A Review on the Biotechnological Applications of the Operational Group Bacillus amyloliquefaciens
Abstract
1. Introduction
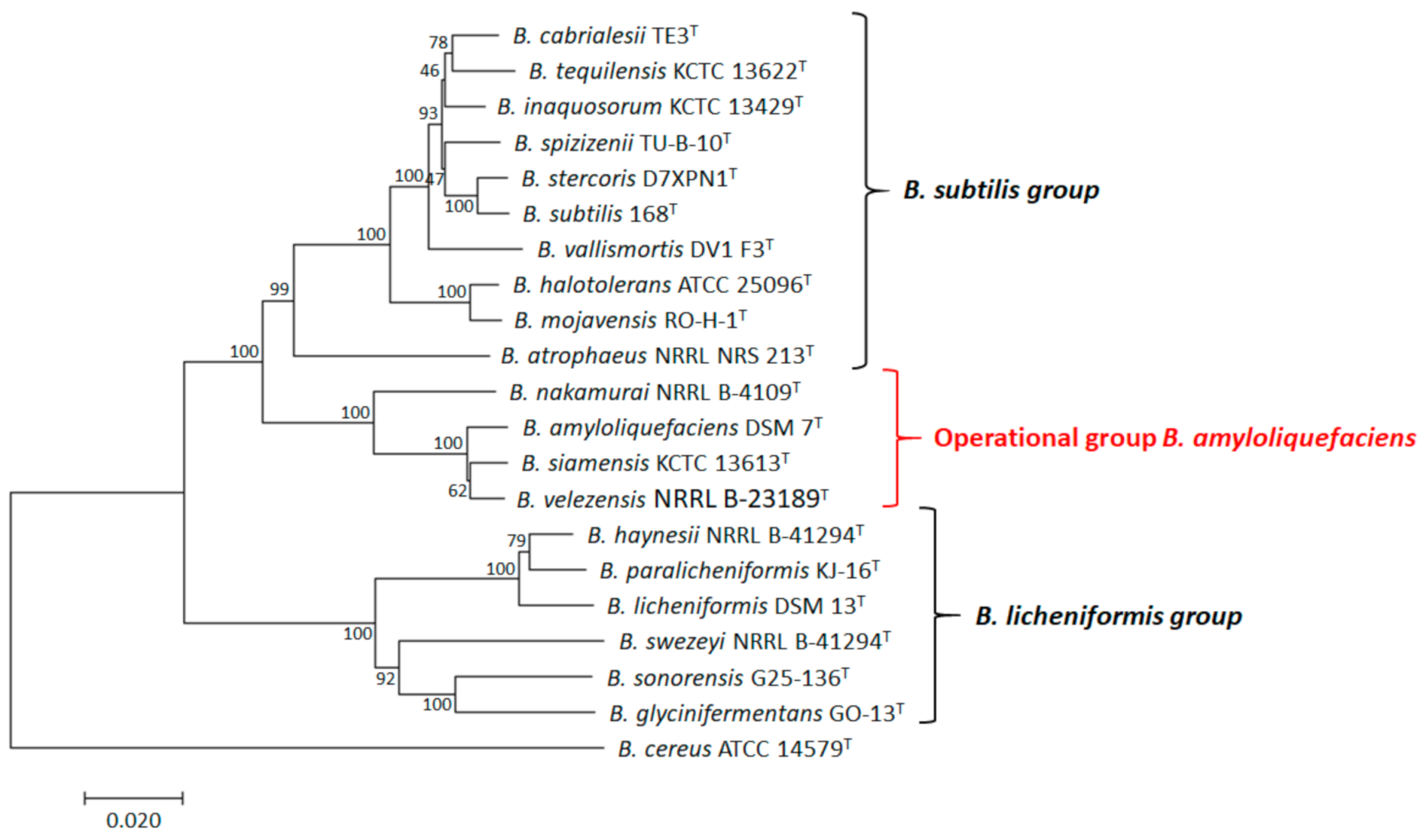
2. An Overview of the OGBa
2.1. Identification and Characterization
2.2. Ecology, Isolation and Cultivation
2.3. Genome and Its Arrangement
3. The Importance and Applications of the OGBa
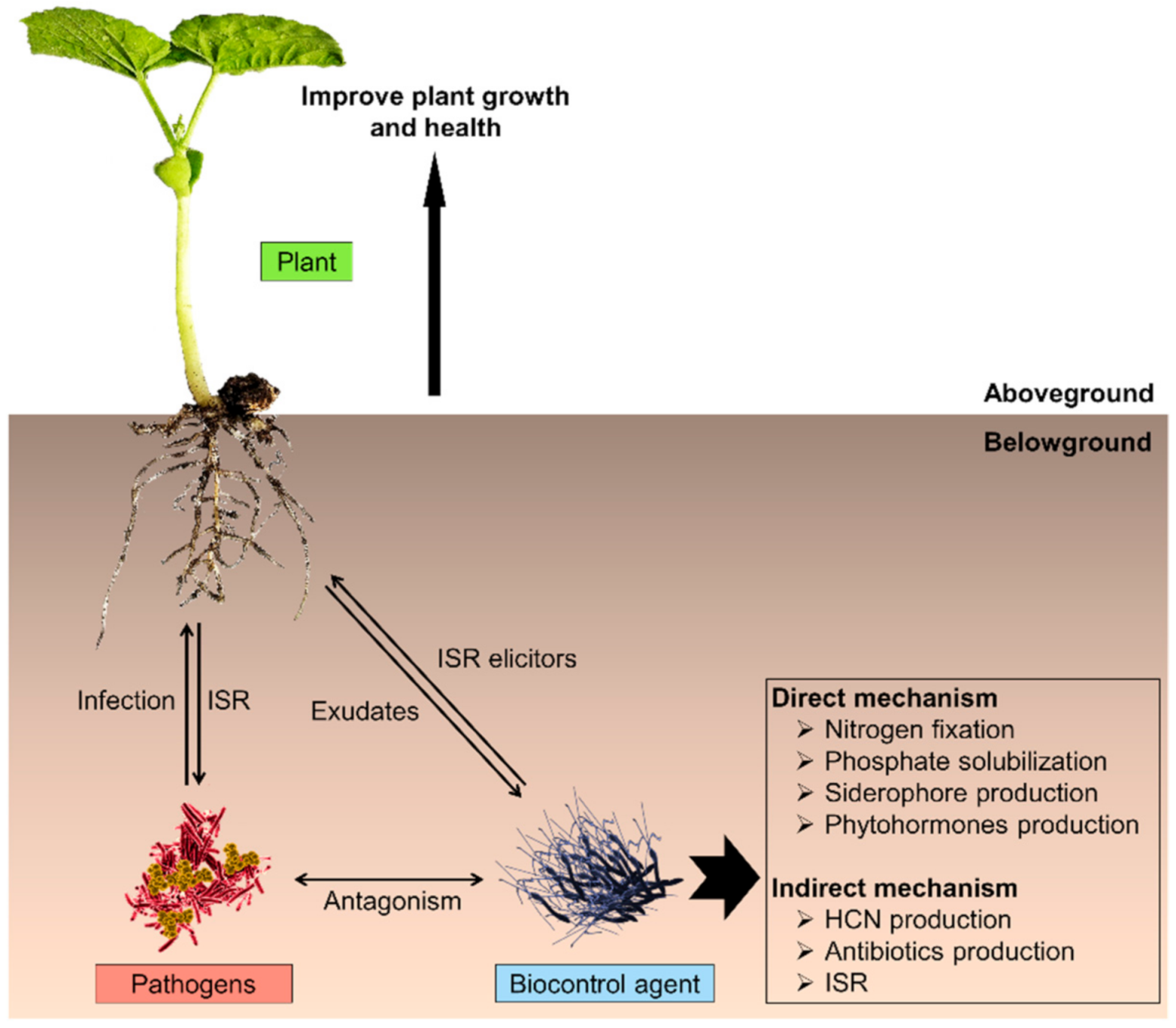
3.1. Plant Growth Promoters and Biocontrol Agents in Agriculture
3.2. Source of Commercial Enzymes
3.3. Antimicrobial Compounds Producer
3.4. Potential as Probiotics
3.5. Potential as Bioremediation Agents
4. Concluding Remark and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Priest, F.G.; Goodfellow, M.; Shute, L.A.; Berkeley, R.C.W. Bacillus amyloliquefaciens sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 1987, 37, 69–71. [Google Scholar] [CrossRef]
- Fukumoto, J. Studies on the production of bacterial amylase. I. Isolation of bacteria secreting potent amylases and their distribution. Nippon. Nogeikagaku Kaishi 1943, 19, 487–503. [Google Scholar]
- Berkeley, R.C.W.; Logan, N.A.; Shute, L.A.; Capey, A.G. Identification of Bacillus species. In Methods in Microbiology; Academic Press: Landon, UK, 1984; Volume 16, pp. 292–328. [Google Scholar]
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the B. subtilis species complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and ‘Bacillus oryzicola’are later heterotypic synonyms of Bacillus velezensis based on phylogenomic. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Bowman, M.J.; Rooney, A.P. Iturinic lipopeptide diversity in the Bacillus subtilis species group—Important antifungals for plant disease biocontrol applications. Front. Microbiol. 2019, 10, 1794. [Google Scholar] [CrossRef] [PubMed]
- Ye, M.; Tang, X.; Yang, R.; Zhang, H.; Li, F.; Tao, F.; Li, F.; Wang, Z. Characteristics and application of a novel species of Bacillus: Bacillus velezensis. ACS Chem. Biol. 2018, 13, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Rabbee, M.F.; Sarafat Ali, M.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K.H. Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed]
- Auch, A.F.; von Jan, M.; Klenk, H.P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef]
- Connor, N.; Sikorski, J.; Rooney, A.P.; Kopac, S.; Koeppel, A.F.; Burger, A.; Cole, S.G.; Perry, E.B.; Krizanc, D.; Field, N.C.; et al. Ecology of speciation in the genus Bacillus. Appl. Environ. Microbiol. 2010, 76, 1349–1358. [Google Scholar] [CrossRef]
- Borriss, R.; Chen, X.H.; Rueckert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C.; et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM7T and FZB42T: A proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus based on complete genome sequence comparisons. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786–1801. [Google Scholar] [CrossRef]
- Chun, J.; Bae, K.S. Phylogenetic analysis of Bacillus subtilis and related taxa based on partial gyrA gene sequences. Antonie Van Leeuwenhoek 2000, 78, 123–127. [Google Scholar] [CrossRef]
- Reva, O.N.; Dixelius, C.; Meijer, J.; Priest, F.G. Taxonomic characterization and plant colonizing abilities of some bacteria related to Bacillus amyloliquefaciens and Bacillus subtilis. FEMS Microbiol. Ecol. 2004, 48, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Ngalimat, M.S.; Sabri, S. Taxonomic note: Speciation within the operational group Bacillus amyloliquefaciens based on comparative phylogenies of housekeeping genes. Asia-Pac. J. Mol. Biol. Biotechnol. 2020, 28, 19–26. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Sumpavapol, P.; Tongyonk, L.; Tanasupawat, S.; Chokesajjawatee, N.; Luxananil, P.; Visessanguan, W. Bacillus siamensis sp. nov., isolated from salted crab (poo-khem) in Thailand. Int. J. Syst. Evol. Microbiol. 2010, 60, 2364–2370. [Google Scholar] [CrossRef]
- Ruiz-García, C.; Béjar, V.; Martinez-Checa, F.; Quesada, E. Bacillus velezensis sp nov., a surfactant-producing bacterium isolated from the river velez in malaga, southern Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 191–195. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Saunders, L.P.; Schisler, D.A.; Leathers, T.D.; Naeem, N.; Cohan, F.M.; Rooney, A.P. Bacillus nakamurai sp. nov., a black-pigment-producing strain. Int. J. Syst. Evol. Microbiol. 2016, 66, 2987–2991. [Google Scholar] [CrossRef]
- Liu, B.; Ge, B.; Azhar, N.; Zhao, W.; Cui, H.; Zhang, K. Complete genome sequence of Bacillus methylotrophicus strain NKG-1, isolated from the changbai mountains, China. Genome Announc. 2018, 6, e01454-17. [Google Scholar] [CrossRef]
- Balderas-Ruíz, K.A.; Bustos, P.; Santamaria, R.I.; González, V.; Cristiano-Fajardo, S.A.; Barrera-Ortíz, S.; Mezo-Villalobos, M.; Aranda-Ocampo, S.; Guevara-García, Á.A.; Galindo, E.; et al. Bacillus velezensis 83 a bacterial strain from mango phyllosphere, useful for biological control and plant growth promotion. AMB Express 2020, 10, 163. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Jia, D.; Liu, J.; Wang, J.; Liu, A.; Liu, Z.; Guan, G.; Liu, G.; Luo, J.; et al. Complete genome sequence of Bacillus velezensis JT3-1, a microbial germicide isolated from yak feces. bioRxiv 2019, 1, 555219. [Google Scholar] [CrossRef] [PubMed]
- Nannan, C.; Gillis, A.; Caulier, S.; Mahillon, J. Complete genome sequence of Bacillus velezensis CN026 exhibiting antagonistic activity against Gram-negative foodborne pathogens. Genome Announc. 2018, 6, e01543-17. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, Y.; Qin, Y.; Li, P. Complete genome sequence of Bacillus velezensis LPL-K103, an antifungal cyclic lipopeptide bacillomycin L producer from the surface of lemon. 3 Biotech 2020, 10, 8. [Google Scholar] [CrossRef]
- Zeng, Q.; Xie, J.; Li, Y.; Chen, X.; Wang, Q. Draft genome sequence of an endophytic biocontrol bacterium, Bacillus velezensis PG12, isolated from apple fruit. Microbiol. Resour. Announc. 2019, 8, e00468-19. [Google Scholar] [CrossRef] [PubMed]
- Gamez, R.M.; Rodríguez, F.; Bernal, J.F.; Agarwala, R.; Landsman, D.; Mariño-Ramírez, L. Genome sequence of the banana plant growth-promoting rhizobacterium Bacillus amyloliquefaciens BS006. Genome Announc. 2015, 3, e01391-15. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Shi, H.; Heng, J.; Wang, D.; Bian, K. Antimicrobial, plant growth-promoting and genomic properties of the peanut endophyte Bacillus velezensis LDO2. Microbiol. Res. 2019, 218, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Schofield, B.J.; Skarshewski, A.; Lachner, N.; Ouwerkerk, D.; Klieve, A.V.; Dart, P.; Hugenholtz, P. Near complete genome sequence of the animal feed probiotic, Bacillus amyloliquefaciens H57. Stand. Genomic Sci. 2016, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Kang, X.; Xi, H.; Liu, C.; Xue, Y. Complete genome sequence of the endophytic biocontrol strain Bacillus velezensis CC09. Genome Announc. 2016, 4, e01048-16. [Google Scholar] [CrossRef] [PubMed]
- Geng, W.; Cao, M.; Song, C.; Xie, H.; Liu, L.; Yang, C.; Feng, J.; Zhang, W.; Jin, Y.; Du, Y.; et al. Complete genome sequence of Bacillus amyloliquefaciens LL3, which exhibits glutamic acid-independent production of poly-γ-glutamic acid. J. Bacteriol. 2011, 193, 3393–3394. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Wang, R.; Sun, D.; Sun, L.; Wang, Y.; Pu, Y.; Fang, Z.; Xu, D.; Liu, Y.; Ye, R.; et al. Complete genome of Bacillus velezensis CMT-6 and comparative genome analysis reveals lipopeptide diversity. Biochem. Genet. 2020, 58, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Khalaf, E.M.; Raizada, M.N. Draft genome sequences of Bacillus and Paenibacillus species isolated from seeds of Citrullus lanata (watermelon), Cucurbita moschata (butternut squash), and Cucurbita pepo L. var pepo L. (pumpkin). Microbiol. Resour. Announc. 2020, 9, e00727-20. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhou, Z.; Han, Y.; Wang, Z.; Fan, J.; Xiao, H. Isolation and identification of antifungal peptides from Bacillus BH072, a novel bacterium isolated from honey. Microbiol. Res. 2013, 168, 598–606. [Google Scholar] [CrossRef]
- Ngalimat, M.S.; Rahman, R.N.Z.R.A.; Yusof, M.T.; Syahir, A.; Sabri, S. Characterisation of bacteria isolated from the stingless bee, Heterotrigona itama, honey, bee bread and propolis. PeerJ 2019, 7, e7478. [Google Scholar] [CrossRef] [PubMed]
- Zulkhairi Amin, F.A.; Sabri, S.; Ismail, M.; Chan, K.W.; Ismail, N.; Mohd Esa, N.; Mohd Lila, M.A.; Zawawi, N. Probiotic properties of Bacillus strains isolated from stingless bee (Heterotrigona itama) honey collected across Malaysia. Int. J. Environ. Res. Public Health 2020, 17, 278. [Google Scholar] [CrossRef]
- Kalinowski, J.; Ahrens, B.; Al-Dilaimi, A.; Winkler, A.; Wibberg, D.; Schleenbecker, U.; Rückert, C.; Wölfel, R.; Grass, G. Isolation and whole genome analysis of endospore-forming bacteria from heroin. Forensic Sci. Int. Genet. 2018, 32, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.B.Y.; Junqueira, A.C.M.; Uchida, A.; Purbojati, R.W.; Houghton, J.N.I.; Chénard, C.; Wong, A.; Kolundžija, S.; Clare, M.E.; Kushwaha, K.K.; et al. Genome sequence of Bacillus velezensis SGAir0473, isolated from tropical air collected in Singapore. Genome Announc. 2018, 6, e00642-18. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Cui, P.; Chen, X. Complete genome of Bacillus sp. Pc3 isolated from the Antarctic seawater with antimicrobial activity. Mar. Genomics 2015, 20, 1–2. [Google Scholar] [CrossRef]
- Agersø, Y.; Stuer-Lauridsen, B.; Bjerre, K.; Jensen, M.G.; Johansen, E.; Bennedsen, M.; Brockmann, E.; Nielsen, B. Antimicrobial susceptibility testing and tentative epidemiological cutoff values for five Bacillus species relevant for use as animal feed additives or for plant protection. Appl. Enviromental Microbiol. 2018, 84, e01108-18. [Google Scholar]
- Pan, H.Q.; Li, Q.L.; Hu, J.C. The complete genome sequence of Bacillus velezensis 9912D reveals its biocontrol mechanism as a novel commercial biological fungicide agent. J. Biotechnol. 2017, 247, 25–28. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhou, J.; Tang, Y.; Ma, Q.; Zhang, J.; Ji, C.; Zhao, L. Characterization and genome analysis of a zearalenone-degrading Bacillus velezensis strain ANSB01E. Curr. Microbiol. 2020, 77, 273–278. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Xu, G.; Jin, Y.; Sun, C.; Zhou, L.; Lin, G.; Xu, R.; Wei, L.; Fei, H.; Wang, D.; et al. Isolation and characterization of Bacillus sp. GFP-2, a novel Bacillus strain with antimicrobial activities, from whitespotted bamboo shark intestine. AMB Express 2018, 8, 84. [Google Scholar] [CrossRef]
- Molinatto, G.; Franzil, L.; Steels, S.; Puopolo, G.; Pertot, I.; Ongena, M. Key impact of an uncommon plasmid on Bacillus amyloliquefaciens subsp. plantarum S499 developmental traits and lipopeptide production. Front. Microbiol. 2017, 8, 17. [Google Scholar] [CrossRef]
- Feng, J.; Gu, Y.; Wang, J.; Song, C.; Yang, C.; Xie, H.; Zhang, W.; Wang, S. Curing the plasmid pMC1 from the poly (γ-glutamic acid) producing Bacillus amyloliquefaciens LL3 strain using plasmid incompatibility. Appl. Biochem. Biotechnol. 2013, 171, 532–542. [Google Scholar] [CrossRef]
- Chun, B.H.; Kim, K.H.; Jeong, S.E.; Jeon, C.O. Genomic and metabolic features of the Bacillus amyloliquefaciens group– B. amyloliquefaciens, B. velezensis, and B. siamensis– revealed by pan-genome analysis. Food Microbiol. 2019, 77, 146–157. [Google Scholar] [CrossRef]
- Belbahri, L.; Chenari Bouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Petrovova, E.; Oszako, T.; Cherrad, S.; Vacher, S.; et al. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, B.K.; Solanki, M.K.; Pandey, A.K.; Prabha, S.; Kumar, P.; Kumari, B. Bacillus as plant growth promoting rhizobacteria (PGPR): A promising green agriculture technology. In Plant Health under Biotic Stress; Springer Nature Singapore Pte Ltd.: Singapore, 2019; pp. 219–236. [Google Scholar]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42—A Review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef] [PubMed]
- Sibponkrung, S.; Kondo, T.; Tanaka, K.; Tittabutr, P.; Boonkerd, N.; Yoshida, K.I.; Teaumroong, N. Co-inoculation of Bacillus velezensis strain S141 and Bradyrhizobium strains promotes nodule growth and nitrogen fixation. Microorganisms 2020, 8, 678. [Google Scholar] [CrossRef]
- Kim, S.Y.; Song, H.; Sang, M.K.; Weon, H.Y.; Song, J. The Complete genome sequence of Bacillus velezensis strain GH1-13 reveals agriculturally beneficial properties and a unique plasmid. J. Biotechnol. 2017, 259, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Jin, Q.; Lin, Y.; Lu, W.; Li, S.; Zhou, C.; Jin, J.; Jiang, Q.; Ling, L.; Xiao, M. Cell-free fermentation broth of Bacillus velezensis strain S3-1 improves pak choi nutritional quality and changes the bacterial community structure of the rhizosphere soil. Front. Microbiol. 2020, 11, 2043. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, D.; Qi, G.; Mao, Z.; Hu, X.; Du, B.; Liu, K.; Ding, Y. Effects of Bacillus velezensis FKM10 for promoting the growth of Malus hupehensis rehd. and inhibiting Fusarium verticillioides. Front. Microbiol. 2020, 10, 2889. [Google Scholar] [CrossRef]
- Kumar, A.; Kumar, R.; Kumari, M.; Goldar, S. Enhancement of plant growth by using PGPR for a sustainable agriculture: A review. Int. J. Curr. Microbiol. Appl. Sci. 2020, 9, 152–165. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Uhl, J.; Grosch, R.; Alquéres, S.; Pittroff, S.; Dietel, K.; Schmitt-Kopplin, P.; Borriss, R.; Hartmann, A. Cyclic lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol. Plant-Microbe Interact. 2015, 28, 984–995. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Li, Q.; Xu, Z.; Zhang, N.; Shen, Q.; Zhang, R. Responses of beneficial Bacillus amyloliquefaciens SQR9 to different soilborne fungal pathogens through the alteration of antifungal compounds production. Front. Microbiol. 2014, 5, 636. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The Gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef]
- Doornbos, R.F.; van Loon, L.C.; Bakker, P.A. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere. a review. Agron. Sustain. Dev. 2012, 32, 227–243. [Google Scholar] [CrossRef]
- Erlacher, A.; Cardinale, M.; Grosch, R.; Grube, M.; Berg, G. The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome. Front. Microbiol. 2014, 5, 175. [Google Scholar] [CrossRef]
- Wang, S.; Wu, H.; Qiao, J.; Ma, L.; Liu, J.; Xia, Y.; Gao, X. Molecular mechanism of plant growth promotion and induced systemic resistance to Tobacco Mosaic Virus by Bacillus spp. J. Microbiol. Biotechnol. 2009, 19, 1250–1258. [Google Scholar] [CrossRef]
- Jeong, H.; Jeong, D.E.; Kim, S.H.; Song, G.C.; Park, S.Y.; Ryu, C.M.; Park, S.H.; Choi, S.K. Draft genome sequence of the plant growth-promoting bacterium Bacillus siamensis KCTC 13613T. J. Bacteriol. 2012, 194, 4148–4149. [Google Scholar] [CrossRef]
- Laird, M.; Piccoli, D.; Weselowski, B.; McDowell, T.; Renaud, J.; MacDonald, J.; Yuan, Z.C. Surfactin-producing Bacillus velezensis 1B-23 and Bacillus sp. 1D-12 protect tomato against bacterial canker caused by Clavibacter michiganensis subsp. michiganensis. J. Plant Pathol. 2019, 102, 451–458. [Google Scholar] [CrossRef]
- Douriet-Gámez, N.R.; Maldonado-Mendoza, I.E.; Ibarra-Laclette, E.; Blom, J.; Calderón-Vázquez, C.L. Genomic analysis of Bacillus sp. strain B25, a biocontrol agent of maize pathogen Fusarium verticillioides. Curr. Microbiol. 2018, 75, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Dutta, S.; Surovy, M.Z.; Gupta, D.R.; Mahmud, N.U.; Chanclud, E.; Win, J.; Kamoun, S.; Islam, T. Genomic analyses reveal that biocontrol of wheat blast by Bacillus spp. may be linked with production of antimicrobial compounds and induced systemic resistance in host plants. Figshare 2018, 17, 1–48. [Google Scholar]
- Cheng, M.; Xu, Q.; Li, Y.; Qin, H.; Chen, J. Antifungal activity and identification of active compounds of Bacillus amyloliquefaciens subsp. plantarum against Botryosphaeria dothidea. For. Pathol. 2016, 46, 561–568. [Google Scholar] [CrossRef]
- Köberl, M.; White, R.A.; Erschen, S.; Spanberger, N.; El-Arabi, T.F.; Jansson, J.K.; Berg, G. Complete genome sequence of Bacillus amyloliquefaciens strain Co1-6, a plant growth-promoting rhizobacterium of Calendula officinalis. Genome Announc. 2015, 3, e00862-15. [Google Scholar] [CrossRef]
- Zhang, J.X.; Gu, Y.B.; Chi, F.M.; Ji, Z.R.; Wu, J.Y.; Dong, Q.L.; Zhou, Z.S. Bacillus amyloliquefaciens GB1 can effectively control apple valsa canker. Biol. Control 2015, 88, 1–7. [Google Scholar] [CrossRef]
- Ma, J.; Liu, H.; Liu, K.; Wang, C.; Li, Y.; Hou, Q.; Yao, L.; Cui, Y.; Zhang, T.; Wang, H.; et al. Complete genome sequence of Bacillus velezensis GQJK49, a plant growth- promoting rhizobacterium with antifungal activity. Genome Announc. 2017, 5, e00922-17. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Jeon, B.J.; Han, J.W.; Park, M.Y.; Kang, S.A.; Kim, B.S. Evaluation of the endophytic nature of Bacillus amyloliquefaciens strain GYL4 and its efficacy in the control of anthracnose. Pest Manag. Sci. 2016, 72, 1529–1536. [Google Scholar] [CrossRef]
- Jia, Z.; Jin, W.; Huang, Y.; Song, S. Complete genome sequence of Bacillus subtilis J-5, a potential biocontrol agent. Genome Announc. 2017, 5, e00275-17. [Google Scholar] [CrossRef] [PubMed]
- Jing, R.; Li, N.; Wang, W.; Liu, Y. An endophytic strain JK of genus Bacillus isolated from the seeds of super hybrid rice (Oryza sativa L., Shenliangyou 5814) has antagonistic activity against rice blast pathogen. Microb. Pathog. 2020, 147, 104422. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Cui, J.; Jia, X.; Wang, W. Complete genome sequence of Bacillus velezensis L-1, which has antagonistic activity against pear diseases. Genome Announc. 2017, 5, e01271-17. [Google Scholar] [CrossRef]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A Comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef]
- Lee, S.Y.; Kim, B.Y.; Ahn, J.H.; Song, J.; Seol, Y.J.; Kim, W.G.; Weon, H.Y. Draft genome sequence of the biocontrol bacterium Bacillus amyloliquefaciens strain M27. J. Bacteriol. 2012, 194, 6934–6935. [Google Scholar] [CrossRef]
- Zhang, Y.; Gao, X.; Wang, S.; Zhu, C.; Li, R.; Shen, Q. Application of Bacillus velezensis NJAU-Z9 enhanced plant growth associated with efficient rhizospheric colonization monitored by QPCR with primers designed from the whole genome sequence. Curr. Microbiol. 2018, 75, 1574–1583. [Google Scholar] [CrossRef]
- Yuan, J.; Raza, W.; Shen, Q.; Huang, Q. Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl. Environ. Microbiol. 2012, 78, 5942–5944. [Google Scholar] [CrossRef]
- Cheffi, M.; Bouket, A.C.; Alenezi, F.N.; Luptakova, L.; Belka, M.; Vallat, A.; Rateb, M.E.; Tounsi, S.; Triki, M.A.; Belbahri, L. Olea Europaea L. Root endophyte Bacillus velezensis OEE1 counteracts oomycete and fungal harmful pathogens and harbours a large repertoire of secreted and volatile metabolites and beneficial functional genes. Microorganisms 2019, 7, 314. [Google Scholar] [CrossRef] [PubMed]
- Rathna, V. Exploiting the Bio Control Potentiality of Bacillus velezensis P42 and A6 Strains Against Important Bacterial Wilt and Early Blight Diseases of Tomato. Master’s Thesis, University of Agricultural Sciences, GKVK, Bengaluru, India, 2018. [Google Scholar]
- Nelson, B.A.; Ramaiya, P.; de Leon, A.L.; Kumar, R.; Crinklaw, A.; Jolkovsky, E.; Crane, J.M.; Bergstrom, G.C.; Rey, M.W. Complete genome sequence for the Fusarium head blight antagonist Bacillus amyloliquefaciens strain TrigoCor 1448. Genome Announc. 2014, 2, e00219-14. [Google Scholar] [CrossRef]
- Niazi, A.; Manzoor, S.; Asari, S.; Bejai, S.; Meijer, J.; Bongcam-Rudloff, E. Genome analysis of Bacillus amyloliquefaciens subsp. plantarum UCMB5113: A rhizobacterium that improves plant growth and stress management. PLoS ONE 2014, 9, e104651. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Hsiang, T.; Zhou, Y.; Zhou, J. Draft genome sequence of Bacillus amyloliquefaciens XK-4-1, a plant growth-promoting endophyte with antifungal activity. Genome Announc. 2015, 3, e01306-15. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Xie, X.; Zhao, Y.; Shi, Y.; Chai, A.; Li, L.; Li, B. Whole-genome analysis of Bacillus velezensis ZF2, a biocontrol agent that protects Cucumis sativus against corynespora leaf spot diseases. 3 Biotech 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- David Paul Raj, R.S.; Beena Kanimozhi, R.; Gomez, L.A.; Rohini, S. Evaluation of biocontrol efficacy of herbal and bioformulations against root rot pathogen Fusarium solani in tomato. Int. J. Recent Technol. Eng. 2019, 8, 5619–5625. [Google Scholar]
- De Curtis, F.; Ianiri, G.; Raiola, A.; Ritieni, A.; Succi, M.; Tremonte, P.; Castoria, R. Integration of biological and chemical control of brown rot of stone fruits to reduce disease incidence on fruits and minimize fungicide residues in juice. Crop Prot. 2019, 119, 158–165. [Google Scholar] [CrossRef]
- Pethybridge, S.J.; Gugino, B.K.; Kikkert, J.R. Efficacy of Double Nickel LC (Bacillus amyloliquefaciens D747 Strain) for management of white mold in snap and dry bean. Plant Health Prog. 2019, 20, 61–66. [Google Scholar] [CrossRef]
- Burkett-Cadena, M.; Kokalis-Burelle, N.; Lawrence, K.S.; Van Santen, E.; Kloepper, J.W. Suppressiveness of root-knot nematodes mediated by rhizobacteria. Biol. Control 2008, 47, 55–59. [Google Scholar] [CrossRef]
- Chen, X.H.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth–promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.Y.; Min, Y.; Wang, K.M.; Wan, Z.Y.; Zhang, Z.G.; Cao, C.X.; Zhou, R.H.; Jiang, A.B.; Liu, C.J.; Zhang, G.Y.; et al. Draft genome sequence of Bacillus amyloliquefaciens HB-26. Stand. Genomic Sci. 2014, 9, 775–782. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Devaraj, K.; Aathika, S.; Periyasamy, K.; Periyaraman, P.M.; Palaniyandi, S.; Subramanian, S. Production of thermostable multiple enzymes from Bacillus amyloliquefaciens KUB29. Nat. Prod. Res. 2019, 33, 1674–1677. [Google Scholar] [CrossRef]
- Kalawong, R.; Wakayama, M.; Anuntalabhochai, S.; Wongsawad, C.; Sangwijit, K. Comparison and characterization of purified cellulase and xylanase from Bacillus amyloliquefaciens CX1 and Bacillus subtilis B4. Chiang Mai J. Sci. 2018, 45, 92–105. [Google Scholar]
- Farhat-Khemakhem, A.; Blibech, M.; Boukhris, I.; Makni, M.; Chouayekh, H. Assessment of the potential of the multi-enzyme producer Bacillus amyloliquefaciens US573 as alternative feed additive. J. Sci. Food Agric. 2018, 98, 1208–1215. [Google Scholar] [CrossRef]
- Sewalt, V.; Shanahan, D.; Gregg, L.; La Marta, J.; Carrillo, R. The Generally Recognized as Safe (GRAS) process for industrial microbial enzymes. Ind. Biotechnol. 2016, 12, 295–302. [Google Scholar] [CrossRef]
- Prajapati, V.S.; Ray, S.; Narayan, J.; Joshi, C.C.; Patel, K.C.; Trivedi, U.B.; Patel, R.M. Draft genome sequence of a thermostable, alkaliphilic α-amylase and protease producing Bacillus amyloliquefaciens strain KCP2. 3 Biotech 2017, 7, 372. [Google Scholar] [CrossRef]
- Meier, M.J.; Dodge, A.; Beaudette, L.A. Draft genome sequence of the industrially significant bacterium Bacillus amyloliquefaciens NRRL 942 Matthew. Microbiol. Resour. Announc. 2018, 7, e01374-18. [Google Scholar]
- Montor-Antonio, J.J.; Sachman-Ruiz, B.; Lozano, L.; del Moral, S. Draft genome sequence of Bacillus amyloliquefaciens JJC33M, isolated from sugarcane soils in the papaloapan region, Mexico. Genome Announc. 2015, 3, e01519-14. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Gu, W.; Xu, H.-Y.; Yang, G.L.; Shan, X.F.; Chen, G.; Wang, C.F.; Qian, A.D. Complete genome sequence of Bacillus velezensis 157 isolated from Eucommia ulmoides with pathogenic bacteria inhibiting and lignocellulolytic enzymes production by SSF. 3 Biotech 2018, 8, 114. [Google Scholar] [CrossRef] [PubMed]
- Gong, G.; Kim, S.; Lee, S.M.; Woo, H.M.; Park, T.H.; Um, Y. Complete genome sequence of Bacillus sp. 275, producing extracellular cellulolytic, xylanolytic and ligninolytic enzymes. J. Biotechnol. 2017, 254, 59–62. [Google Scholar] [CrossRef]
- Hassan, M. The Role of Pectin Utilization in Root Colonization and Plant Growth-Promotion by Bacillus amyloliquefaciens subsp. plantarum (Bap). Master’s Thesis, Auburn University, Auburn, ME, USA, 2016. [Google Scholar]
- Das, R.; Liang, Z.; Li, G.; Mai, B.; An, T. Genome sequence of a spore-laccase forming, BPA-degrading Bacillus sp. GZB isolated from an electronic-waste recycling site reveals insights into BPA degradation pathways. Arch. Microbiol. 2019, 201, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.Y.; Chun, B.H.; Moon, J.Y.; Yeo, S.H.; Jeon, C.O. Complete genome sequence of Bacillus methylotrophicus JJ-D34 isolated from deonjang, a korean traditional fermented soybean paste. J. Biotechnol. 2016, 219, 36–37. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Yang, L.; Li, X.; Li, H.; Tu, Z.; Wang, X. Genome sequencing, purification, and biochemical characterization of a strongly fibrinolytic enzyme from Bacillus amyloliquefaciens Jxnuwx-1 isolated from chinese traditional douchi. J. Gen. Appl. Microbiol. 2019. in Press. [Google Scholar] [CrossRef] [PubMed]
- Marasini, D.; Cornell, C.R.; Oyewole, O.; Sheaff, R.J.; Fakhr, M.K. The whole-genome sequence of Bacillus velezensis strain SB1216 isolated from the great salt plains of Oklahoma reveals the presence of a novel extracellular RNase with antitumor activity. Genome Announc. 2017, 5, e01343-17. [Google Scholar] [CrossRef]
- Song, P.; Xu, X.; Jiang, L.; Zhang, R.; Wang, J.; Xu, Q.; Li, S. Genome sequence of Bacillus subtilis SPZ1, an evolved strain for higher uptake rate of tributyrin. Genome Announc. 2013, 1, e00511-13. [Google Scholar] [CrossRef]
- Zhai, L.; Ren, R.; Meng, D.; Tian, Q.; Guan, Z.; Cai, Y.; Liao, X. Comparison of aminotransferases of three Bacillus strains Bacillus altitudinis W3, Bacillus velezensis SYBC H47, and Bacillus amyloliquefaciens YP6 via genome analysis and bioinformatics. J. Appl. Genet. 2019, 60, 427–430. [Google Scholar] [CrossRef]
- Wu, L.; Li, X.; Ma, L.; Blom, J.; Wu, H.; Gu, Q.; Borriss, R.; Gao, X. The “pseudo-pathogenic” effect of plant growth-promoting bacilli on starchy plant storage organs is due to their α-amylase activity which is stimulating endogenous opportunistic pathogens. Appl. Microbiol. Biotechnol. 2020, 104, 2701–2714. [Google Scholar] [CrossRef]
- Clatworthy, A.E.; Pierson, E.; Hung, D.T. Targeting virulence: A new paradigm for antimicrobial therapy. Nat. Chem. Biol. 2007, 3, 541–548. [Google Scholar] [CrossRef]
- Fazle Rabbee, M.; Baek, K.H. Antimicrobial activities of lipopeptides and polyketides of Bacillus velezensis for agricultural applications. Molecules 2020, 25, 4973. [Google Scholar] [CrossRef]
- Perlman, D.; Bodanszey, M. Biosynthesis of peptide antibiotics. Annu. Rev. Microbiol. 1971, 40, 449–464. [Google Scholar] [CrossRef]
- Hancock, R.E.; Chapple, D.S. Peptide antibiotics. Antimicrob. Agents Chemother. 1999, 43, 1317–1323. [Google Scholar] [CrossRef] [PubMed]
- Grady, E.N.; MacDonald, J.; Ho, M.T.; Weselowski, B.; McDowell, T.; Solomon, O.; Renaud, J.; Yuan, Z.C. Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol. 2019, 19, 5. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.H.; Lu, Y.Q.; Ye, Z.W.; Zheng, Q.W.; Wei, T.; Lin, J.F.; Guo, L.Q. Genomics-guided discovery and structure identification of cyclic lipopeptides from the Bacillus siamensis JFL15. PLoS ONE 2018, 13, e0202893. [Google Scholar] [CrossRef]
- Abdelhamid, A.G.; Hussein, W.E.; Gerst, M.M.; Yousef, A.E. Draft genome sequence of Bacillus velezensis OSY-GA1, which encodes multiple antimicrobial metabolites and expresses antimicrobial activity against foodborne pathogens. Microbiol. Resour. Announc. 2019, 8, e01725-18. [Google Scholar] [CrossRef] [PubMed]
- Huffman, J.; Gerber, R.; Du, L. Recent advancements in the biosynthetic mechanisms for polyketide-derived mycotoxins. Biopolymers 2010, 93, 764–776. [Google Scholar] [CrossRef] [PubMed]
- Weissman, K.J. Introduction to polyketide biosynthesis. Methods Enzymol. 2009, 459, 3–16. [Google Scholar] [PubMed]
- Gomes, E.S.; Schuch, V.; Lemos, E.G.D.M. Biotechnology of polyketides: New breath of life for the novel antibiotic genetic pathways discovery through metagenomics. Brazilian J. Microbiol. 2013, 44, 1007–1034. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Chun, B.H.; Jeon, H.H.; Kim, Y.B.; Lee, S.H. Complete genome sequence of Bacillus velezensis YJ11-1-4, a strain with broadspectrum antimicrobial activity, isolated from traditional korean fermented soybean paste. Genome Announc. 2017, 5, e01352-17. [Google Scholar] [CrossRef] [PubMed]
- Zheng, C.J.; Lee, S.; Lee, C.H.; Kim, W.G. Macrolactins O–R, glycosylated 24-membered lactones from Bacillus sp. AH159-1. J. Nat. Prod. 2007, 70, 1632–1635. [Google Scholar] [CrossRef] [PubMed]
- Kechagia, M.; Basoulis, D.; Konstantopoulou, S.; Dimitriadi, D.; Gyftopoulou, K.; Skarmoutsou, N.; Fakiri, E.M. Health benefits of probiotics: A review. ISRN Nutr. 2013, 2013, 7. [Google Scholar] [CrossRef] [PubMed]
- Ryu, M.S.; Yang, H.J.; Jeong, S.J.; Seo, J.W.; Ha, G.; Jeong, S.Y.; Jeong, D.Y. Characteristic study and optimization of culture conditions for Bacillus amyloliquefaciens SRCM 100731 as probiotic resource for companion animal. Microbiol. Soc. Korea 2018, 54, 384–397. [Google Scholar]
- Brown, S.; Dart, P. Testing Hay Treated with Mould-Inhibiting, Biocontrol Inoculum; Rural Industries Research and Development Corporation: Canberra, Australia, 2005.
- Pereira, J.Q.; Ritter, A.C.; Cibulski, S.; Brandelli, A. Functional genome annotation depicts probiotic properties of Bacillus velezensis FTC01. Gene 2019, 713, 143971. [Google Scholar] [CrossRef] [PubMed]
- Karlyshev, A.V.; Melnikov, V.G.; Chistyakov, V.A. Draft genome sequence of Bacillus amyloliquefaciens B-1895. Genome Announc. 2014, 2, e00633-14. [Google Scholar] [CrossRef]
- Golovko, G.; Zipelt, L.; Karpenko, G.; Chistyakov, V.; Sazykina, M.; Kolenko, M. Method for Growth of Young Azov-Chernomorskaya Royal Fish in Ponds. RU Patent No. 2,376,755, 23 July 2008. [Google Scholar]
- AlGburi, A.; Volski, A.; Cugini, C.; Walsh, E.M.; Chistyakov, V.A.; Mazanko, M.S.; Bren, A.B.; Dicks, L.M.T.; Chikindas, M.L. Safety properties and probiotic potential of Bacillus subtilis KATMIRA1933 and Bacillus amyloliquefaciens B-1895. Adv. Microbiol. 2016, 6, 432–452. [Google Scholar] [CrossRef]
- Yi, Y.; Zhang, Z.; Zhao, F.; Liu, H.; Yu, L.; Zha, J.; Wang, G. Probiotic potential of Bacillus velezensis JW: Antimicrobial activity against fish pathogenic bacteria and immune enhancement effects on Carassius auratus. Fish Shellfish Immunol. 2018, 78, 322–330. [Google Scholar] [CrossRef]
- Gao, X.Y.; Liu, Y.; Miao, L.L.; Li, E.W.; Sun, G.X.; Liu, Y.; Liu, Z.P. Characterization and mechanism of anti-Aeromonas salmonicida activity of a marine probiotic strain, Bacillus velezensis V4. Appl. Microbiol. Biotechnol. 2017, 101, 3759–3768. [Google Scholar] [CrossRef] [PubMed]
- Llario, F.; Romano, L.A.; Rodilla, M.; Sebastiá-Frasquet, M.T.; Poersch, L.H. Application of Bacillus amyloliquefaciens as probiotic for Litopenaeus vannamei (Boone, 1931) cultivated in a biofloc system. Iran. J. Fish. Sci. 2020, 19, 904–920. [Google Scholar]
- Lin, Y.S.; Saputra, F.; Chen, Y.C.; Hu, S.Y. Dietary administration of Bacillus amyloliquefaciens R8 reduces hepatic oxidative stress and enhances nutrient metabolism and immunity against Aeromonas hydrophila and Streptococcus agalactiae in zebrafish (Danio rerio). Fish Shellfish Immunol. 2019, 86, 410–419. [Google Scholar] [CrossRef]
- Al-Deriny, S.H.; Dawood, M.A.; Abou Zaid, A.A.; Wael, F.; Paray, B.A.; Van Doan, H.; Mohamed, R.A. The synergistic effects of Spirulina platensis and Bacillus amyloliquefaciens on the growth performance, intestinal histomorphology, and immune response of Nile tilapia (Oreochromis niloticus). Aquacult. Repot. 2020, 17, 100390. [Google Scholar] [CrossRef]
- Chauhan, A.; Singh, R. Probiotics in aquaculture: A promising emerging alternative approach. Symbiosis 2019, 77, 99–113. [Google Scholar] [CrossRef]
- Azrin, N.A.R.; Yuzine, E.; Ina-Salwany, M.Y.; Karim, M. The Efficacy of Potential Probiont Bacillus amyloliquefaciens Strain L11 in Protecting Artemia Nauplii and Blue Crab Juveniles against Vibrio harveyi Infection. J. Pure Appl. Microbiol. 2019, 13, 923–931. [Google Scholar]
- Abatenh, E.; Gizaw, B.; Tsegaye, Z.; Wassie, M. The role of microorganisms in bioremediation-a review. Open J. Environ. Biol. 2017, 1, 38–46. [Google Scholar] [CrossRef]
- Meng, D.; Zhai, L.X.; Tian, Q.P.; Guan, Z.B.; Cai, Y.J.; Liao, X.R. Complete genome sequence of Bacillus amyloliquefaciens YP6, a plant growth rhizobacterium efficiently degrading a wide range of organophosphorus pesticides. J. Integr. Agric. 2019, 18, 2668–2672. [Google Scholar] [CrossRef]

| Characterization | B. amyloliquefaciens | B. siamensis | B. velezensis | B. nakamurai | |
|---|---|---|---|---|---|
| Type Strain | DSM 7T / ATCC 23350T / FT | KCTC 13613T / PD-A10T / BCC 22614T | NRRL B-23189T / CR-502T / CECT 5686T / LMG 22478T | NRRL B-41091T / CCUG 68786T | |
| Isolation Source | Soil and industrial α-amylase fermentations | Salted crab (poo-khem) in Thailand | Brackish water sample from the river Velez at Torredelmar in Ma’laga, southern Spain | Soil in Tierra del Fuego, Argentina | |
| Size | 0.7–0.9 × 1.8–3.0 µm | 0.3–0.6 × 1.5–3.5 µm | 0.5 × 1.5–3.5 µm | 0.74–0.93 × 1.39–2.04 µm | |
| Endospore | Oval spores are central or paracentral in unswollen sporangia | Ellipsoidal spores are central or sub-terminal positions in swollen sporangia | Ellipsoidal spores are paracentral or sub-terminal positions in unswollen sporangia | Ellipsoidal spores are central in unswollen sporangia | |
| G + C Content (mol %) | 44.6 | 41.4 | 46.1–46.4 | 43.8 | |
| Growth Temperature | Optimal growth temperature is 30–40 °C. No growth occurs below 15 °C or above 50 °C. | Optimal growth temperature is 37 °C. Growth occurs at 4 °C and 55 °C. | Grow within the temperature range of 15–45 °C | Grow within the temperature range of 17–50 °C, with an optimum of 37 °C | |
| NaCl Resistance (w/v) | Growth occurs with 0–10% NaCl | Growth occurs with 0–14% NaCl | Growth occurs with 0–12% NaCl | Growth occurs with 0–9% NaCl | |
| Substrate Utilization | Tyrosine | - | - | - | + |
| Citrate | + | - | - | + | |
| Fermentation (acid) | Lactose | + | + | + | - |
| Trehalose | + | - | + | + | |
| Reference | [1] | [16] | [17] | [18] | |
| PGPB Strain | Disease and Pathogen | Plant Species | Reference |
|---|---|---|---|
| B. siamensis KCTC 13613 | R. solani Botrytis cinerea Micrococcus luteus | Arabidopsis thaliana | [59] |
| B. velezensis 83 | Anthracnose disease | Zea mays A. thaliana | [20] |
| B. velezensis 1B-23 | Clavibacter michiganensis subsp. michiganensis | Solanum lycopersicum | [60] |
| B. velezensis B25 | Fusarium verticillioides | Z. mays | [61] |
| B. velezensis BTLK6A | Magnaporthe oryzae Triticum | Triticum aestivum | [62] |
| B. velezensis BTS 4 | |||
| B. velezensis CC09 | Powdery mildew disease | T. aestivum | [28] |
| B. velezensis CGMCC 11640 | Botryosphaeria dothidea | Carya cathayensis | [63] |
| B. velezensis Co1-6 | Verticillium dahliae R. solani Fusarium culmorum Ralstonia solanacearum | Matricaria chamomilla | [64] |
| B. velezensis GB1 | Valsa mali | Malus domestica | [65] |
| B. velezensis GH1-13 | Fusarium fujikuroi R. solani Xanthmonas oryzae | Oryza sativa | [49] |
| B. velezensis GQJK49 | F. solani | Lycium barbarum L. | [66] |
| B. velezensis GYL4 | Anthracnose disease | Cucumis sativus L. cv. Chunsim | [67] |
| B. velezensis J-5 | B. cinerea | S. lycopersicum | [68] |
| B. velezensis JK | M. oryzae | O. sativa | [69] |
| B. velezensis L-1 | Botryosphaeria berengeriana | Pyrus communis | [70] |
| B. velezensis LM2303 | Fusarium graminearum | T. aestivum | [71] |
| B. velezensis M27 | Sclerotinia sclerotiorum | Lactuca sativa L. | [72] |
| B. velezensis NJAU-Z9 | Fusarium oxysporum f. sp. niveum Ralstonia solanacearum | Capsicum annuum L. | [73] |
| B. velezensis NJN-6 | F. oxysporum f. sp. cubense | Musa sp. | [74] |
| B. velezensis OEE1 | F. solani | Olea europaea L. | [75] |
| B. velezensis P42 | Bacterial wilt and early blight diseases | S. lycopersicum | [76] |
| B. velezensis PG12 | Apple ring rot disease | Malus domestica | [24] |
| B. velezensis TrigoCor1448 | Fusarium head blight disease | T. aestivum | [77] |
| B. velezensis UCMB5113 | Alternaria brassicae B. cinerea Leptosphaeria maculans Verticillium longisporum | Brassica napus | [78] |
| B. velezensis XK-4-1 | Verticillium wilt disease | Gossypium sp. | [79] |
| B. velezensis ZF2 | Corynespora leaf spot diseases | C. sativus | [80] |
| Bacterial Strain | Commercial Product | Company | Description |
|---|---|---|---|
| B. velezensis QST 713 (previously B. subtilis QST 713) | SERENADE Max | Bayer Crop Science, previously AgraQuest | EPA-registered biofungicide. Controls and suppresses fungal pathogens on foliage and in the soil |
| SERENADE SOIL® | Bayer Crop Science, previously AgraQuest | EPA-registered biofungicide for food crops | |
| CEASE® | BioWorks, Inc., Victor, New York, U.S.A. | Aqueous suspension biofungicide for leafy and fruiting vegetables, herbs and spices, and ornamentals | |
| B. velezensis FZB42 (previously B. amyloliquefaciens FZB42) | RhizoVital® 42 | ABiTEP GmbH, Berlin, Germany | Biofertilizer, plant-growth-promoting activity, provides protection against various soil-borne diseases |
| FZB24® TB | ABiTEP GmbH, Berlin, Ger-many | Plant growth-promoting agent for plant strengthening | |
| Taegro® | Syngenta, Basel, previously Novozyme, Davis, California, and Earth Biosciences | EPA-registered biofungicide for use in North America | |
| B. velezensis GB03 (previously B. subtilis GB03) | Kodiak™ | Bayer Crop Science, North Carolina, NC | EPA-registered biological seed treatment fungicide with demonstrable PGR activity. Efficient in cotton, beans, and vegetables |
| Companion | Growth Products Ltd., White Plains, NY | EPA-registered biofungicide that prevents and controls plant diseases | |
| B. velezensis D747 (previously B. amyloliquefaciens D747) | Double Nickel 55™ | Certis Columbia, MD, U.S.A. | EPA-registered biofungicide for control or suppression of fungal and bacterial plant |
| Amylo-X® | Certis Columbia, MD USA/Intrachem Bio Italia SpA | Biocontrol of Botrytis and other fungal diseases of grapes, strawberries, and vegetables, and bacterial diseases, such as fire blight in pome fruit and PSA in kiwi fruit |
| Bacterial Species | Enzymes | Reference |
|---|---|---|
| B. amyloliquefaciens KCP2 | α-amylase and protease | [91] |
| B. amyloliquefaciens NRRL 942 | α-amylase | [92] |
| B. siamensis JJC33M | α-amylase | [93] |
| B. velezensis 157 | α-amylase, cellulase, xylanase and pectinase | [94] |
| B. velezensis 275 | Cellulase, xylanase, peroxidase, and laccase | [95] |
| B. velezensis AP194 | Pectinase | [96] |
| B. velezensis AP214 | Pectinase | [96] |
| B. velezensis GZB | Laccase | [97] |
| B. velezensis JJ-D34 | α-amylase, protease and cellulase | [98] |
| B. velezensis Jxnuwx-1 | Protease | [99] |
| B. velezensis SB1216 | Barnase | [100] |
| B. velezensis SPZ1 | Lipase | [101] |
| B. velezensis SYBC H47 | Aminotransferase | [102] |
| B. velezensis ZL918 | α-amylase | [103] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ngalimat, M.S.; Yahaya, R.S.R.; Baharudin, M.M.A.-a.; Yaminudin, S.M.; Karim, M.; Ahmad, S.A.; Sabri, S. A Review on the Biotechnological Applications of the Operational Group Bacillus amyloliquefaciens. Microorganisms 2021, 9, 614. https://doi.org/10.3390/microorganisms9030614
Ngalimat MS, Yahaya RSR, Baharudin MMA-a, Yaminudin SM, Karim M, Ahmad SA, Sabri S. A Review on the Biotechnological Applications of the Operational Group Bacillus amyloliquefaciens. Microorganisms. 2021; 9(3):614. https://doi.org/10.3390/microorganisms9030614
Chicago/Turabian StyleNgalimat, Mohamad Syazwan, Radin Shafierul Radin Yahaya, Mohamad Malik Al-adil Baharudin, Syafiqah Mohd. Yaminudin, Murni Karim, Siti Aqlima Ahmad, and Suriana Sabri. 2021. "A Review on the Biotechnological Applications of the Operational Group Bacillus amyloliquefaciens" Microorganisms 9, no. 3: 614. https://doi.org/10.3390/microorganisms9030614
APA StyleNgalimat, M. S., Yahaya, R. S. R., Baharudin, M. M. A.-a., Yaminudin, S. M., Karim, M., Ahmad, S. A., & Sabri, S. (2021). A Review on the Biotechnological Applications of the Operational Group Bacillus amyloliquefaciens. Microorganisms, 9(3), 614. https://doi.org/10.3390/microorganisms9030614









