Mab_3083c Is a Homologue of RNase J and Plays a Role in Colony Morphotype, Aggregation, and Sliding Motility of Mycobacterium abscessus
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Condition
2.2. Transposon Mutagenesis
2.3. Cloning of Mab_3083c and Msmeg_2685 and Creation of Mab_3083cD89K,H90A Mutant
2.4. Sliding Motility Assay
2.5. Aggregation Assay
2.6. Lipid Extraction and Thin-Layer Chromatography (TLC)
2.7. Intracellular Survival Assay
2.8. Lysozyme and H2O2 Susceptibility Assay
2.9. Sequence Alignment
2.10. Statistical Analysis
3. Results
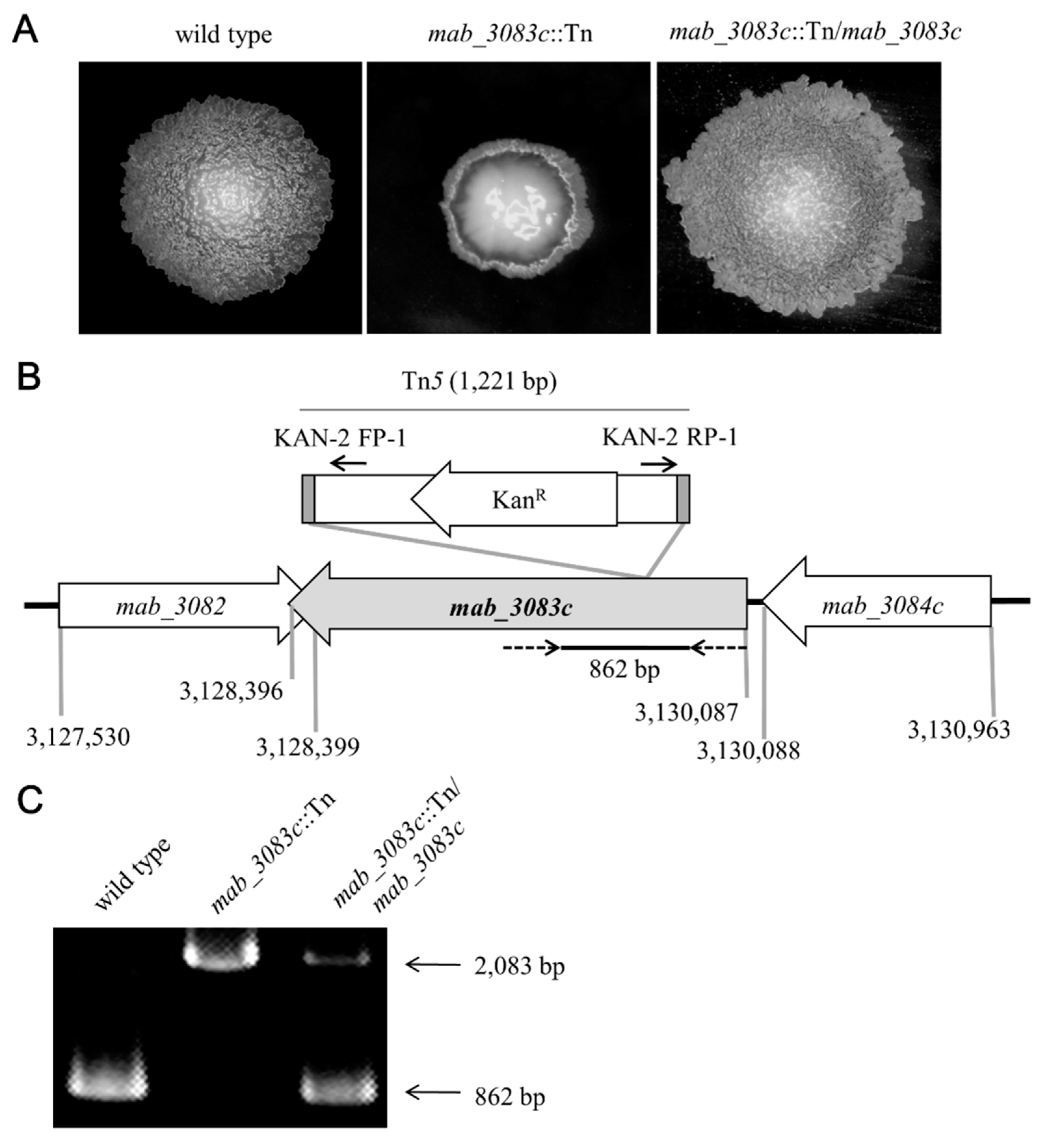
3.1. Characterization of the Mab_3083c::Tn Mutant
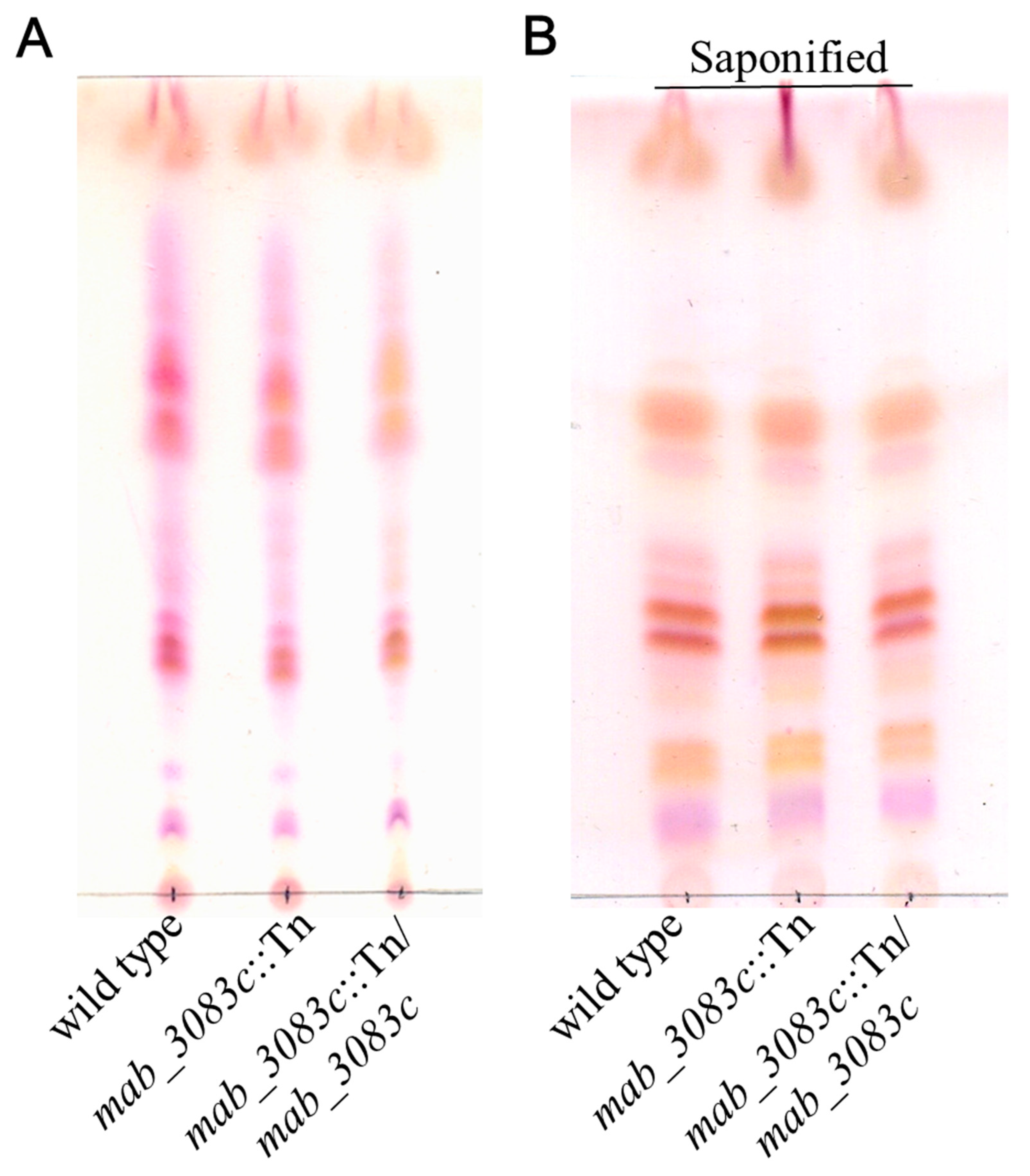
3.2. No Association of GPL with Colony Morphotype Switching of the mab_3083c::Tn Mutant
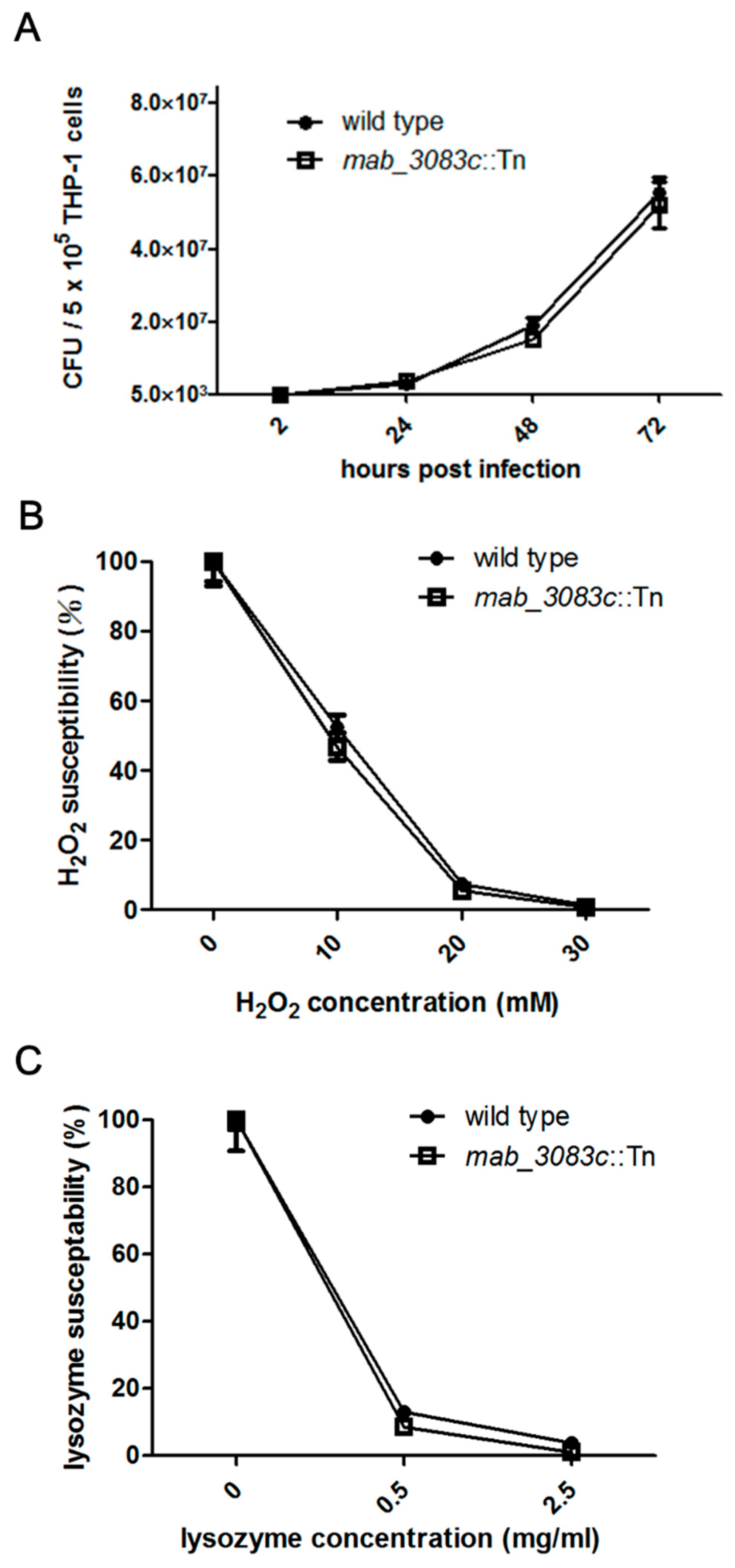
3.3. No Association of Mab_3083c with Intracellular Survival and Susceptibility to Hydrogen Peroxide and Lysozyme of M. abscessus
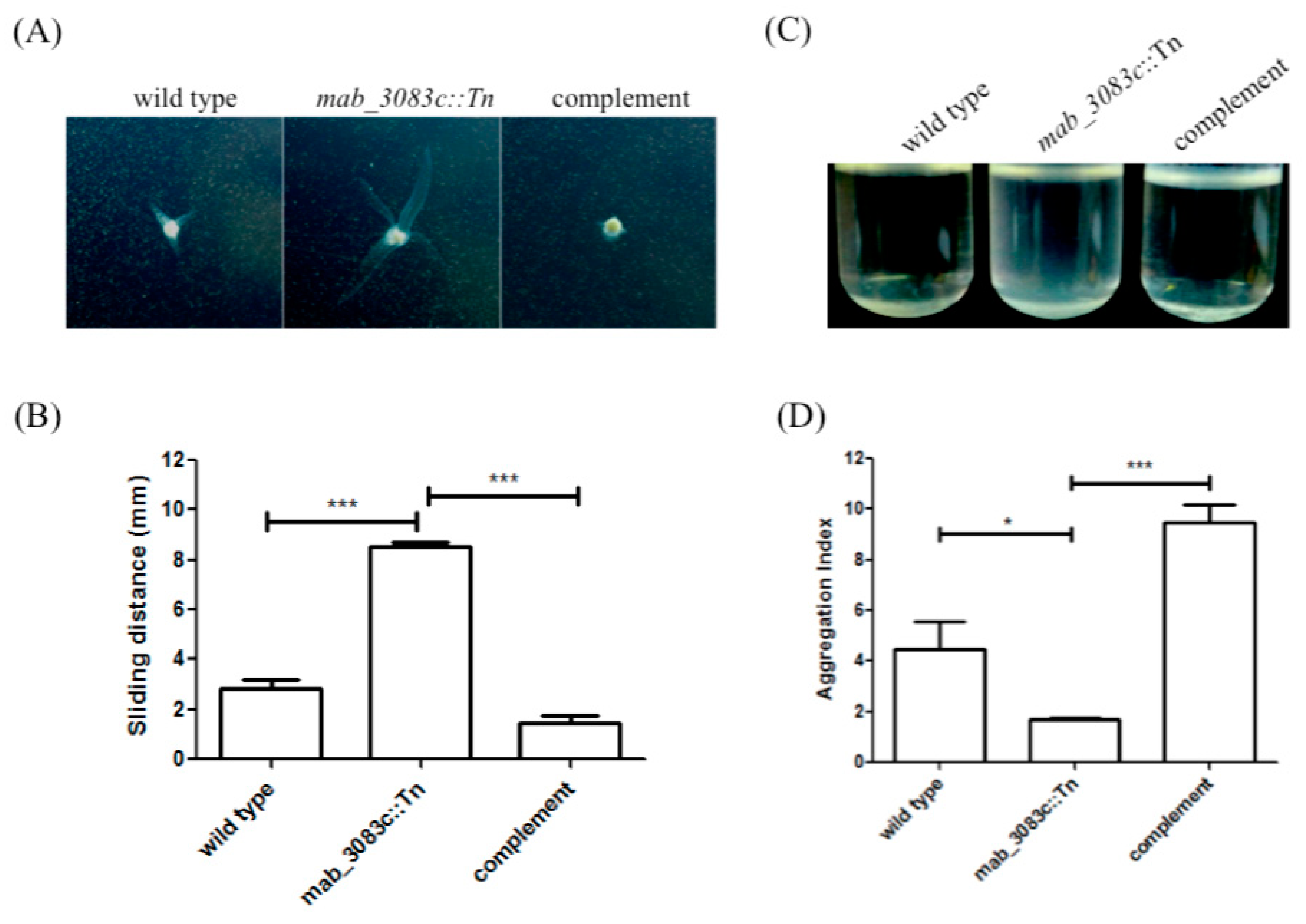
3.4. Higher Sliding Motility and Less Aggregation Capability of the Mab_3083c::Tn Mutant
3.5. Identification of Mab_3083c as a Homologue of Ribonuclease J
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Falkinham, J.O., III. Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 1996, 9, 177–215. [Google Scholar] [CrossRef] [PubMed]
- McShane, P.J.; Glassroth, J. Pulmonary disease due to nontuberculous mycobacteria: Current state and new insights. Chest 2015, 148, 1517–1527. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.E.; Girard, W.M.; Wallace, R.J. Clinical features of pulmonary disease caused by rapidly growing mycobacteria: An analysis of 154 patients. Am. Rev. Respir. Dis. 1993, 147, 1271–1278. [Google Scholar] [CrossRef] [PubMed]
- Griffith, D.E.; Aksamit, T.; Iseman, M.; Olivier, K.; Ruoss, S.; Von Reyn, C.F.; Wallace, R.J.; Winthrop, K.; Brown-Elliott, B.A.; Catanzaro, A.; et al. An official ATS/IDSA statement: Diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am. J. Respir. Crit. Care Med. 2007, 175, 367–416. [Google Scholar] [CrossRef] [PubMed]
- Nessar, R.; Cambau, E.; Reyrat, J.M.; Murray, A.; Gicquel, B. Mycobacterium abscessus: A new antibiotic nightmare. J. Antimicrob. Chemother. 2012, 67, 810–818. [Google Scholar] [CrossRef] [PubMed]
- Sethiya, J.P.; Sowards, M.A.; Jackson, M.; North, E.J. MmpL3 inhibition: A new approach to treat nontuberculous mycobacterial infections. Int. J. Mol. Sci. 2020, 21, 6202. [Google Scholar] [CrossRef]
- Olivier, K.N.; Weber, D.J.; Wallace, R.J., Jr.; Edwards, L.J.; Chakraborti, S.; Knowles, M.R.; Faiz, A.R.; Lee, J.-H.; Zhang, Y.; Brown-Elliot, B.A.; et al. Nontuberculous mycobacteria. I: Amulticenter prevalence study in cystic fibrosis. Am. J. Respir. Crit. Care Med. 2003, 167, 828–834. [Google Scholar] [CrossRef]
- Roux, A.-L.; Catherinot, E.; Gutierrez, C.; Vincent, V.; Fauroux, B.; Rottman, M.; Guillemot, D.; Gaillard, J.-L.; Jean-Louis Herrmann for the OMA Group; Ripoll, F.; et al. Multicenter study of prevalence of nontuberculous mycobacteria in patients with cystic fibrosis in france. J. Clin. Microbiol. 2009, 47, 4124–4128. [Google Scholar] [CrossRef]
- Huang, W.-C.; Chiou, C.-S.; Chen, J.-H.; Shen, G.-H. Molecular epidemiology of Mycobacterium abscessus infections in a subtropical chronic ventilatory setting. J. Med. Microbiol. 2010, 59, 1203–1211. [Google Scholar] [CrossRef][Green Version]
- Huang, H.-L.; Cheng, M.-H.; Lu, P.-L.; Shu, C.-C.; Wang, J.-Y.; Wang, J.-T.; Chong, I.-W.; Lee, L.-N. Epidemiology and predictors of NTM pulmonary infection in Taiwan-A retrospective, five-year multicenter study. Sci. Rep. 2017, 7, 16300. [Google Scholar] [CrossRef] [PubMed]
- Wallace, R.J., Jr. The clinical presentation, diagnosis, and therapy of cutaneous and pulmonary infections due to the rapidly growing mycobacteria, M. fortuitum and M. chelonae. Clin. Chest Med. 1989, 10, 419–429. [Google Scholar]
- Li, B.; Ye, M.; Guo, Q.; Zhang, Z.; Yang, S.; Ma, W.; Yu, F.; Chu, H. Determination of MIC distribution and mechanisms of decreased susceptibility to bedaquiline among clinical isolates of mycobacterium abscessus. Antimicrob. Agents Chemother. 2018, 62. [Google Scholar] [CrossRef]
- Sha, W.; Weng, X.-H.; Xiao, H.-P.; He, G.-J. Investigation of drug-resistance to rifampin and rpoB gene sequence analysis of Mycobacterium abscessus. Zhonghua Jiehe He Huxi Zazhi. Chin. J. Tuberc. Respir. Dis. 2003, 26, 544–547. [Google Scholar]
- Alcaide, F.; Pfyffer, G.E.; Telenti, A. Role of embB in natural and acquired resistance to ethambutol in mycobacteria. Antimicrob. Agents Chemother. 1997, 41, 2270–2273. [Google Scholar] [CrossRef] [PubMed]
- Jarand, J.; Levin, A.; Zhang, L.; Huitt, G.; Mitchell, J.D.; Daley, C.L. Clinical and microbiologic outcomes in patients receiving treatment for mycobacterium abscessus pulmonary disease. Clin. Infect. Dis. 2011, 52, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Zaidi, S.; Elidemir, O.; Heinle, J.; McKenzie, E.; Schecter, M.; Kaplan, S.; Dishop, M.; Kearney, D.; Mallory, G. Mycobacterium abscessusin cystic fibrosis lung transplant recipients: Report of 2 cases and risk for recurrence. Transpl. Infect. Dis. 2009, 11, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.; Ardito, F.; Fiscarelli, E.; La Sorda, M.; D’Argenio, P.; Ricciotti, G.; Fadda, G. Fatal pulmonary infection due to multidrug-resistant mycobacterium abscessus in a patient with cystic fibrosis. J. Clin. Microbiol. 2001, 39, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.C.; Gan, A.W.; Yam, A.; Tan, A.B.; Tay, S.C. Mycobacterium abscessus hand infections in immunocompetent fish handlers: Case report. J. Hand Surg. 2010, 35, 1142–1145. [Google Scholar] [CrossRef]
- Fregnan, G.B.; Smith, D.W. Description of various colony forms of mycobacteria. J. Bacteriol. 1962, 83, 819–827. [Google Scholar] [CrossRef]
- Recht, J.; Martínez, A.; Torello, S.; Kolter, R. Genetic analysis of sliding motility inmycobacterium smegmatis. J. Bacteriol. 2000, 182, 4348–4351. [Google Scholar] [CrossRef]
- Howard, S.T.; Rhoades, E.; Recht, J.; Pang, X.; Alsup, A.; Kolter, R.; Lyons, C.R.; Byrd, T.F. Spontaneous reversion of Mycobacterium abscessus from a smooth to a rough morphotype is associated with reduced expression of glycopeptidolipid and reacquisition of an invasive phenotype. Microbiology 2006, 152, 1581–1590. [Google Scholar] [CrossRef]
- Deshayes, C.; Laval, F.; Montrozier, H.; Daffé, M.; Etienne, G.; Reyrat, J.-M. A glycosyltransferase involved in biosynthesis of triglycosylated glycopeptidolipids in mycobacterium smegmatis: Impact on surface properties. J. Bacteriol. 2005, 187, 7283–7291. [Google Scholar] [CrossRef]
- Catherinot, E.; Roux, A.-L.; Rottman, M.; Gaillard, J.-L.; Herrmann, J.-L.; Macheras, E.; Hubert, D.; Matmar, M.; Dannhoffer, L.; Chinet, T.; et al. Acute respiratory failure involving an R variant of mycobacterium abscessus. J. Clin. Microbiol. 2008, 47, 271–274. [Google Scholar] [CrossRef]
- Byrd, T.F.; Lyons, C.R. Preliminary characterization of aMycobacterium abscessus mutant in human and murine models of infection. Infect. Immun. 1999, 67, 4700–4707. [Google Scholar] [CrossRef]
- Ripoll, F.; Deshayes, C.; Pasek, S.; Laval, F.; Beretti, J.-L.; Biet, F.; Risler, J.-L.; Daffé, M.; Etienne, G.; Gaillard, J.-L.; et al. Genomics of glycopeptidolipid biosynthesis in Mycobacterium abscessus and M. chelonae. Bmc Genom. 2007, 8, 114. [Google Scholar] [CrossRef]
- Gutiérrez, A.V.; Viljoen, A.; Ghigo, E.; Herrmann, J.-L.; Kremer, L. Glycopeptidolipids, a double-edged sword of the mycobacterium abscessus complex. Front. Microbiol. 2018, 9, 1145. [Google Scholar] [CrossRef] [PubMed]
- Rhoades, E.R.; Archambault, A.S.; Greendyke, R.; Hsu, F.-F.; Streeter, C.; Byrd, T.F. Mycobacterium abscessusGlycopeptidolipids mask underlying cell wall phosphatidyl-myo-inositol mannosides blocking induction of human macrophage TNF-α by preventing interaction with TLR2a. J. Immunol. 2009, 183, 1997–2007. [Google Scholar] [CrossRef] [PubMed]
- Tsai, S.-H.; Shen, G.-H.; Lin, C.-H.; Liau, J.-R.; Lai, H.-C.; Hu, S.-T. Mab_3168c, a putative acetyltransferase, enhances adherence, intracellular survival and antimicrobial resistance of mycobacterium abscessus. PLoS ONE 2013, 8, e67563. [Google Scholar] [CrossRef]
- Li, H.; Havens, W.M.; Nibert, M.L.; Ghabrial, S.A. RNA sequence determinants of a coupled termination-reinitiation strategy for downstream open reading frame translation in helminthosporium victoriae virus 190s and other victoriviruses (family totiviridae). J. Virol. 2011, 85, 7343–7352. [Google Scholar] [CrossRef]
- Martínez, A.; Torello, S.; Kolter, R. Sliding motility in mycobacteria. J. Bacteriol. 1999, 181, 7331–7338. [Google Scholar] [CrossRef] [PubMed]
- Naka, T.; Nakata, N.; Fujiwara, N.; Maeda, S.; Yamamoto, R.; Doe, M.; Mizuno, S.; Niki, M.; Kobayashi, K.; Ogura, H.; et al. Structure and host recognition of serotype 13 glycopeptidolipid from mycobacterium intracellulare. J. Bacteriol. 2011, 193, 5766–5774. [Google Scholar] [CrossRef][Green Version]
- Chen, C.-C.; Tsai, S.-H.; Lu, C.-C.; Hu, S.-T.; Wu, T.-S.; Huang, T.-T.; Saïd-Sadier, N.; Ojcius, D.M.; Lai, H.-C. Activation of an NLRP3 inflammasome restricts mycobacterium kansasii infection. PLoS ONE 2012, 7, e36292. [Google Scholar] [CrossRef]
- Shin, D.-M.; Jeon, B.-Y.; Friedman, R.L.; Jo, E.-K.; Lee, H.-M.; Jin, H.S.; Yuk, J.-M.; Song, C.-H.; Lee, S.-H.; Lee, Z.-W.; et al. Mycobacterium tuberculosis eis regulates autophagy, inflammation, and cell death through redox-dependent signaling. PLoS Pathog. 2010, 6, e1001230. [Google Scholar] [CrossRef]
- Taverniti, V.; Forti, F.; Ghisotti, D.; Putzer, H. Mycobacterium smegmatis RNase J is a 5′-3′ exo-/endoribonuclease and both RNase J and RNase E are involved in ribosomal RNA maturation. Mol. Microbiol. 2011, 82, 1260–1276. [Google Scholar] [CrossRef]
- Hunt-Serracin, A.C.; Parks, B.J.; Boll, J.; Boutte, C.C. Mycobacterium abscessus cells have altered antibiotic tolerance and surface glycolipids in artificial cystic fibrosis sputum medium. Antimicrob. Agents Chemother. 2019, 63. [Google Scholar] [CrossRef]
- Halloum, I.; Carrère-Kremer, S.; Blaise, M.; Viljoen, A.; Bernut, A.; Le Moigne, V.; Vilchèze, C.; Guérardel, Y.; Lutfalla, G.; Herrmann, J.-L.; et al. Deletion of a dehydratase important for intracellular growth and cording renders rough Mycobacterium abscessus avirulent. Proc. Nat. Acad. Sci. USA 2016, 113, E4228–E4237. [Google Scholar] [CrossRef]
- Davidson, L.B.; Nessar, R.; Kempaiah, P.; Perkins, D.J.; Byrd, T.F. Mycobacterium abscessus glycopeptidolipid prevents respiratory epithelial TLR2 signaling as measured by HβD2 gene expression and IL-8 release. PLoS ONE 2011, 6, e29148. [Google Scholar] [CrossRef]
- Figaro, S.; Durand, S.; Gilet, L.; Cayet, N.; Sachse, M.; Condon, C. Bacillus subtilis mutants with knockouts of the genes encoding ribonucleases RNAse Y and rnase J1 are viable, with major defects in cell morphology, sporulation, and competence. J. Bacteriol. 2013, 195, 2340–2348. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Leong, V.; Ortega, J.; Elliot, M.A. Development, antibiotic production, and ribosome assembly in streptomyces venezuelae are impacted by RNAse J and RNAse III deletion. J. Bacteriol. 2014, 196, 4253–4267. [Google Scholar] [CrossRef]
- Bugrysheva, J.V.; Scott, J.R. Regulation of virulence gene expression in Streptococcus pyogenes: Determinants of differential mRNA decay. Rna Biol. 2010, 7, 569–572. [Google Scholar] [CrossRef][Green Version]
- Madhugiri, R.; Evguenieva-Hackenberg, E. RNase J is involved in the 5′-end maturation of 16S rRNA and 23S rRNA inSinorhizobium meliloti. FEBS Lett. 2009, 583, 2339–2342. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Condon, C.; Bechhofer, D.H. Regulated RNA stability in the Gram positives. Curr. Opin. Microbiol. 2011, 14, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Mohanty, B.K.; Kushner, S.R. Regulation of mRNA decay in bacteria. Annu. Rev. Microbiol. 2016, 70, 25–44. [Google Scholar] [CrossRef]
- Even, S.; Pellegrini, O.; Zig, L.; Labas, V.; Vinh, J.; Brechemmier-Baey, D.; Putzer, H. Ribonucleases J1 and J2: Two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005, 33, 2141–2152. [Google Scholar] [CrossRef]
- Dorléans, A.; De La Sierra-Gallay, I.L.; Piton, J.; Zig, L.; Gilet, L.; Putzer, H.; Condon, C. Molecular basis for the recognition and cleavage of RNA by the bifunctional 5′–3′ exo/Endoribonuclease RNAse J. Structure 2011, 19, 1252–1261. [Google Scholar] [CrossRef]
- Mäder, U.; Zig, L.; Kretschmer, J.; Homuth, G.; Putzer, H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol. Microbiol. 2008, 70, 183–196. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, T.-Y.; Tsai, S.-H.; Chen, J.-W.; Wang, Y.-C.; Hu, S.-T.; Chen, Y.-Y. Mab_3083c Is a Homologue of RNase J and Plays a Role in Colony Morphotype, Aggregation, and Sliding Motility of Mycobacterium abscessus. Microorganisms 2021, 9, 676. https://doi.org/10.3390/microorganisms9040676
Liu T-Y, Tsai S-H, Chen J-W, Wang Y-C, Hu S-T, Chen Y-Y. Mab_3083c Is a Homologue of RNase J and Plays a Role in Colony Morphotype, Aggregation, and Sliding Motility of Mycobacterium abscessus. Microorganisms. 2021; 9(4):676. https://doi.org/10.3390/microorganisms9040676
Chicago/Turabian StyleLiu, Ting-Yu, Sheng-Hui Tsai, Jenn-Wei Chen, Yu-Ching Wang, Shiau-Ting Hu, and Yih-Yuan Chen. 2021. "Mab_3083c Is a Homologue of RNase J and Plays a Role in Colony Morphotype, Aggregation, and Sliding Motility of Mycobacterium abscessus" Microorganisms 9, no. 4: 676. https://doi.org/10.3390/microorganisms9040676
APA StyleLiu, T.-Y., Tsai, S.-H., Chen, J.-W., Wang, Y.-C., Hu, S.-T., & Chen, Y.-Y. (2021). Mab_3083c Is a Homologue of RNase J and Plays a Role in Colony Morphotype, Aggregation, and Sliding Motility of Mycobacterium abscessus. Microorganisms, 9(4), 676. https://doi.org/10.3390/microorganisms9040676






