference to tea and rice crops. Plant. Growth Promot. Rhizobacteria Sustain. Stress Manag. 2019, 65–110. [Google Scholar] [CrossRef]
- Rahma, H.; Nurbailis; Kristina, N. Characterization and potential of plant growth-promoting rhizobacteria on rice seedling growth and the effect on Xanthomonas oryzae pv. oryzae. Biodiversitas 2019, 20, 3654–3661. [Google Scholar] [CrossRef]
- Wubneh, W.Y.; Bayu, F.A. Assessment of diseases on rice (Oriza sativa L.) in major growing fields of Pawe district, northwestern Ethiopia. World Sci. News 2016, 42, 13–23. [Google Scholar]
- Rajarajeswari, N.V.L.; Muralidharan, K. Assessments of farm yield and district production loss from Bacterial Leaf Blight epidemics in rice. Crop. Prot. 2006, 25, 244–252. [Google Scholar] [CrossRef]
- Wonni, I.; Hutin, M.; Ouedrago, L.; Somda, I.; Verdier, V.; Szurek, B. Evaluation of elite rice varieties unmasks new sources of Bacterial Blight and Leaf Streak resistance for Africa. Rice Res. Open Access 2016, 4, 162. [Google Scholar] [CrossRef]
- Nandakumar, R.; Shahjahan, A.K.M.; Yuan, X.L.; Dickstein, E.R.; Groth, D.E.; Clark, C.A.; Cartwright, R.D.; Rush, M.C. Burkholderia glumae and B. gladioli cause Bacterial Panicle Blight in rice in the southern United States. Plant. Dis. 2009, 93, 896–905. [Google Scholar] [CrossRef] [PubMed]
- Niño-Liu, D.O.; Ronald, P.C.; Bogdanove, A.J. Xanthomonas oryzae pathovars: Model pathogens of a model crop. Mol. Plant. Pathol. 2006, 7, 303–324. [Google Scholar] [CrossRef]
- Zhou-qi, C.; Bo, Z.; Guan-lin, X.; Bin, L.; Shi-wen, H. Research status and prospect of Burkholderia glumae, the pathogen causing Bacterial Panicle Blight. Rice Sci. 2016, 23, 111–118. [Google Scholar] [CrossRef]
- Doni, F.; Suhaimi, N.S.M.; Mohamed, Z.; Ishak, N.; Mispan, M.S. Pantoea: A newly identified causative agent for Leaf Blight Disease in rice. J. Plant. Dis. Prot. 2019, 126, 491–494. [Google Scholar] [CrossRef]
- Von Bodman, S.B.; Bauer, W.D.; Coplin, D.L. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2003, 41, 455–482. [Google Scholar] [CrossRef]
- Hugouvieux-Cotte-Pattat, N.; Condemine, G.; Shevchik, V.E. Bacterial Pectate lyases, structural and functional diversity. Environ. Microbiol. Rep. 2014, 6, 427–440. [Google Scholar] [CrossRef]
- Aslam, S.N.; Newman, M.; Erbs, G.; Morrissey, K.L.; Chinchilla, D.; Boller, T.; Jensen, T.T.; De Castro, C.; Ierano, T.; Molinaro, A.; et al. Bacterial polysaccharides suppress induced innate immunity by calcium chelation. Curr. Biol. 2008, 18, 1078–1083. [Google Scholar] [CrossRef]
- Zakaria, L.; Misman, N. The pathogen and control management of Rice Blast Disease. Malays. J. Microbiol. 2018, 14, 705–714. [Google Scholar] [CrossRef]
- Mizobuchi, R.; Fukuoka, S.; Tsuiki, C.; Tsushima, S.; Sato, H. Evaluation of major japanese rice cultivars for resistance to bacterial grain rot caused by Burkholderia glumae and identification of standard cultivars for resistance. Breed. Sci. 2018, 68, 413–419. [Google Scholar] [CrossRef]
- van Esse, H.P.; Reuber, T.L.; van der Does, D. Genetic modification to improve disease resistance in crops. New Phytol. 2020, 225, 70–86. [Google Scholar] [CrossRef]
- Gnanamanickam, S.S.; Candole, B.L.; Mew, T.W. Influence of soil factors and cultural practice on biological control of Sheath Blight of rice with antagonistic bacteria. Plant. Soil 1992, 144, 67–75. [Google Scholar] [CrossRef]
- Howard, R.J. Cultural control of plant diseases: A historical perspective. Can. J. Plant. Pathol. 1996, 18, 145–150. [Google Scholar] [CrossRef]
- Obi, V.I. Activity of botanical extracts on paddy rice pathogen. Am. J. Sustain. Agric. 2012, 6, 66–73. [Google Scholar]
- Naqvi, S.A.H.; Umar, U.; Hasnain, A.; Rehman, A.; Perveen, R. Effect of botanical extracts: A potential biocontrol agent for Xanthomonas oryzae pv. oryzae, causing Bacterial Leaf Blight disease of rice. Pakistan J. Agricltural Res. 2018, 32, 59–72. [Google Scholar] [CrossRef]
- Lakshmanan, V.; Thangaraj, M.; Ponnusamy, B.; Santhiraka, S.; Kannan, R.; Regunathan, U.; Selvaraj, S. Antibacterial activity of Rhizophora apiculata leaf extract for the management of rice Bacterial Blight disease. Plant. Pathol. J. 2019, 18, 39–46. [Google Scholar] [CrossRef][Green Version]
- Khan, J.A.; Siddiq, R.; Arshad, H.M.I.; Anwar, H.S.; Saleem, K.; Jamil, F.F. Chemical control of Bacterial Leaf Blight of rice caused by Xanthomonas oryzae pv. oryzae. Pak. J. Phytopathol. 2012, 24, 97–100. [Google Scholar]
- Qudsia, H.; Akhter, M.; Riaz, A.; Haider, Z.; Mahmood, A. Comparative efficacy of different chemical treatments for paddy Blast, Brown Leaf Spot and Bacterial Leaf Blight diseases in rice (Oryza sativa L.). Appl. Microbiol. Open Access 2017, 3, 138. [Google Scholar] [CrossRef]
- Mahmood, T.; White, F. Disease resistance and susceptibility genes to bacterial blight of rice. In Protecting Rice Grains in the Post-Genomic Era; IntechOpen: London, UK, 2019. [Google Scholar]
- Wang, L.; Makino, S.; Subedee, A.; Bogdanove, A.J. Novel candidate virulence factors in rice pathogen Xanthomonas oryzae pv. oryzicola as revealed by mutational analysis. Appl. Environ. Microbiol. 2007, 73, 8023–8027. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Gao, Y.; Wang, J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-seq technique. PLoS ONE 2012, 7, e30646. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Tariq, R.; Ji, Z.; Wei, Z.; Zheng, K.; Mishra, R.; Zhao, K. Transcriptome analysis of a rice cultivar reveals the differentially expressed genes in response to wild and mutant strains of Xanthomonas oryzae pv. oryzae. Sci. Rep. 2019, 9, 3757. [Google Scholar] [CrossRef]
- Bangratz, M.; Wonni, I.; Kini, K.; Sondo, M.; Brugidou, C.; Béna, G.; Gnacko, F.; Barro, M.; Koebnik, R.; Silué, D.; et al. Design of a new multiplex PCR assay for rice pathogenic bacteria detection and its application to infer disease incidence and detect co-infection in rice fields in Burkina Faso. PLoS ONE 2020, 15, e0232115. [Google Scholar] [CrossRef]
- Lang, J.M.; Pérez-Quintero, A.L.; Koebnik, R.; DuCharme, E.; Sarra, S.; Doucoure, H.; Keita, I.; Ziegle, J.; Jacobs, J.M.; Oliva, R.; et al. A pathovar of Xanthomonas oryzae infecting wild grasses provides insight into the evolution of pathogenicity in rice agroecosystems. Front. Plant. Sci. 2019, 10, 507. [Google Scholar] [CrossRef]
- Wamishe, Y.; Cartwright, R.; Lee, F. Management of rice diseases. In Rice Production Handbook; University of Arkansas Coop Extension Service: Little Rock, AR, USA, 2001; pp. 87–100. [Google Scholar]
- Liu, H.; Qiu, H.; Zhao, W.; Cui, Z.; Ibrahim, M.; Jin, G.; Li, B.; Zhu, B.; Xie, G.L. Genome sequence of the plant pathogen Pseudomonas syringae pv. panici LMG 2367. J. Bacteriol. 2012, 194, 5693–5694. [Google Scholar] [CrossRef][Green Version]
- Li, B.; Liu, B.; Yu, R.; Tao, Z.; Wang, Y.; Xie, G.; Li, H.; Sun, G. Bacterial Brown Stripe of rice in soil-less culture system caused by Acidovorax avenae subsp. avenae in China. J. Gen. Plant. Pathol. 2011, 77, 64–67. [Google Scholar] [CrossRef]
- Ishiyama, S. Studies on Bacterial Leaf Blight of rice. Rep. Agric. Exp. Stn. 1922, 45, 233–261. [Google Scholar]
- Kini, K.; Agnimonhan, R.; Afolabi, O.; Soglonou, B.; Silué, D.; Koebnik, R. First report of a new Bacterial Leaf Blight of rice caused by Pantoea ananatis and Pantoea stewartii in Togo. Plant. Dis. 2017, 101, 241. [Google Scholar] [CrossRef]
- Azizi, M.M.F.; Ismail, S.I.; Hata, E.M.; Zulperi, D.; Ina-Salwany, M.Y.; Abdullah, M.A.F. First report of Pantoea stewartii subsp. indologenes causing Leaf Blight on rice in Malaysia. Plant. Dis. 2019, 103, 1407. [Google Scholar] [CrossRef]
- Lee, H.B.; Hong, J.P.; Kim, S.B. First report of Leaf Blight caused by Pantoea agglomerans on rice in Korea. Plant. Dis. 2010, 94, 1372. [Google Scholar] [CrossRef]
- OEPP/EPPO. Xanthomonas oryzae; EPPO Bulletin: Hoboken, NJ, USA, 2007; Volume 37, pp. 543–553. [Google Scholar]
- Kuwata, H. Pseudomonas syringae pv. oryze pv. nov. causal agent of bacterial Halo Blight of rice. Ann. Phytopathol. Soc. Jpn. 1985, 51, 212–218. [Google Scholar] [CrossRef]
- Tanii, A.; Miyajima, K.; Akita, T. The Sheath Brown Rot disease of rice plant and its causal bacterium, Pseudomonas fuscovaginae. Ann. Phytopathol. Soc. Jpn. 1976, 42, 540–548. [Google Scholar] [CrossRef]
- Zeigler, R.S.; Alvarez, E. Characteristics of Pseudomonas spp. causing grain discoloration and Sheath Rot of rice, and associated pseudomonad epiphytes. Plant. Dis. 1990, 74, 917–922. [Google Scholar] [CrossRef]
- Azegami, K. Bacterial Palea Browning, a new disease of rice caused by Erwinia herbicola. Bull. Natl. Inst. Agric. Sci. Ser. C 1983, 37, 1–12. [Google Scholar]
- Yan, H.; Yu, S.H.; Xie, G.L.; Fang, W.; Su, T.; Li, B. Grain discoloration of rice caused by Pantoea ananatis (synonym Erwinia uredovora) in China. Plant. Dis. 2010, 94, 482. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; Li, C.; Tan, K.; Liu, C.; Xu, X.; Zhang, S.; Wang, X.; Zhao, J.; Xiang, W. Characterization, phylogenetic analyses, and pathogenicity of Enterobacter cloacae on rice seedlings in Heilongjiang Province, China. Plant. Dis. 2020, 104, 1601–1609. [Google Scholar] [CrossRef] [PubMed]
- Goto, M. Bacterial Foot Rot of rice caused by a strain of Erwinia chrysanthemi. Phytopathology 1979, 69, 213–216. [Google Scholar] [CrossRef]
- Pu, X.M.; Zhou, J.N.; Lin, B.R.; Shen, H.F. First report of Bacterial Foot Rot of rice caused by a Dickeya zeae in China. Plant. Dis. 2012, 96, 1818. [Google Scholar] [CrossRef]
- García-Salamanca, A.; Molina-Henares, M.A.; van Dillewijn, P.; Solano, J.; Pizarro-Tobías, P.; Roca, A.; Duque, E.; Ramos, J.L. Bacterial diversity in the rhizosphere of maize and the surrounding carbonate-rich bulk soil. Microb. Biotechnol. 2013, 6, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Baptist, F.; Aranjuelo, I.; Legay, N.; Lopez-Sangil, L.; Molero, G.; Rovira, P.; Nogués, S. Rhizodeposition of organic carbon by plants with contrasting traits for resource acquisition: Responses to different fertility regimes. Plant. Soil 2015, 394, 391–406. [Google Scholar] [CrossRef]
- Huang, X.F.; Chaparro, J.M.; Reardon, K.F.; Zhang, R.; Shen, Q.; Vivanco, J.M. Rhizosphere interactions: Root exudates, microbes, and microbial communities. Botany 2014, 92, 267–275. [Google Scholar] [CrossRef]
- Zhou, D.; Huang, X.F.; Chaparro, J.M.; Badri, D.V.; Manter, D.K.; Vivanco, J.M.; Guo, J. Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant. Soil 2016, 401, 259–272. [Google Scholar] [CrossRef]
- Kumari, B.; Mallick, M.A.; Solanki, M.K. Plant growth promoting rhizobacteria (PGPR): Modern prospects for sustainable agriculture. In Plant Health under Biotic Stress; Springer Nature Singapore Pte Ltd.: Singapore, 2019; pp. 109–127. [Google Scholar]
- Verma, M.; Mishra, J.; Arora, N.K. Plant growth-promoting rhizobacteria: Diversity and applications. In Environmental Biotechnology: For Sustainable Future; Springer Nature Singapore Pte Ltd.: Singapore, 2019; pp. 129–173. [Google Scholar]
- Kumar, A.; Kumar, R.; Kumari, M.; Goldar, S. Enhancement of plant growth by using PGPR for a sustainable agriculture: A review. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 152–165. [Google Scholar] [CrossRef]
- Kloepper, J.W. Plant growth-promoting rhizobacteria (other systems). In Azospirillum/Plant Associations; CRC Press: Boca Raton, FL, USA, 1994; pp. 137–166. [Google Scholar]
- Tabassum, B.; Khan, A.; Tariq, M.; Ramzan, M.; Iqbal Khan, M.S.; Shahid, N.; Aaliya, K. Bottlenecks in commercialisation and future prospects of PGPR. Appl. Soil Ecol. 2017, 121, 102–117. [Google Scholar] [CrossRef]
- Kashyap, B.K.; Solanki, M.K.; Pandey, A.K.; Prabha, S.; Kumar, P.; Kumari, B. Bacillus as plant growth promoting rhizobacteria (PGPR): A promising green agriculture technology. In Plant Health under Biotic Stress; Springer Nature Singapore Pte Ltd.: Singapore, 2019; pp. 219–236. [Google Scholar]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Aeron, A.; Kumar, S.; Pandey, P.; Maheshwari, D.K. Emerging role of plant growth promoting rhizobacteria in agrobiology. In Bacteria in Agrobiology: Crop Ecosystems; Springer: Berlin/Heidelberg, Germany, 2011; pp. 1–36. [Google Scholar]
- Amara, U.; Khalid, R.; Hayat, R. Soil bacteria and phytohormones for sustainable crop production. In Bacterial Metabolites in Sustainable Agroecosystem; Springer: Chem, Switzerland, 2015; pp. 87–103. [Google Scholar]
- Hedden, P.; Phillips, A.L. Gibberellin metabolism: New insights revealed by the genes. Trends Plant. Sci. 2000, 5, 523–530. [Google Scholar] [CrossRef]
- Eskew, D.L.; Focht, D.D.; Ting, I.P. Nitrogen fixation, denitrification, and pleomorphic growth in a highly pigmented Spirillum lipoferum. Appl. Environ. Microbiol. 1977, 34, 582–585. [Google Scholar] [CrossRef]
- Sajjad Mirza, M.; Ahmad, W.; Latif, F.; Haurat, J.; Bally, R.; Normand, P.; Malik, K.A. Isolation, Partial characterization, and the effect of plant growth-promoting bacteria (PGPB) on micro-propagated sugarcane in vitro. Plant. Soil 2001, 237, 47–54. [Google Scholar] [CrossRef]
- Hardy, R.W.F.; Burns, R.C.; Holsten, R.D. Applications of the acetylene-ethylene assay for measurement of nitrogen fixation. Soil Biol. Biochem. 1973, 5, 47–81. [Google Scholar] [CrossRef]
- Pikovskaya, R.I. Mobilization of phosphorus in soil in connection with vital activity of some microbial species. Mikrobiologiya 1948, 17, 362–370. [Google Scholar]
- Mahmoud, A.; Abd-Alla, M. Siderophore production by some microorganisms and their effect on bradyrhizobium-mung bean symbiosis. Int. J. Agric. Biol. 2001, 3, 157–162. [Google Scholar]
- Gutierrez, C.K.; Matsui, G.Y.; Lincoln, D.E.; Lovell, C.R. Production of the phytohormone indole-3-acetic acid by estuarine species of the genus Vibrio. Appl. Environ. Microbiol. 2009, 75, 2253–2258. [Google Scholar] [CrossRef]
- Gordon, S.A.; Weber, R.P. Colorimetric estimation of indoleacetic acid. Anal. Biochem. 1951, 26, 192–195. [Google Scholar] [CrossRef]
- Tien, T.M.; Gaskins, M.H.; Hubbell, D.H. Plant growth substances produced by Azospirillum brasilense and their effect on the growth of pearl millet (Pennisetum americanum L.). Appl. Environ. Microbiol. 1979, 37, 1016–1024. [Google Scholar] [CrossRef]
- Wilson, P.W.; Knight, S.G. Experiments in Bacterial Physiology; Burgess Publishing Co.: Minneapolis, MN, USA, 1952. [Google Scholar]
- Gonzalez-Lopez, J.; Salmeron, V.; Martinez-Toledo, M.V.; Ballesteros, F.; Ramos-Cormenzana, A. Production of auxins, gibberellins and cytokinins by Azotobacter vinelandh ATCC 12837 in chemically-defined media and dialysed soil media. Soil Biol. Biochem. 1986, 18, 119–120. [Google Scholar] [CrossRef]
- Borrow, A.; Brian, P.W.; Chester, V.E.; Curtis, P.J.; Hemming, H.G.; Henehan, C.; Jeffreys, E.G.; Lloyd, P.B.; Nixon, I.S.; Norris, G.L.F.; et al. Gibberellic acid, a metabolic product of the fungus Gibberella fujikuroi: Some observations on its production and isolation. J. Sci. Food Agric. 1955, 6, 340–348. [Google Scholar] [CrossRef]
- Dworkin, M.; Foster, J.W. Experiments with some microorganisms which utilize ethane and hydrogen. J. Bacteriol. 1958, 75, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Honma, M.; Smmomura, T. Metabolism of 1-aminocyclopropane-1-carboxylic acid. Agric. Biol. Chem. 1978, 42, 1825–1831. [Google Scholar]
- Bakker, A.W.; Schippers, B. Microbial cyanide production in the rhizosphere in relation to potato yield reduction and Pseudomonas spp-mediated plant growth-stimulation. Soil Biol. Biochem. 1987, 19, 451–457. [Google Scholar] [CrossRef]
- Lorck, H. Production of hydrocyanic acid by bacteria. Physiol. Plant. 1948, 1, 142–146. [Google Scholar] [CrossRef]
- Pollock, H.M.; Barry, A.L.; Gavan, T.L.; Fuchs, P.C.; Hansen, S.; Thornsberry, C.L.; Frankel, H.; Forsythe, S.B. Selection of a reference lot of mueller-hinton agar. J. Clin. Microbiol. 1986, 24, 1–6. [Google Scholar] [CrossRef]
- Balouiri, M.; Sadiki, M.; Ibnsouda, S.K. Methods for in vitro evaluating antimicrobial activity: A review. J. Pharm. Anal. 2016, 6, 71–79. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 169, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Flury, P.; Vesga, P.; Péchy-Tarr, M.; Aellen, N.; Dennert, F.; Hofer, N.; Kupferschmied, K.P.; Kupferschmied, P.; Metla, Z.; Ma, Z.; et al. Antimicrobial and insecticidal: Cyclic lipopeptides and hydrogen cyanide produced by plant-beneficial Pseudomonas strains CHA0, CMR12a, and PCL1391 contribute to insect killing. Front. Microbiol. 2017, 8, 100. [Google Scholar] [CrossRef]
- Arshad, M.; Saleem, M.; Hussain, S. Perspectives of bacterial ACC deaminase in phytoremediation. Trends Biotechnol. 2007, 25, 356–362. [Google Scholar] [CrossRef]
- Glick, B.R. Plant growth-promoting bacteria: Mechanisms and applications. Scientifica 2012, 2012. [Google Scholar] [CrossRef]
- Zahir, Z.A.; Munir, A.; Asghar, H.N.; Shaharoona, B.; Arshad, M. Effectiveness of rhizobacteria containing ACC deaminase for growth promotion of peas (Pisum sativum) under drought conditions. J. Microbiol. Biotechnol. 2008, 18, 958–963. [Google Scholar] [PubMed]
- Etesami, H. Plant growth promotion and suppression of fungal pathogens in rice (Oryza sativa L.) by plant growth-promoting bacteria. In Field Crops: Sustainable Management by PGPR; Springer: Cham, Switzerland, 2019; pp. 351–383. [Google Scholar]
- Li, Y.; Li, X.; Jia, D.; Liu, J.; Wang, J.; Liu, A.; Liu, Z.; Guan, G.; Liu, G.; Luo, J.; et al. Complete genome sequence and antimicrobial activity of Bacillus velezensis JT3-1, a microbial germicide isolated from yak feces. 3 Biotech. 2020, 10, 231. [Google Scholar] [CrossRef]
- Ngalimat, M.S.; Raja Abd Rahman, R.N.Z.; Yusof, M.T.; Syahir, A.; Sabri, S. Characterisation of bacteria isolated from the stingless bee, Heterotrigona itama, honey, bee bread and propolis. PeerJ 2019, 7, e7478. [Google Scholar] [CrossRef]
- Liu, J.; Cui, X.; Liu, Z.; Guo, Z.; Yu, Z.; Yao, Q.; Sui, Y.; Jin, J.; Liu, X.; Wang, G. The diversity and geographic distribution of cultivable Bacillus-like bacteria across black soils of northeast China. Front. Microbiol. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Ducrest, P.J.; Pfammatter, S.; Stephan, D.; Vogel, G.; Thibault, P.; Schnyder, B. Rapid detection of Bacillus ionophore cereulide in food products. Sci. Rep. 2019, 9, 5814. [Google Scholar] [CrossRef]
- Kuebutornye, F.K.A.; Abarike, E.D.; Lu, Y. A review on the application of Bacillus as probiotics in aquaculture. Fish. Shellfish Immunol. 2019, 87, 820–828. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd Allah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef]
- Shakeel, M.; Rais, A.; Hassan, M.N.; Hafeez, F.H. Root associated Bacillus sp. improves growth, yield and zinc translocation for basmati rice (Oryza sativa) varieties. Front. Microbiol. 2015, 6, 1286. [Google Scholar] [CrossRef] [PubMed]
- Elekhtyar, N. Efficiency of Pseudomonas fluorescens as plant growth-promoting rhizobacteria (PGPR) for the enhancement of seedling vigor, nitrogen uptake, yield and its attributes of rice (Oryza sativa L.). Int. J. Sci. Res. Agric. Sci. 2016, 2, 57–67. [Google Scholar]
- Win, K.T.; Win, A.Z.O.; Ohkama-Ohtsu, N.; Yokoyama, T. Bacillus pumilus strain TUAT-1 and nitrogen application in nursery phase promote growth of rice plants under field conditions. Agronomy 2018, 8, 216. [Google Scholar] [CrossRef]
- Nautiyal, C.S.; Srivastava, S.; Chauhan, P.S.; Seem, K.; Mishra, A.; Sopory, S.K. Plant growth-promoting bacteria Bacillus amyloliquefaciens NBRISN13 modulates gene expression profile of leaf and rhizosphere community in rice during salt stress. Plant. Physiol. Biochem. 2013, 66, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Bist, V.; Srivastava, S.; Singh, P.C.; Trivedi, P.K.; Asif, M.H.; Chauhan, P.S.; Nautiyal, C.S. Unraveling aspects of Bacillus amyloliquefaciens mediated enhanced production of rice under biotic stress of Rhizoctonia solani. Front. Plant. Sci. 2016, 7, 587. [Google Scholar] [CrossRef]
- Shultana, R.; Kee Zuan, A.T.; Yusop, M.R.; Saud, H.M. Characterization of salt-tolerant plant growth-promoting rhizobacteria and the effect on growth and yield of saline-affected rice. PLoS ONE 2020, 15, e0238537. [Google Scholar] [CrossRef]
- Shultana, R.; Tan Kee Zuan, A.; Yusop, M.R.; Mohd Saud, H.; Ayanda, A.F. Effect of salt-tolerant bacterial inoculations on rice seedlings differing in salt-tolerance under saline soil conditions. Agronomy 2020, 10, 1030. [Google Scholar] [CrossRef]
- Aswathy, S.K.; Sridar, R.; Sivakumar, U. Mitigation of drought in rice by a phyllosphere bacterium Bacillus altitudinis FD48. African, J. Microbiol. Res. 2017, 11, 1614–1625. [Google Scholar] [CrossRef]
- Rais, A.; Jabeen, Z.; Shair, F.; Hafeez, F.Y.; Hassan, M.N. Bacillus spp. a bio-control agent enhances the activity of antioxidant defense enzymes in rice against Pyricularia oryzae. PLoS ONE 2017, 12, e0187412. [Google Scholar] [CrossRef] [PubMed]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip. 2017, 31, 446–459. [Google Scholar] [CrossRef]
- Jin, P.; Wang, Y.; Tan, Z.; Liu, W.; Miao, W. Antibacterial activity and rice-induced resistance, mediated by C15surfactin A, in controlling rice disease caused by Xanthomonas oryzae pv. oryzae. Pestic. Biochem. Physiol. 2020, 169, 104669. [Google Scholar] [CrossRef] [PubMed]
- Belbahri, L.; Chenari Bouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Petrovova, E.; Oszako, T.; Cherrad, S.; Vacher, S.; et al. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Chithrashree; Udayashankar, A.C.; Chandra Nayaka, S.; Reddy, M.S.; Srinivas, C. Plant growth-promoting rhizobacteria mediate induced systemic resistance in rice against Bacterial Leaf Blight caused by Xanthomonas oryzae pv. oryzae. Biol. Control. 2011, 59, 114–122. [Google Scholar] [CrossRef]
- Jayaraj, J.; Anand, A.; Muthukrishnan, S.; Punja, Z. Pathogenesis-Related Proteins and Their Roles in Resistance to Fungal Pathogens; Food Products Press: New York, NY, USA, 2004. [Google Scholar]
- Fira, D.; Dimkić, I.; Berić, T.; Lozo, J.; Stanković, S. Biological control of plant pathogens by Bacillus species. J. Biotechnol. 2018, 285, 44–55. [Google Scholar] [CrossRef]
- Wu, G.; Liu, Y.; Xu, Y.; Zhang, G.; Shen, Q.; Zhang, R. Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant. Microbe Interact. 2018, 31, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, A.; Hassan, M.N.; Imran, M.; Iqbal, M.; Majeed, S.; Brader, G.; Sessitsch, A.; Hafeez, F.Y. Biocontrol activity of surfactin a purified from Bacillus NH-100 and NH-217 against Rice Bakanae Disease. Microbiol. Res. 2018, 209, 1–13. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; De Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A.; Joris, B.; Arpigny, J.L.; Thonart, P. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 2007, 9, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Jourdan, E.; Henry, G.; Duby, F.; Dommes, J.; Barthelemy, J.P.; Thonart, P.; Ongena, M.A.R.C. Insights into the defense-related events occurring in plant cells following perception of surfactin-type lipopeptide from Bacillus subtilis. Mol. Plant. Microbe Interact. 2009, 22, 456–468. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice pathogens. Sci. Rep. 2015, 5, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Anayo, O.F.; Scholastica, E.C.; Peter, O.C.; Nneji, U.G.; Obinna, A.; Mistura, L.O. The beneficial roles of Pseudomonas in medicine, industries, and environment: A review. In Pseudomonas Aeruginosa-An Armory Within; IntechOpen: London, UK, 2016. [Google Scholar]
- Diggle, S.P.; Whiteley, M. Microbe profile: Pseudomonas aeruginosa: Opportunistic pathogen and lab rat. Microbiology 2020, 166, 30–33. [Google Scholar] [CrossRef]
- Morris, C.E.; Sands, D.C.; Vinatzer, B.A.; Glaux, C.; Guilbaud, C.; Buffière, A.; Yan, S.; Dominguez, H.; Thompson, B.M. The life history of the plant pathogen Pseudomonas syringae is linked to the water cycle. ISME J. 2008, 2, 321–334. [Google Scholar] [CrossRef]
- Kandaswamy, R.; Ramasamy, M.K.; Palanivel, R.; Balasundaram, U. Impact of Pseudomonas putida RRF3 on the root transcriptome of rice plants: Insights into defense response, secondary metabolism and root exudation. J. Biosci. 2019, 44, 98. [Google Scholar] [CrossRef]
- David, B.V.; Chandrasehar, G.; Selvam, P.N. Pseudomonas fluorescens: A plant-growth-promoting rhizobacterium (PGPR) with potential role in biocontrol of pests of crops. In Crop Improvement through Microbial Biotechnology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 221–243. [Google Scholar]
- Saikia, R.; Kumar, R.; Arora, D.K.; Gogoi, D.K.; Azad, P. Pseudomonas aeruginosa inducing rice resistance against Rhizoctonia solani: Production of salicylic acid and peroxidases. Folia Microbiol. 2006, 51, 375–380. [Google Scholar] [CrossRef]
- Yasmin, S.; Hafeez, F.Y.; Mirza, M.S.; Rasul, M.; Arshad, H.M.I.; Zubair, M.; Iqbal, M. Biocontrol of Bacterial Leaf Blight of rice and profiling of secondary metabolites produced by rhizospheric Pseudomonas aeruginosa BRp3. Front. Microbiol. 2017, 8, 1895. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez-Barranquero, J.A.; Cazorla, F.M.; de Vicente, A. Pseudomonas syringae pv. syringae associated with mango trees, a particular pathogen within the “hodgepodge” of the Pseudomonas syringae complex. Front. Plant. Sci. 2019, 10, 570. [Google Scholar] [PubMed]
- Hirano, S.S.; Upper, C.D. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiol. Mol. Biol. Rev. 2000, 64, 624–653. [Google Scholar] [CrossRef] [PubMed]
- Passera, A.; Compant, S.; Casati, P.; Maturo, M.G.; Battelli, G.; Quaglino, F.; Antonielli, L.; Salerno, D.; Brasca, M.; Toffolatti, S.L.; et al. Not just a pathogen? description of a plant-beneficial Pseudomonas syringae strain. Front. Microbiol. 2019, 10, 1409. [Google Scholar] [CrossRef] [PubMed]
- Lavakush; Yadav, J.; Verma, J.P. Isolation and characterization of effective plant growth promoting rhizobacteria from rice rhizosphere of indian soil. Asian J. Biol. Sci. 2012, 5, 294–303. [Google Scholar]
- Habibi, S.; Djedidi, S.; Ohkama-Ohtsu, N.; Sarhadi, W.A.; Kojima, K.; Rallos, R.V.; Ramirez, M.D.A.; Yamaya, H.; Sekimoto, H.; Yokoyama, T. Isolation and screening of indigenous plant growth-promoting rhizobacteria from different rice cultivars in afghanistan soils. Microbes Environ. 2019, 34, 347–355. [Google Scholar] [CrossRef]
- Awasthi, S.; Chauhan, R.; Dwivedi, S.; Srivastava, S.; Srivastava, S.; Tripathi, R.D. A consortium of alga (Chlorella vulgaris) and bacterium (Pseudomonas putida) for amelioration of arsenic toxicity in rice: A promising and feasible approach. Environ. Exp. Bot. 2018, 150, 115–126. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.B. Characterization of root-associated bacteria from paddy and its growth-promotion efficacy. 3 Biotech. 2014, 4, 325–330. [Google Scholar] [CrossRef][Green Version]
- Lingaiah, S.; Umesha, S. Pseudomonas fluorescens inhibits the Xanthomonas oryzae pv. oryzae, the Bacterial Leaf Blight pathogen in rice. Can. J. Plant. Prot. 2013, 1, 147–153. [Google Scholar]
- Velusamy, P.; Ebenezar Immanuel, J.; Gnanamanickam, S.S.; Thomashow, L. Biological control of rice Bacterial Blight by plant-associated bacteria producing 2,4-diacetylphloroglucinol. Can. J. Microbiol. 2006, 52, 56–65. [Google Scholar] [CrossRef]
- Jha, C.K.; Aeron, A.; Patel, B.V.; Dinesh, K. Bacteria in Agrobiology: Plant. Growth Responses; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Kumar, V.; Jain, L.; Jain, S.K.; Chaturvedi, S.; Kaushal, P. Bacterial endophytes of rice (Oryza sativa L.) and their potential for plant growth promotion and antagonistic activities. S. Afr. J. Bot. 2020, 134, 50–63. [Google Scholar] [CrossRef]
- Chen, W.L.; Li, D.B.; Ge, Q.X. A study on Enterobacter cloacae B8, Bacillus subtilis B826 and their antagonistic substance to Xanthomonas campestris pv. oryzae. Acta Agric. Univ. Zhejiangensis 1990, 16 (Suppl. 2), 61–67. [Google Scholar]
- Hongfei, C.W.; Debao, L. Study on the coloninzation of Enterobacter cloacae B8x on rice leaves and its control of rice Bacterial Leaf Blight (Xanthomonas oryzae pv. oryzae). J. Agric. Biotechnol. 1994, 2. [Google Scholar]
- Pramanik, K.; Mitra, S.; Sarkar, A.; Soren, T.; Maiti, T.K. Characterization of a Cd2+-resistant plant growth promoting rhizobacterium (Enterobacter sp.) and its effects on rice seedling growth promotion under Cd2+-stress in vitro. Agric. Nat. Resour. 2018, 52, 215–221. [Google Scholar] [CrossRef]
- Pramanik, K.; Mitra, S.; Sarkar, A.; Maiti, T.K. Alleviation of phytotoxic effects of cadmium on rice seedlings by cadmium resistant PGPR strain Enterobacter aerogenes MCC 3092. J. Hazard. Mater. 2018, 351, 317–329. [Google Scholar] [CrossRef]
- Pattnaik, S.; Dash, D.; Mohapatra, S.; Pattnaik, M.; Marandi, A.K.; Das, S.; Samantaray, D.P. Improvement of rice plant productivity by native Cr(VI) reducing and plant growth promoting soil bacteria Enterobacter cloacae. Chemosphere 2020, 240, 124895. [Google Scholar] [CrossRef]
- Liu, Y.; Tan, H.; Cao, L.; Zhang, R. Rice sprout endophytic Enterobacter sp. SE-5 could improve tolerance of mature rice plants to salt or Cd2+ in soils. Arch. Agron. Soil Sci. 2020, 66, 873–883. [Google Scholar] [CrossRef]
- Sarkar, A.; Ghosh, P.K.; Pramanik, K.; Mitra, S.; Soren, T.; Pandey, S.; Mondal, M.H.; Maiti, T.K. A halotolerant Enterobacter sp. displaying ACC deaminase activity promotes rice seedling growth under salt stress. Res. Microbiol. 2018, 169, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Cao, L.; Tan, H.; Zhang, R. Surface display of ACC deaminase on endophytic enterobacteriaceae strains to increase saline resistance of host rice sprouts by regulating plant ethylene synthesis. Microb. Cell Fact. 2017, 16, 214. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, S.; Zaheer, A.; Aiysha, D.; Abdulla Malik, K.; Mehnaz, S. Actinomycetes: A source of industrially important enzymes. J. Proteomics Bioinform. 2017, 10, 316–319. [Google Scholar] [CrossRef]
- Quinn, G.A.; Banat, A.M.; Abdelhameed, A.M.; Banat, I.M. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J. Med. Microbiol. 2020, 69, 1040–1048. [Google Scholar] [CrossRef]
- Chater, K.F. Streptomyces inside-out: A new perspective on the bacteria that provide us with antibiotics. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 761–768. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.L.; Tan, L.T.H.; Law, J.W.F.; Chan, K.G.; Duangjai, A.; Saokaew, S.; Pusparajah, P.; Mutalib, N.S.A.; Khan, T.M.; Goh, B.H.; et al. Focused review: Cytotoxic and antioxidant potentials of mangrove-derived Streptomyces. Front. Microbiol. 2017, 8, 2065. [Google Scholar] [CrossRef] [PubMed]
- Ngalimat, M.S.; Raja Abd Rahman, R.N.Z.; Yusof, M.T.; Amir Hamzah, A.S.; Zawawi, N.; Sabri, S. A review on the association of bacteria with stingless bees. Sains Malays. 2020, 49, 1853–1863. [Google Scholar] [CrossRef]
- Subramaniam, G.; Thakur, V.; Saxena, R.K.; Vadlamudi, S.; Purohit, S.; Kumar, V.; Rathore, A.; Chitikineni, A.; Varshney, R.K. Complete genome sequence of sixteen plant growth promoting Streptomyces strains. Sci. Rep. 2020, 10, 10294. [Google Scholar] [CrossRef] [PubMed]
- Viaene, T.; Langendries, S.; Beirinckx, S.; Maes, M.; Goormachtig, S. Streptomyces as a plant’s best friend? FEMS Microbiol. Ecol. 2016, 92, fiw119. [Google Scholar] [CrossRef]
- Ferrer, C.M.; Olivete, E.; Orias, S.L.; Rocas, M.R.; Juan, S.; Dungca, J.Z.; Mahboob, T.; Barusrux, S.; Nissapatorn, V. A review on Streptomyces spp. as plant-growth promoting bacteria (PGPB). Asian J. Pharmacogn. 2018, 2, 32–40. [Google Scholar]
- Vurukonda, S.S.; Giovanardi, D.; Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef]
- Amaresan, N.; Kumar, K.; Naik, J.H.; Bapatla, K.G.; Mishra, R.K. Streptomyces in plant growth promotion: Mechanisms and role. In New and Future Developments in Microbial Biotechnology and Bioengineering; Elsevier: Amsterdan, The Netherlands, 2018; pp. 125–135. [Google Scholar]
- Suárez-Moreno, Z.R.; Vinchira-Villarraga, D.M.; Vergara-Morales, D.I.; Castellanos, L.; Ramos, F.A.; Guarnaccia, C.; Degrassi, G.; Venturi, V.; Moreno-Sarmiento, N. Plant-growth promotion and biocontrol properties of three Streptomyces spp. isolates to control bacterial rice pathogens. Front. Microbiol. 2019, 10, 290. [Google Scholar] [CrossRef]
- Betancur, L.A.; Naranjo-Gaybor, S.J.; Vinchira-Villarraga, D.M.; Moreno-Sarmiento, N.C.; Maldonado, L.A.; Suarez-Moreno, Z.R.; Acosta-González, A.; Padilla-Gonzalez, G.F.; Puyana, M.; Castellanos, L.; et al. Marine actinobacteria as a source of compounds for phytopathogen control: An integrative metabolic-profiling/bioactivity and taxonomical approach. PLoS ONE 2017, 12, e0170148. [Google Scholar] [CrossRef]
- Muangham, S.; Pathom-aree, W.; Duangmal, K. Melanogenic actinomycetes from rhizosphere soil-antagonistic activity against Xanthomonas oryzae and plant-growth-promoting traits. Can. J. Microbiol. 2015, 61, 164–170. [Google Scholar] [CrossRef]
- Ham, Y.; Kim, T.J. Anthranilamide from Streptomyces spp. inhibited Xanthomonas oryzae biofilm formation without affecting cell growth. Appl. Biol. Chem. 2018, 61, 673–680. [Google Scholar] [CrossRef]
- Hata, E.M.; Sijam, K.; Ahmad, Z.A.M.; Yusof, M.T.; Azman, N.A. In vitro antimicrobial assay of actinomycetes in rice against Xanthomonas oryzae pv. oryzicola and as potential plant growth promoter. Braz. Arch. Biol. Technol. 2015, 58, 821–832. [Google Scholar] [CrossRef]
- Shang, N.N.; Zhang, Z.; Huang, J.P.; Wang, L.; Luo, J.; Yang, J.; Peng, T.; Yan, Y.; Ma, Y.T.; Huang, S.X. Glycosylated piericidins from an endophytic Streptomyces with cytotoxicity and antimicrobial activity. J. Antibiot. 2018, 71, 672–676. [Google Scholar] [CrossRef]
- Nanjundan, J.; Ramasamy, R.; Ponnusamy, M. Optimization of culture conditions for antimicrobial metabolites production by Streptomyces sp. against Bacterial Leaf Blight pathogen Xanthomonas oryzae pv. oryzae. Int. J. Chem. Stud. 2019, 7, 1187–1191. [Google Scholar]
- Ilsan, N.A.; Nawangsih, A.A.; Wahyudi, A.T. Rice phyllosphere actinomycetes as biocontrol agent of Bacterial Leaf Blight disease on rice. Asian J. Plant. Pathol. 2016, 10, 1–8. [Google Scholar] [CrossRef][Green Version]
- Jaivel, N.; Rajesh, R.; Velmurugan, D.; Marimuthu, P. Antimicrobial activity of a novel secondary metabolite from Streptomyces sp. and molecular docking studies against Bacterial Leaf Blight pathogen of rice. Int. J. Curr. Microbiol. Appl. Sci. 2017, 6, 2861–2870. [Google Scholar] [CrossRef]
- Betancur, L.A.; Forero, A.M.; Romero-Otero, A.; Sepúlveda, L.Y.; Moreno-Sarmiento, N.C.; Castellanos, L.; Ramos, F.A. Cyclic tetrapeptides from the marine strain Streptomyces sp. PNM-161a with activity against rice and yam phytopathogens. J. Antibiot. 2019, 72, 744–751. [Google Scholar] [CrossRef] [PubMed]
- El Karkouri, A.; El Hassani, F.Z.; El Mzibri, M.; Benlemlih, M.; El Hassouni, M. Isolation and identification of an actinomycete strain with a biocontrol effect on the phytopathogenic Erwinia chrysanthemi 3937VIII responsible for soft rot disease. Ann. Microbiol. 2010, 60, 263–268. [Google Scholar] [CrossRef]
- Aeny, T.N.; Prasetyo, J.; Suharjo, R.; Dirmawati, S.R.; Efri; Niswati, A. Short communication: Isolation and identification of actinomycetes potential as the antagonist of Dickeya zeae pineapple soft rot in lampung, indonesia. Biodiversitas 2018, 19, 2052–2058. [Google Scholar]
- Huang, K.T.; Misatq, T.; Asuyama, H. Selective toxicity of blasticidin S to Piricularia oryzae and Pellicularia sasakii. J. Antibiot. Ser. A 1963, 17, 71–74. [Google Scholar]
- Copping, L.G.; Duke, S.O. Natural products that have been used commercially as crop protection agents. Pest. Manag. Sci. 2007, 63, 524–554. [Google Scholar] [CrossRef]
- Isono, K.; Nagatsu, J.; Kawashima, Y.; Suzuki, S. Studies on polyoxins, antifungal antibiotics. Agric. Biol. Chem. 1965, 29, 848–854. [Google Scholar]
- Yang, P.W.; Li, M.G.; Zhao, J.Y.; Zhu, M.Z.; Shang, H.; Li, J.R.; Cui, X.L.; Huang, R.; Wen, M.L. Oligomycins A and C, major secondary metabolites isolated from the newly isolated strain Streptomyces diastaticus. Folia Microbiol. 2010, 55, 10–16. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. A review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Cordovez, V.; Carrion, V.J.; Etalo, D.W.; Mumm, R.; Zhu, H.; Van Wezel, G.P.; Raaijmakers, J.M. Diversity and functions of volatile organic compounds produced by Streptomyces from a disease-suppressive soil. Front. Microbiol. 2015, 6, 1081. [Google Scholar] [CrossRef] [PubMed]
- Citron, C.; Barra, L.; Wink, J.; Dickschat, J.S. Volatiles from nineteen recently genome sequenced actinomycetes. Org. Biomol. Chem. 2015, 13, 2673. [Google Scholar] [CrossRef] [PubMed]
- Cavite, H.J.M.; Mactal, A.G.; Evangelista, E.V.; Cruz, J.A. Growth and yield response of upland rice to application of plant growth-promoting rhizobacteria. J. Plant. Growth Regul. 2020, 1–15. [Google Scholar] [CrossRef]
- Yanni, Y.G.; Abd El-Fattah, F.K. Towards integrated biofertilization management with free living and associative dinitrogen fixers for enhancing rice performance in the Nile Delta. Symbiosis 1999, 27, 319–331. [Google Scholar]
- Sarkar, A.; Pramanik, K.; Mitra, S.; Soren, T.; Maiti, T.K. Enhancement of growth and salt tolerance of rice seedlings by ACC deaminase-producing Burkholderia sp. MTCC 12259. J. Plant. Physiol. 2018, 231, 434–442. [Google Scholar] [CrossRef]
- Dar, A.I.; Saleem, F.; Ahmad, M.; Tariq, M.; Khan, A.; Ali, A.; Tabassum, B.; Ali, Q.; Khan, G.A.; Rashid, B.; et al. Characterization and efficiency assessment of PGPR for enhancement of rice (Oryza sativa L.) Yield. Av. Life Sci. 2014, 2, 38–45. [Google Scholar]
- Yasmin, S.; Zaka, A.; Imran, A.; Zahid, M.A.; Yousaf, S.; Rasul, G.; Arif, M.; Mirza, M.S. Plant growth promotion and suppression of Bacterial Leaf Blight in rice by inoculated bacteria. PLoS ONE 2016, 11, e0160688. [Google Scholar] [CrossRef]
- Chennappa, G.; Naik, M.K.; Adkar-Purushothama, C.R.; Amaresh, Y.S.; Sreenivasa, M.Y. PGP potential, abiotic stress tolerance and antifungal activity of Azotobacter strains isolated from paddy soils. In Indian Journal of Experimental Biology; NISCAIR-CSIR: New Delhi, India, 2016; Volume 54, pp. 322–331. [Google Scholar]
- Bal, H.B.; Nayak, L.; Das, S.; Adhya, T.K. Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant. Soil 2013, 366, 93–105. [Google Scholar] [CrossRef]
- de Souza, R.; Beneduzi, A.; Ambrosini, A.; da Costa, P.B.; Meyer, J.; Vargas, L.K.; Schoenfeld, R.; Passaglia, L.M.P. The effect of plant growth-promoting rhizobacteria on the growth of rice (Oryza sativa L.) cropped in southern brazilian fields. Plant. Soil 2013, 366, 585–603. [Google Scholar] [CrossRef]
- James, E.K.; Gyaneshwar, P.; Barraquio, W.L.; Mathan, N.; Ladha, J.K. Endophytic diazotrophs associated with rice. In The Quest for Nitrogen Fixation in Rice; International Rice Research Institute: Los Baños, Philippines, 2000; pp. 119–140. [Google Scholar]
- Azman, N.A.; Sijam, K.; Hata, E.M.; Othman, R.; Saud, H.M. Screening of bacteria as antagonist against Xanthomonas oryzae pv. oryzae, the causal agent of Bacterial Leaf Blight of paddy and as plant growth promoter. J. Exp. Agric. Int. 2017, 16, 1–15. [Google Scholar] [CrossRef][Green Version]
- Kumar, U.; Vithal, L.; Annapurna, K. Antagonistic potential and functional diversity of endo and rhizospheric bacteria of Basmati rice. Oryza 2013, 50, 162–168. [Google Scholar]
- Singh, D.P.; Singh, H.B.; Prabha, R. Omics-driven approaches in plant-microbe interaction. In Microbial Inoculants in Sustainable Agricultural Productivity; Springer: New Delhi, Delhi India, 2016; pp. 61–84. [Google Scholar]
- Rasul, M.; Yasmin, S.; Hakim, S.; Zaheer, A.; Mirza, B.; Mirza, M.S. Metagenomic analysis of bacterial community associated with rhizosphere and phyllosphere of Basmati rice. bioRxiv 2020. [Google Scholar] [CrossRef]
- Imchen, M.; Kumavath, R.; Vaz, A.B.M.; Góes-Neto, A.; Barh, D.; Ghosh, P.; Kozyrovska, N.; Podolich, O.; Azevedo, V. 16S rRNA gene amplicon based metagenomic signatures of rhizobiome community in rice field during various growth stages. Front. Microbiol. 2019, 10, 2103. [Google Scholar] [CrossRef]
- Aamir, M.; Rai, K.; Zehra, A.; Dubey, M.K.; Kumar, S.; Shukla, V.; Upadhyay, R.S. Microbial bioformulation-based plant biostimulants: A plausible approach toward next generation of sustainable agriculture. In Microbial Endophytes; Woodhead Publishing: Cambridge, UK, 2020; pp. 195–225. [Google Scholar]
- Prathap, M.; Ranjitha Kumari, B.D. Bioformulation in biological control for plant diseases—A review. Int. J. Biotech. Trends Technol. 2017, 7, 1–8. [Google Scholar]
- Stojanović, S.S.; Karabegović, I.; Beškoski, V.; Nikolić, N.; Lazić, M. Bacillus based microbial formulations: Optimization of the production process. Hem. Ind. 2019, 73, 169–182. [Google Scholar] [CrossRef]
- Mishra, J.; Arora, N.K. Bioformulations for plant growth promotion and combating phytopathogens: A sustainable approach. In Bioformulations: For Sustainable Agriculture; Springer: New Delhi, Delhi India, 2016; pp. 3–33. [Google Scholar]
- Singh, S.; Gupta, G.; Khare, E.; Behal, K.K.; Arora, N.K. Effect of enrichment material on the shelf life and field efficiency of bioformulation of Rhizobium sp. and P-solubilizing Pseudomonas fluorescens. Sci. Res. Report. 2014, 4, 44–50. [Google Scholar]
- Ajeng, A.A.; Abdullah, R.; Ling, T.C.; Ismail, S.; Lau, B.F.; Ong, H.C.; Chew, K.W.; Show, P.L.; Chang, J.S. Bioformulation of biochar as a potential inoculant carrier for sustainable agriculture. Environ. Technol. Innov. 2020, 20, 101168. [Google Scholar] [CrossRef]
- Brahmaprakash, G.P.; Sahu, P.K.; Lavanya, G.; Gupta, A.; Nair, S.S.; Gangaraddi, V. Role of Additives in Improving Efficiency of Bioformulation for Plant Growth and Development. In Frontiers in Soil and Environmental Microbiology; CRC Press: Boca Raton, FL, USA, 2020; pp. 1–10. [Google Scholar]
- Ijaz, M.; Ali, Q.; Ashraf, S.; Kamran, M.; Rehman, A. Development of future bio-formulations for sustainable agriculture. In Microbiome in Plant Health and Disease; Springer Nature Singapore Pte Ltd.: Singapore, 2019; pp. 421–446. [Google Scholar]
- Chakraborty, A.P. Carrier based bioformulations of PGPR- characteristics, shelf life and application in improving health status of crop plants—A mini review. Int. J. Res. Rev. 2020, 7, 88–98. [Google Scholar]
- Hastuti, R.D.; Lestari, Y.; Saraswati, R.; Suwanto, A. Capability of Streptomyces spp. in controlling Bacterial Leaf Blight disease in rice plants. Am. J. Agric. Biol. Sci. 2012, 7, 217–223. [Google Scholar]
- Chakraborty, S.; Tumpa, F.H.; Khokon, M.A.R. Development of formulation of fluorescent pseudomonads and its evaluation on bio-management of blast of rice. Arch. Phytopathol. Plant. Prot. 2020, 1–22. [Google Scholar] [CrossRef]
- Chandra, D.; Sharma, A.K. Field evaluation of consortium of bacterial inoculants producing ACC Deaminase on growth, nutrients and yield components of rice and wheat. J. Crop. Sci. Biotechnol. 2020, 1–13. [Google Scholar] [CrossRef]
- Fatima, T.; Mishra, I.; Verma, R.; Arora, N.K. Mechanisms of halotolerant plant growth promoting Alcaligenes sp. involved in salt tolerance and enhancement of the growth of rice under salinity stress. 3 Biotech. 2020, 10, 361. [Google Scholar] [CrossRef]
- Prathuangwong, S.; Chuaboon, W.; Chatnaparat, T.; Kladsuwan, L.; Shoorin, M.; Kasem, S. Induction of disease and drought resistance in rice by Pseudomonas fluorescens SP007s. Chiang Mai Univ. J. Nat. Sci. 2012, 11, 45–56. [Google Scholar]
- Kumar, V. Characterization, bio-formulation development and shelf-life studies of locally isolated bio-fertilizer strains. Octa J. Environ. Res. 2014, 2, 32–37. [Google Scholar]
- Mącik, M.; Gryta, A.; Frąc, M. Biofertilizers in agriculture: An overview on concepts, strategies and effects on soil microorganisms. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2020; Volume 162, pp. 31–87. [Google Scholar]
- Uribe, D.; Sánchez-Nieves, J.; Vanegas, J. Role of microbial biofertilizers in the development of a sustainable agriculture in the tropics. In Soil Biology and Agriculture in the Tropics; Springer: Berlin/Heidelberg, Germany, 2010; pp. 235–250. [Google Scholar]
- Paul, N.; Cruz, P.C.; Aguilar, E.A.; Badayos, R.B.; Hafele, S. Evaluation of biofertilizers in cultured rice. J. Biofertil. Biopestic. 2013, 4, 133. [Google Scholar]
- Sana, N.; Bajwa, R.; Javaid, A.; Shoaib, A. Effect of biopower application on weed growth and yield of rice. Planta Daninha 2017, 35, e017164872. [Google Scholar] [CrossRef][Green Version]
- Soumare, A.; Diedhiou, A.G.; Thuita, M.; Hafidi, M. Exploiting biological nitrogen fixation: A route towards a sustainable agriculture. Plants 2020, 9, 1011. [Google Scholar] [CrossRef]
- Arora, N.K. Agricultural sustainability and food security. Environ. Sustain. 2018, 1, 217–219. [Google Scholar] [CrossRef]
- Santos, V.B.; Araújo, A.S.F.; Leite, L.F.C.; Nunes, L.A.P.L.; Melo, W.J. Soil microbial biomass and organic matter fractions during transition from conventional to organic farming systems. Geoderma 2012, 170, 227–231. [Google Scholar] [CrossRef]
- Patra, B.; Singh, J. A review: Usage of biofertilizer in cereal crops. Curr. J. Appl. Sci. Technol. 2019, 36, 1–8. [Google Scholar] [CrossRef]
- Tyagi, S.; Naresh, R.K.; Prakash, S.; Yadav, G.; Tiwari, S.; Rawat, B.; Tiwari, S.; Joshi, A.; Tyagi, A.; Sharma, N. Conservation agriculture, biofertilizers and biopesticides: A holistic approach for agricultural sustainability and food security: A review. Int. J. Chem. Stud. 2019, 7, 3036–3046. [Google Scholar]
- Singh, T.B.; Ali, A.; Prasad, M.; Yadav, A.; Shrivastav, P.; Goyal, D.; Dantu, P.K. Role of organic fertilizers in improving soil fertility. In Contaminants in Agriculture; Springer: Cham, Switzerland, 2020; pp. 61–77. [Google Scholar]
- Gopalakrishnan, S.; Sathya, A.; Rajendran Vijayabharathi, R.; Srinivas, V. Formulations of plant growth- promoting microbes for field applications. In Microbial Inoculants in Sustainable Agricultural Productivity; Springer: New Delhi, India, 2016; pp. 239–251. [Google Scholar]
- FAO. Guidelines for the Export, Shipment, Import and Release of Biological Control Agents and Other Beneficial Organisms (International Standard for Phutosanitary Measures No. 3), 2nd ed.; FAO: Rome, Italy, 2005.
- Kang, Y.; Shen, M.; Xia, D.; Ye, K.; Zhao, Q.; Hu, J. Caution of Intensified spread of antibiotic resistance genes by inadvertent introduction of beneficial bacteria into soil. Acta Agric. Scand. Sect. B Soil Plant. Sci. 2017, 67, 576–582. [Google Scholar] [CrossRef]
- Mouhamad, R.S.; Alabboud, M. Plant growth-promoting bacteria as a natural resource for sustainable rice production under the soil salinity, wastewater, and heavy metal stress. In Plant Stress Physiology; IntechOpen: London, UK, 2020. [Google Scholar]
- Fernando, W.D.; Nakkeeran, S.; Zhang, Y. Biosynthesis of antibiotics by PGPR and its relation in biocontrol of plant diseases. In PGPR: Biocontrol and Biofertilization; Springer: Dordrecht, The Netherlands, 2005; pp. 67–109. [Google Scholar]
- Kenawy, A.; Dailin, D.J.; Abo-Zaid, G.A.; Abd Malek, R.; Ambehabati, K.K.; Zakaria, K.H.N.; Sayyed, R.Z.; El Enshasy, H.A. Biosynthesis of antibiotics by PGPR and their roles in biocontrol of plant diseases. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Singapore, 2019; pp. 1–35. [Google Scholar]
- Misra, B.B.; Langefeld, C.; Olivier, M.; Cox, L.A. Integrated omics: Tools, advances and future approaches. J. Mol. Endocrinol. 2019, 62, R21–R45. [Google Scholar] [CrossRef] [PubMed]
- Park, S.J.; Onizuka, S.; Seki, M.; Suzuki, Y.; Iwata, T.; Nakai, K. A systematic sequencing-based approach for microbial contaminant detection and functional inference. BMC Biol. 2019, 17, 72. [Google Scholar] [CrossRef] [PubMed]
| Mechanisms | Media | Descriptions | References | |
|---|---|---|---|---|
| Direct | ||||
| Nitrogen fixation | Nitrogen-free (NF) agar | Nitrogen fixation was observed qualitatively by the blue coloration around the colonies | [80] | |
| Malate (NFM) semisolid medium | Acetylene production was quantified on a gas chromatograph equipped with a Porapak Q column and a H2-flame ionization detector (FID) | [81,82] | ||
| Phosphate solubilization | Pikovskaya’s agar | Phosphate solubilization was determined qualitatively by the formation of halo zones around the colonies | [83] | |
| Siderophore production | Chrome azurol S (CAS) agar | Siderophore production was observed qualitatively by the yellow halo coloration around the colonies | [84] | |
| Phytohormones production | IAA | Nutrient broth medium supplemented with L- tryptophan | IAA production was determined using colorimetric methods and quantified on HPLC using ethyl acetate oxidation method | [85,86,87] |
| Cytokinins | Burk’s medium | Cytokinin production was determined using colorimetric methods | [88,89] | |
| Gibberellins | Nutrient broth medium | Gibberellin production was determined using colorimetric methods | [90] | |
| Indirect | ||||
| ACC deaminase production | Dworkin and Foster’s (DF) salts medium | Colonies growing on the DF agar were taken as ACC deaminase producers and ACC deaminase activity was determined using colorimetric method | [91,92] | |
| HCN production | Nutrient broth supplemented with 4.4 g/L of glycine | HCN production was observed qualitatively by the changes in the filter paper color from yellow to orange-brown | [93,94] | |
| Antibiotics production | Mueller Hinton (MH) medium | Screening of antimicrobial activity was observed using diffusion methods | [95,96] | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngalimat, M.S.; Mohd Hata, E.; Zulperi, D.; Ismail, S.I.; Ismail, M.R.; Mohd Zainudin, N.A.I.; Saidi, N.B.; Yusof, M.T. Plant Growth-Promoting Bacteria as an Emerging Tool to Manage Bacterial Rice Pathogens. Microorganisms 2021, 9, 682. https://doi.org/10.3390/microorganisms9040682
Ngalimat MS, Mohd Hata E, Zulperi D, Ismail SI, Ismail MR, Mohd Zainudin NAI, Saidi NB, Yusof MT. Plant Growth-Promoting Bacteria as an Emerging Tool to Manage Bacterial Rice Pathogens. Microorganisms. 2021; 9(4):682. https://doi.org/10.3390/microorganisms9040682
Chicago/Turabian StyleNgalimat, Mohamad Syazwan, Erneeza Mohd Hata, Dzarifah Zulperi, Siti Izera Ismail, Mohd Razi Ismail, Nur Ain Izzati Mohd Zainudin, Noor Baity Saidi, and Mohd Termizi Yusof. 2021. "Plant Growth-Promoting Bacteria as an Emerging Tool to Manage Bacterial Rice Pathogens" Microorganisms 9, no. 4: 682. https://doi.org/10.3390/microorganisms9040682
APA StyleNgalimat, M. S., Mohd Hata, E., Zulperi, D., Ismail, S. I., Ismail, M. R., Mohd Zainudin, N. A. I., Saidi, N. B., & Yusof, M. T. (2021). Plant Growth-Promoting Bacteria as an Emerging Tool to Manage Bacterial Rice Pathogens. Microorganisms, 9(4), 682. https://doi.org/10.3390/microorganisms9040682








