Influence of Temperature and Sulfate Concentration on the Sulfate/Sulfite Reduction Prokaryotic Communities in the Tibetan Hot Springs
Abstract
1. Introduction
2. Methods and Materials
2.1. Field Measurements and Sample Collection
2.2. Geochemistry Analyses
2.3. DNA Isolation, PCR Amplification, and Phylogenetic Analyses
2.4. Quantitative Polymerase Chain Reaction (qPCR)
3. Results
3.1. Geochemical Characteristics of the Studied Hot Springs
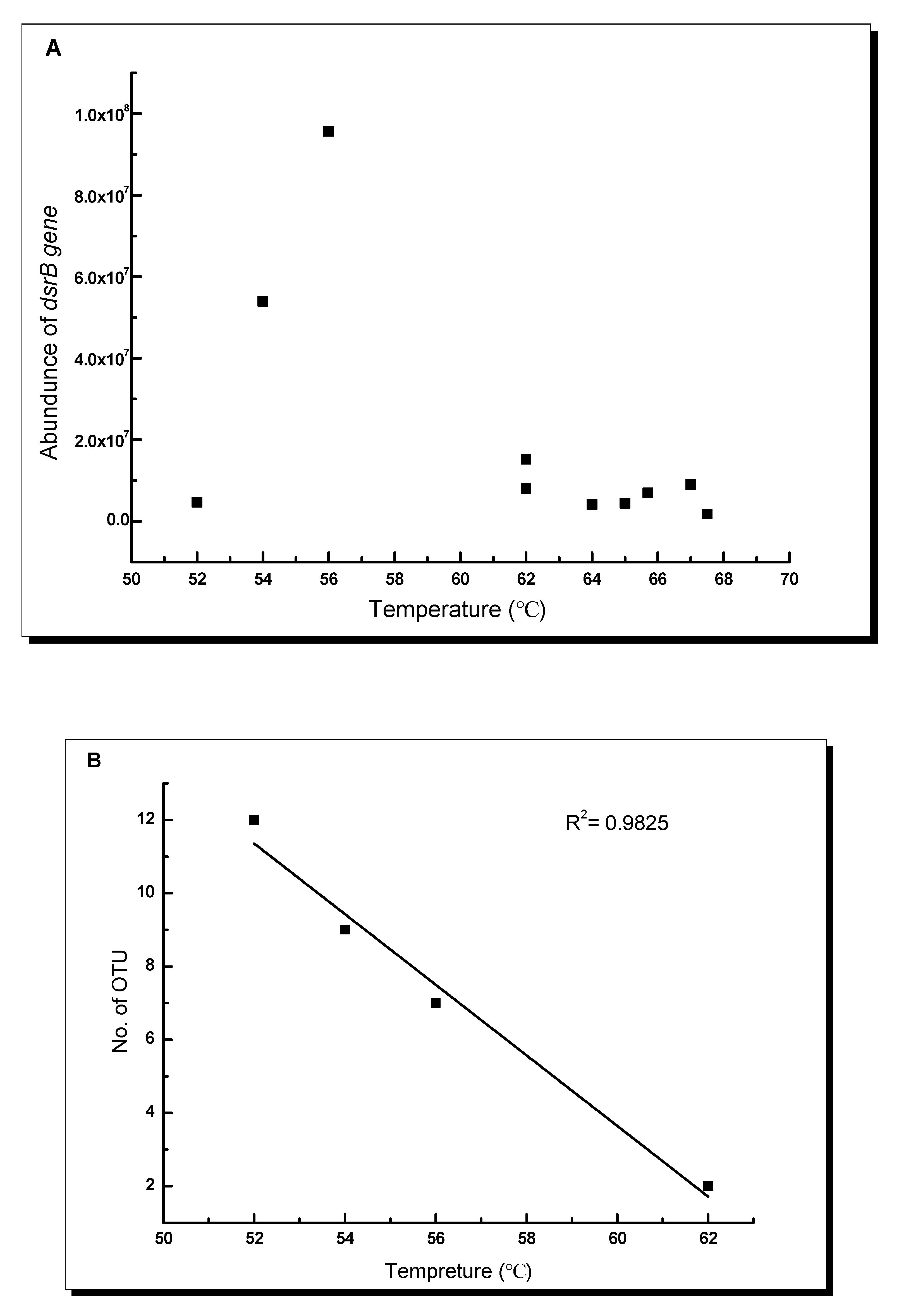
3.2. dsrB Gene Diversity in the Studied Hot Springs
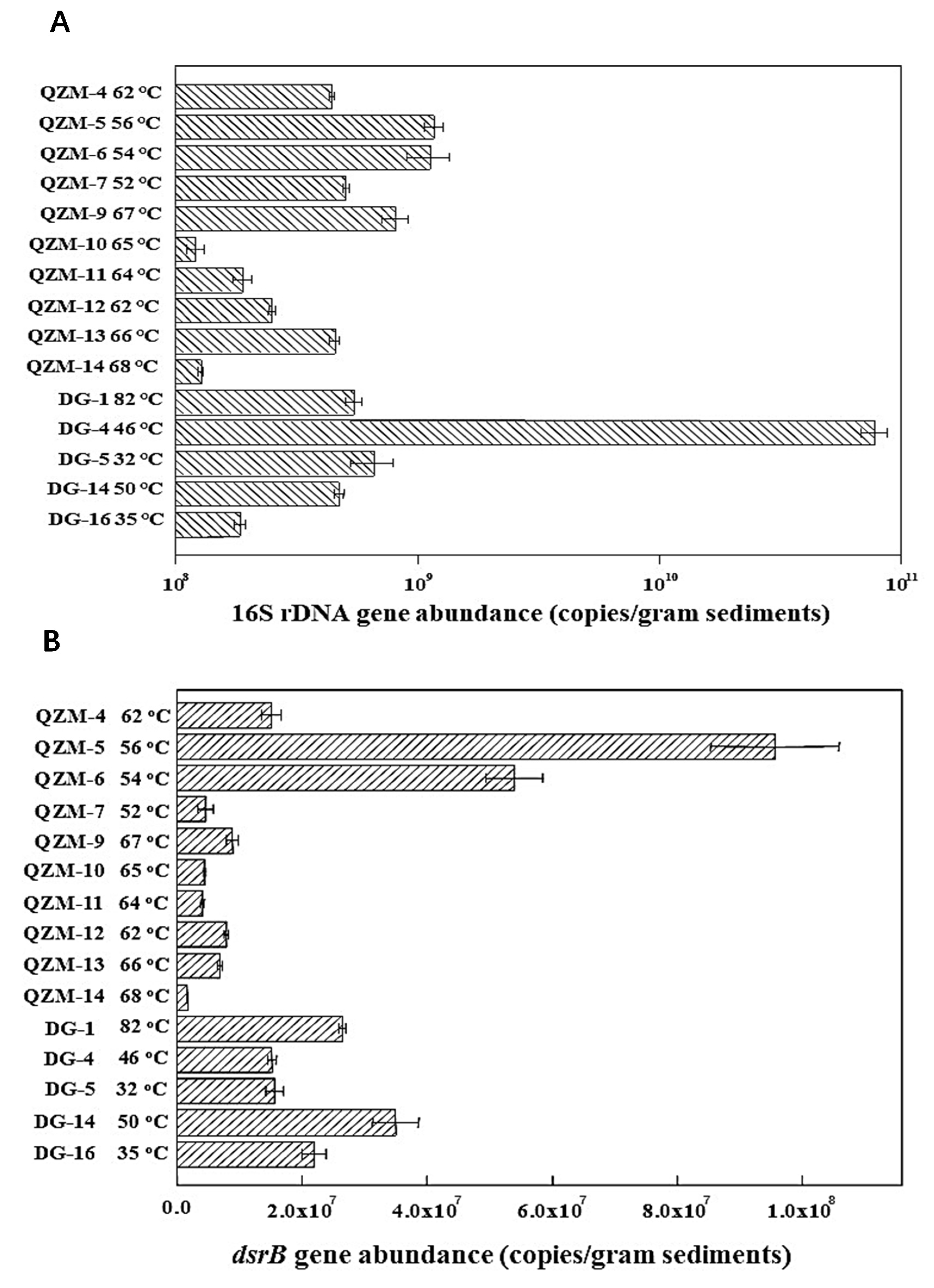
3.3. 16S rRNA and dsrB Gene Abundance in the Studied Hot Springs
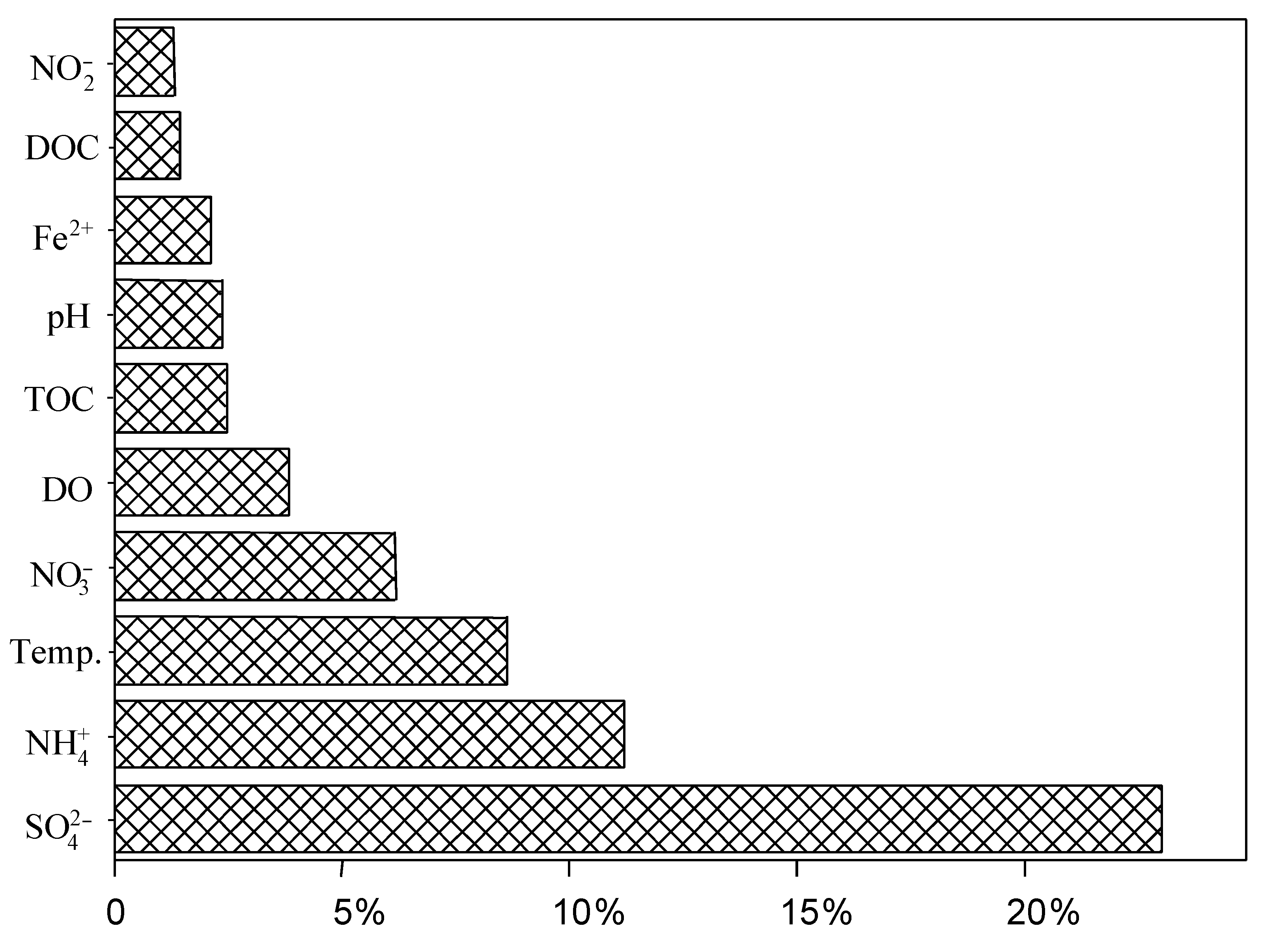
3.4. Impact of Environmental Factors on the Distribution of dsrB Gene in the Studied Hot Springs
4. Discussion
4.1. SRP Diversity in the Tibetan Hot Springs
4.2. Temperature Response of dsrB Gene Diversity and Abundance in the Tibetan Hot Springs
4.3. Sulfate Concentration Was Important Factor Shaping the Distribution of dsrB Gene Diversity in the Tibetan Hot Springs
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cameron, E.M. Sulphate and sulphate reduction in early Precambrian oceans. Nature 1982, 296, 145–148. [Google Scholar] [CrossRef]
- Friedrich, M.W. Phylogenetic Analysis Reveals Multiple Lateral Transfers of Adenosine-5′-Phosphosulfate Reductase Genes among Sulfate-Reducing Microorganisms. J. Bacteriol. 2002, 184, 278. [Google Scholar] [CrossRef] [PubMed]
- Klenk, H.P.; Clayton, R.A.; Tomb, J.F.; White, O.; Nelson, K.E.; Ketchum, K.A.; Dodson, R.J.; Gwinn, M.; Hickey, E.K.; Peterson, J.D.; et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 1997, 390, 364–370. [Google Scholar] [CrossRef]
- Anantharaman, K.; Hausmann, B.; Jungbluth, S.P.; Kantor, R.S.; Lavy, A.; Warren, L.A.; Rappé, M.S.; Pester, M.; Loy, A.; Thomas, B.C.; et al. Expanded diversity of microbial groups that shape the dissimilatory sulfur cycle. ISME J. 2018, 12, 1715–1728. [Google Scholar] [CrossRef] [PubMed]
- Trudinger, P.; Chambers, L.; Smith, J. Low-temperature sulphate reduction: Biological versus abiological. Can. J. Earth Sci. 1985, 22, 1910–1918. [Google Scholar] [CrossRef]
- Rickard, D. Microbial sulfate reduction in sediments. Dev. Sedimentol. 2012, 65, 319–351. [Google Scholar]
- Elderfield, H.; Schultz, A. Mid-ocean ridge hydrothermal fluxes and the chemical composition of the ocean. Annu. Rev. Earth Planet. Sci. 1996, 24, 191–224. [Google Scholar] [CrossRef]
- Andres, R.J.; Kasgnoc, A.D. A time-averaged inventory of subaerial volcanic sulfur emissions. J. Geophys. Res. Atmos. 1998, 103, 25251–25261. [Google Scholar] [CrossRef]
- Picard, A.; Gartman, A.; Girguis, P.R. What do we really know about the role of microorganisms in iron sulfide mineral formation? Front. Earth Sci. 2016, 4, 68. [Google Scholar] [CrossRef]
- Rabus, R.; Hansen, T.A.; Widdel, F. Dissimilatory sulfate-and sulfur-reducing prokaryotes. In The Prokaryotes: Prokaryotic Physiology and Biochemistry; Rosenberg, E., Ed.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 309–404. [Google Scholar] [CrossRef]
- Canfield, D.E.; Stewart, F.J.; Thamdrup, B.; De Brabandere, L.; Dalsgaard, T.; Delong, E.F.; Revsbech, N.P.; Ulloa, O. A cryptic sulfur cycle in oxygen-minimum-zone waters off the Chilean coast. Science 2010, 330, 1375–1378. [Google Scholar] [CrossRef]
- Boetius, A.; Ravenschlag, K.; Schubert, C.J.; Rickert, D.; Widdel, F.; Gieseke, A.; Amann, R.; Jørgensen, B.B.; Witte, U.; Pfannkuche, O. A marine microbial consortium apparently mediating anaerobic oxidation of methane. Nature 2000, 407, 623. [Google Scholar] [CrossRef]
- Wu, G.; Huang, L.; Jiang, H.; Peng, Y.E.; Guo, W.; Chen, Z.; She, W.; Guo, Q.; Dong, H. Thioarsenate formation coupled with anaerobic arsenite oxidation by a sulfate-reducing bacterium isolated from a hot spring. Front. Microbiol. 2017, 8, 1336. [Google Scholar] [CrossRef]
- Wagner, M.; Roger, A.J.; Flax, J.L.; Brusseau, G.A.; Stahl, D.A. Phylogeny of dissimilatory sulfite reductases supports an early origin of sulfate respiration. J. Bacteriol. 1998, 180, 2975–2982. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.; Friedrich, M.; Roger, A.J.; Hugenholtz, P.; Fishbain, S.; Abicht, H.; Blackall, L.L.; Stahl, D.A.; Wagner, M. Multiple lateral transfers of dissimilatory sulfite reductase genes between major lineages of sulfate-reducing prokaryotes. J. Bacteriol. 2001, 183, 6028–6035. [Google Scholar] [CrossRef]
- Li, J.; Purdy, K.J.; Takii, S.; Hayashi, H. Seasonal changes in ribosomal RNA of sulfate-reducing bacteria and sulfate reducing activity in a freshwater lake sediment. FEMS Microbiol. Ecol. 1999, 28, 31–39. [Google Scholar] [CrossRef][Green Version]
- Fishbain, S.; Dillon, J.G.; Gough, H.L.; Stahl, D.A. Linkage of high rates of sulfate reduction in Yellowstone hot springs to unique sequence types in the dissimilatory sulfate respiration pathway. Appl. Environ. Microbiol. 2003, 69, 3663–3667. [Google Scholar] [CrossRef]
- Walker, T.R. Distribution of oxygen, sulfides and optimum temperature for sulfate reduction in Antarctic marine sediments. Pol. Polar Res. 2005, 26, 215–230. [Google Scholar]
- Dhillon, A.; Teske, A.; Dillon, J.; Stahl, D.A.; Sogin, M.L. Molecular characterization of sulfate-reducing bacteria in the Guaymas basin. Appl. Environ. Microbiol. 2003, 69, 2765–2772. [Google Scholar] [CrossRef]
- Leloup, J.; Loy, A.; Knab, N.J.; Borowski, C.; Wagner, M.; Jørgensen, B.B. Diversity and abundance of sulfate-reducing microorganisms in the sulfate and methane zones of a marine sediment, Black Sea. Environ. Microbiol. 2007, 9, 131–142. [Google Scholar] [CrossRef]
- Leloup, J.; Fossing, H.; Kohls, K.; Holmkvist, L.; Borowski, C.; Jørgensen, B.B. Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): Abundance and diversity related to geochemical zonation. Environ. Microbiol. 2009, 11, 1278–1291. [Google Scholar] [CrossRef]
- Quillet, L.; Besaury, L.; Popova, M.; Paisse, S.; Deloffre, J.; Ouddane, B. Abundance, diversity and activity of sulfate-reducing prokaryotes in heavy metal-contaminated sediment from a salt marsh in the Medway Estuary (UK). Mar. Biotechnol. 2012, 14, 363–381. [Google Scholar] [CrossRef]
- Foti, M.; Sorokin, D.Y.; Lomans, B.; Mussman, M.; Zacharova, E.E.; Pimenov, N.V.; Kuenen, J.G.; Muyzer, G. Diversity, activity, and abundance of sulfate-reducing bacteria in saline and hypersaline soda lakes. Appl. Environ. Microbiol. 2007, 73, 2093–2100. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, L.; Prosser, J.I.; He, J. Abundance and community structure of sulfate reducing prokaryotes in a paddy soil of southern China under different fertilization regimes. Soil Biol. Biochem. 2009, 41, 687–694. [Google Scholar] [CrossRef]
- Moreau, J.W.; Zierenberg, R.A.; Banfield, J.F. Diversity of dissimilatory sulfite reductase genes (dsrAB) in a salt marsh impacted by long-term acid mine drainage. Appl. Environ. Microbiol. 2010, 76, 4819–4828. [Google Scholar] [CrossRef]
- Schauer, R.; Roy, H.; Augustin, N.; Gennerich, H.H.; Peters, M.; Wenzhoefer, F.; Amann, R.; Meyerdierks, A. Bacterial sulfur cycling shapes microbial communities in surface sediments of an ultramafic hydrothermal vent field. Environ. Microbiol. 2011, 13, 2633–2648. [Google Scholar] [CrossRef]
- Pester, M.; Knorr, K.-H.; Friedrich, M.; Wagner, M.; Loy, A. Sulfate-reducing microorganisms in wetlands—Fameless actors in carbon cycling and climate change. Front. Microbiol. 2012, 3, 72. [Google Scholar] [CrossRef]
- Li, P.; Li, B.; Webster, G.; Wang, Y.; Jiang, D.; Dai, X.; Jiang, Z.; Dong, H.; Wang, Y. Abundance and diversity of sulfate-reducing bacteria in high arsenic shallow aquifers. Geomicrobiol. J. 2014, 31, 802–812. [Google Scholar] [CrossRef]
- Cho, H.; Hyun, J.H.; You, O.R.; Kim, M.; Kim, S.H.; Choi, D.L.; Green, S.J.; Kostka, J.E. Microbial community structure associated with biogeochemical processes in the Sulfate–Methane Transition Zone (SMTZ) of gas-hydrate-bearing sediment of the Ulleung basin, East Sea. Geomicrobiol. J. 2017, 34, 207–219. [Google Scholar] [CrossRef]
- Momper, L.; Jungbluth, S.P.; Lee, M.D.; Amend, J.P. Energy and carbon metabolisms in a deep terrestrial subsurface fluid microbial community. ISME J. 2017, 2319–2333. [Google Scholar] [CrossRef]
- Konneke, M.; Widdel, F. Effect of growth temperature on cellular fatty acids in sulphate-reducing bacteria. Environ. Microbiol. 2003, 5, 1064–1070. [Google Scholar] [CrossRef]
- Knoblauch, C.; Jørgensen, B.B. Effect of temperature on sulphate reduction, growth rate and growth yield in five psychrophilic sulphate-reducing bacteria from Arctic sediments. Environ. Microbiol. 1999, 1, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Jørgensen, B.B. Mineralisation of organic matter in the sea bed: The role of sulphate reduction. Nature 1982, 296, 643–645. [Google Scholar] [CrossRef]
- Isaksen, M.F.; Jørgensen, B.B. Adaptation of psychrophilic and psychrotrophic sulfate-reducing bacteria to permanently cold marine environments. Appl. Environ. Microbiol. 1996, 62, 408–414. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, H.; Nedwell, D.B. Seasonal temperature as a factor influencing bacterial sulfate reduction in a saltmarsh sediment. Microb. Ecol. 1979, 5, 73–79. [Google Scholar] [CrossRef]
- Robador, A.; Brüchert, V.; Jørgensen, B.B. The impact of temperature change on the activity and community composition of sulfate-reducing bacteria in arctic versus temperate marine sediments. Environ. Microbiol. 2009, 11, 1692–1703. [Google Scholar] [CrossRef] [PubMed]
- Yim, L.C.; Jing, H.; Aitchison, J.C.; Pointing, S.B. Highly diverse community structure in a remote central Tibetan geothermal spring does not display monotonic variation to thermal stress. FEMS Microbiol. Ecol. 2006, 57, 80–91. [Google Scholar] [CrossRef]
- Lau, M.C.; Aitchison, J.C.; Pointing, S.B. Bacterial community composition in thermophilic microbial mats from five hot springs in central Tibet. Extremophiles 2009, 13, 139–149. [Google Scholar] [CrossRef]
- Huang, Q.; Dong, C.Z.; Dong, R.M.; Jiang, H.; Wang, S.; Wang, G.; Fang, B.; Ding, X.; Niu, L.; Li, X. Archaeal and bacterial diversity in hot springs on the Tibetan Plateau, China. Extremophiles 2011, 15, 549–563. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Hou, W.; Dong, H.; Jiang, H.; Huang, L.; Wu, G.; Zhang, C.; Song, Z.; Zhang, Y.; Ren, H.; et al. Control of temperature on microbial community structure in hot springs of the Tibetan Plateau. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Jiang, H.; Dong, H.; Huang, Q.; Yang, J.; Webb, L.; Liu, Q.; Guo, H.; Yuan, S.; Li, P.; et al. Distribution of arsenite-oxidizing bacteria and its correlation with temperature in hot springs of the tibetan-yunnan geothermal zone in western china. Geomicrobiol. J. 2015, 32, 482–493. [Google Scholar] [CrossRef]
- Song, Z.Q.; Wang, F.P.; Zhi, X.Y.; Chen, J.Q.; Zhou, E.M.; Liang, F.; Xiao, X.; Tang, S.K.; Jiang, H.C.; Zhang, C.L.; et al. Bacterial and archaeal diversities in Yunnan and Tibetan hot springs, China. Environ. Microbiol. 2013, 15, 1160–1175. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, G.; Jiang, H.; Yang, J.; She, W.; Khan, I.; Li, W. Abundant and Rare Microbial Biospheres Respond Differently to Environmental and Spatial Factors in Tibetan Hot Springs. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef]
- Geets, J.; Borremans, B.; Diels, L.; Springael, D.; Vangronsveld, J.; Lelie, D.V.D.; Vanbroekhoven, K. DsrB gene-based DGGE for community and diversity surveys of sulfate-reducing bacteria. J. Microbiol. Methods 2006, 66, 194–205. [Google Scholar] [CrossRef]
- Schloss, P.D.; Handelsman, J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005, 71, 1501–1506. [Google Scholar] [CrossRef]
- Jiang, Z.; Li, P.; Jiang, D.; Wu, G.; Dong, H.; Wang, Y.; Li, B.; Wang, Y.; Guo, Q. Diversity and abundance of the arsenite oxidase gene aioA in geothermal areas of Tengchong, Yunnan, China. Extremophiles 2014, 18, 161–170. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.; Ryan, P. PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontol. Electron. 2001, 4, 1–9. [Google Scholar]
- De’ath, G. Boosted trees for ecological modeling and prediction. Ecology 2007, 88, 243–251. [Google Scholar] [CrossRef]
- Jiang, L.; Zheng, Y.; Peng, X.; Zhou, H.; Zhang, C.; Xiao, X.; Wang, F. Vertical distribution and diversity of sulfate-reducing prokaryotes in the Pearl River estuarine sediments, Southern China. FEMS Microbiol. Ecol. 2009, 70, 249–262. [Google Scholar] [CrossRef]
- Klappenbach, J.A.; Saxman, P.R.; Cole, J.R.; Schmidt, T.M. rrndb: The ribosomal RNA operon copy number database. Nucleic Acids Res. 2001, 29, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; Stams, A.J. The ecology and biotechnology of sulphate-reducing bacteria. Nat. Rev. Microbiol. 2008, 6, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Stetter, K.O.; Lauerer, G.; Thomm, M.; Neuner, A. Isolation of extremely thermophilic sulfate reducers: Evidence for a novel branch of archaebacteria. Science 1987, 236, 822–824. [Google Scholar] [CrossRef] [PubMed]
- Stetter, K.O.; Huber, R.; Blöchl, E.; Kurr, M.; Eden, R.D.; Fielder, M.; Cash, H.; Vance, I. Hyperthermophilic archaea are thriving in deep North Sea and Alaskan oil reservoirs. Nature 1993, 365, 743. [Google Scholar] [CrossRef]
- Huber, H.; Jannasch, H.; Rachel, R.; Fuchs, T.; Stetter, K.O. Archaeoglobus veneficus sp. nov., a Novel Facultative Chemolithoautotrophic Hyperthermophilic Sulfite Reducer, Isolated from Abyssal Black Smokers. Syst. Appl. Microbiol. 1997, 20, 374–380. [Google Scholar] [CrossRef]
- Itoh, T.; Suzuki, K.; Nakase, T. Thermocladium modestius gen. nov., sp. nov., a new genus of rod-shaped, extremely thermophilic crenarchaeote. Int. J. Syst. Bacteriol. 1998, 48, 879–887. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Itoh, T.; Suzuki, K.; Sanchez, P.C.; Nakase, T. Caldivirga maquilingensis gen. nov., sp. nov., a new genus of rod-shaped crenarchaeote isolated from a hot spring in the Philippines. Int. J. Syst. Bacteriol. 1999, 49, 1157–1163. [Google Scholar] [CrossRef]
- Andreote, F.D.; Jiménez, D.J.; Chaves, D.; Dias, A.C.; Luvizotto, D.M.; Diniandreote, F.; Fasanella, C.C.; Lopez, M.V.; Baena, S.; Taketani, R.G. The microbiome of Brazilian mangrove sediments as revealed by metagenomics. PLoS ONE 2012, 7, e38600. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, H.F.; Cury, J.C.; Do Carmo, F.L.; Dos Santos, A.L.; Tiedje, J.; Van Elsas, J.D.; Rosado, A.S.; Peixoto, R.S. Mangrove Bacterial Diversity and the Impact of Oil Contamination Revealed by Pyrosequencing: Bacterial Proxies for Oil Pollution. PLoS ONE 2011, 6, e16943. [Google Scholar] [CrossRef] [PubMed]
- Peixoto, R.; Chaer, G.M.; Carmo, F.L.; Araújo, F.V.; Paes, J.E.; Volpon, A.; Santiago, G.A.; Rosado, A.S. Bacterial communities reflect the spatial variation in pollutant levels in Brazilian mangrove sediment. Antonie Van Leeuwenhoek 2011, 99, 341–354. [Google Scholar] [CrossRef]
- Varon-Lopez, M.; Dias, A.C.F.; Fasanella, C.C.; Durrer, A.; Melo, I.S.; Kuramae, E.E.; Andreote, F.D. Sulphur-oxidizing and sulphate-reducing communities in Brazilian mangrove sediments. Environ. Microbiol. 2014, 16, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Bu, N.; Long, X.; Sun, J.; He, C.; Liu, X.; Cui, J.; Liu, D.; Chen, X. Sulfate reducer and sulfur oxidizer respond differentially to the invasion of Spartina alterniflora in estuarine salt marsh of China. Ecol. Eng. 2017, 99, 182–190. [Google Scholar] [CrossRef]
- Scholten, J.C.; Joye, S.B.; Hollibaugh, J.T.; Murrell, J.C. Molecular analysis of the sulfate reducing and archaeal community in a meromictic soda lake (Mono Lake, California) by targeting 16S rRNA, mcrA, apsA, and dsrAB genes. Microb. Ecol. 2005, 50, 29. [Google Scholar] [CrossRef]
- Edwardson, C.F.; Hollibaugh, J.T. Metatranscriptomic analysis of prokaryotic communities active in sulfur and arsenic cycling in Mono Lake, California, USA. ISME J. 2017. [Google Scholar] [CrossRef]
- Hug, K.; Maher, W.A.; Stott, M.B.; Krikowa, F.; Foster, S.; Moreau, J.W. Microbial contributions to coupled arsenic and sulfur cycling in the acid-sulfide hot spring Champagne Pool, New Zealand. Front. Microbiol. 2014, 5, 569. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, Y.; Muramatsu, M.; Imachi, H.; Narihiro, T.; Ohashi, A.; Harada, H.; Hanada, S.; Kamagata, Y. Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus Thermodesulfovibrio. Int. J. Syst. Evol. Microbiol. 2008, 58, 2541–2548. [Google Scholar] [CrossRef]
- Dalcin Martins, P.; Hoyt, D.W.; Bansal, S.; Mills, C.T.; Tfaily, M.; Tangen, B.A.; Finocchiaro, R.G.; Johnston, M.D.; McAdams, B.C.; Solensky, M.J.; et al. Abundant carbon substrates drive extremely high sulfate reduction rates and methane fluxes in Prairie Pothole Wetlands. Glob. Chang. Biol. 2017, 23, 3107–3120. [Google Scholar] [CrossRef] [PubMed]
- Beeder, J.; Torsvik, T.; Lien, T. Thermodesulforhabdus norvegicus gen. nov., sp. nov., a novel thermophilic sulfate-reducing bacterium from oil field water. Arch. Microbiol. 1995, 164, 331–336. [Google Scholar] [CrossRef]
- Henry, E.A.; Devereux, R.; Maki, J.S.; Gilmour, C.C.; Woese, C.R.; Mandelco, L.; Schauder, R.; Remsen, C.C.; Mitchell, R. Characterization of a new thermophilic sulfate-reducing bacterium Thermodesulfovibrio yellowstonii, gen. nov. and sp. nov.: Its phylogenetic relationship to Thermodesulfobacterium commune and their origins deep within the bacterial domain. Arch. Microbiol. 1994, 161, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Sonne-Hansen, J.; Ahring, B.K. Thermodesulfobacterium hveragerdense sp. nov., and Thermodesulfovibrio islandicus sp. nov., two thermophilic sulfate reducing bacteria isolated from a Icelandic hot spring. Syst. Appl. Microbiol. 1999, 22, 559–564. [Google Scholar] [CrossRef]
- Mori, K.; Kim, H.; Kakegawa, T.; Hanada, S. A novel lineage of sulfate-reducing microorganisms: Thermodesulfobiaceae fam. nov., Thermodesulfobium narugense, gen. nov., sp. nov., a new thermophilic isolate from a hot spring. Extrem. Life Under Extrem. Cond. 2003, 7, 283. [Google Scholar] [CrossRef] [PubMed]
- Bhatnagar, S.; Badger, J.H.; Madupu, R.; Khour, H.M.; O’Connor, E.M.; Robb, F.T.; Ward, N.L.; Eisen, J.A. Genome sequence of a sulfate-reducing thermophilic bacterium, Thermodesulfobacterium commune DSM 2178(T) (Phylum Thermodesulfobacteria). Genome Announc. 2015, 3, e01490-14. [Google Scholar] [CrossRef]
- Robador, A.; Müller, A.L.; Sawicka, J.E.; Berry, D.; Hubert, C.; Loy, A.; Jørgensen, B.B.; Brüchert, V. Activity and community structures of sulfate-reducing microorganisms in polar, temperate and tropical marine sediments. ISME J. 2016, 10, 796–809. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [CrossRef] [PubMed]
- Sharp, C.E.; Brady, A.L.; Sharp, G.H.; Grasby, S.E.; Stott, M.B.; Dunfield, P.F. Humboldt’s spa: Microbial diversity is controlled by temperature in geothermal environments. ISME J. 2014, 8, 1166. [Google Scholar] [CrossRef]
- Lozupone, C.; Knight, R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl. Environ. Microbiol. 2005, 71, 8228–8235. [Google Scholar] [CrossRef] [PubMed]
- Vigneron, A.; Cruaud, P.; Alsop, E.; De Rezende, J.R.; Head, I.M.; Tsesmetzis, N. Beyond the tip of the iceberg; a new view of the diversity of sulfite- and sulfate-reducing microorganisms. ISME J. 2018, 12, 2096–2099. [Google Scholar] [CrossRef] [PubMed]
- Rabus, R.; Venceslau, S.S.; Wöhlbrand, L.; Voordouw, G.; Wall, J.D.; Pereira, I.A.C. Chapter Two—A Post-Genomic View of the Ecophysiology, Catabolism and Biotechnological Relevance of Sulphate-Reducing Prokaryotes. Adv. Microb. Physiol. 2015, 66, 55. [Google Scholar]
- Holmer, M.; Storkholm, P. Sulphate reduction and sulphur cycling in lake sediments: A review. Freshw. Biol. 2001, 46, 431–451. [Google Scholar] [CrossRef]
- Pavloudi, C.; Oulas, A.; Vasileiadou, K.; Kotoulas, G.; De Troch, M.; Friedrich, M.W.; Arvanitidis, C. Diversity and abundance of sulfate-reducing microorganisms in a Mediterranean lagoonal complex (Amvrakikos Gulf, Ionian Sea) derived from dsrB gene. Aquat. Microb. Ecol. 2017, 79, 209–219. [Google Scholar] [CrossRef]
- Stevens, T.O.; Mckinley, J.P. Lithoautotrophic Microbia, Ecosystems in Deep Basalt Aquifers. Science 1995, 270, 450–454. [Google Scholar] [CrossRef]
- Chen, M.; Li, X.; He, Y.; Song, N.; Cai, H.; Wang, C.; Li, Y.; Chu, H.; Krumholz, L.R.; Jiang, H. Increasing sulfate concentrations result in higher sulfide production and phosphorous mobilization in a shallow eutrophic freshwater lake. Water Res. 2016, 96, 94–104. [Google Scholar] [CrossRef]


| Site | Characteristic | Sample | GPS Location (N/E) | Altitude (m) | pH | Temp. (°C) | Fe2+ (mg/L) | DO (ug/L) | DOC (mg/L) | TOC | SO42− (mg/L) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Daggyai (DG) | Separated | DG-1 | 85.7506°/29.5985° | 5058 | 8.0 | 82.0 | 0.07 | 116 | 90.46 | 0.852% | 83.4 |
| DG-4 | 85.7509°/29.5982° | 5057 | 6.8 | 45.5 | 0.02 | 2100 | 60.68 | 0.555% | 76.5 | ||
| DG-5 | 85.7509°/29.5982° | 5057 | 7.5 | 32.2 | 0.03 | 3900 | 30.88 | 0.448% | 47.9 | ||
| DG-14 | 85.7492°/29.6018° | 5082 | 7.4 | 50.0 | 0.2 | n.d | 7.83 | 1.180% | 8.4 | ||
| DG-16 | 85.7492°/29.6017° | 5075 | 8.0 | 35.0 | 0.01 | n.d | 9.66 | 0.733% | 62.4 | ||
| Quzhuomu (QZM) | Channel I | QZM-4 | 91.8086°/28.2482° | 4505 | 6.5 | 62.0 | 0.05 | 204 | 30.30 | 1.380% | 327.8 |
| QZM-5 | 91.8086°/28.2482° | 4505 | 6.8 | 56.0 | 0.34 | 1850 | 50.64 | 1.340% | 492.3 | ||
| QZM-6 | 91.8086°/28.2482° | 4505 | 7.0 | 54.0 | 0.29 | 2000 | 27.00 | 3.500% | 460.6 | ||
| QZM-7 | 91.8086°/28.2482° | 4505 | 6.8 | 52.0 | 0.15 | 3100 | 28.00 | 8.380% | 431.9 | ||
| Channel II | QZM-9 | 91.8037°/28.2486° | 4450 | 7.0 | 67.0 | 0.91 | 213 | 36.72 | 1.300% | 461.2 | |
| QZM-10 | 91.8037°/28.2486° | 4450 | 7.0 | 65.0 | 0.57 | 496 | 33.61 | 1.980% | 252.7 | ||
| QZM-11 | 91.8037°/28.2486° | 4450 | 6.8 | 64.0 | 0.57 | 617 | 20.60 | 2.540% | 417.9 | ||
| QZM-12 | 91.8037°/28.2486° | 4450 | 6.8 | 62.0 | 0.52 | 1350 | 48.78 | 1.220% | 381.5 | ||
| Separated | QZM-13 | 91.8034°/28.2485° | 4438 | 6.7 | 65.7 | 0.33 | 561 | 958.60 | 1.020% | 350.2 | |
| QZM-14 | 91.8097°/28.2472° | 4502 | 6.5 | 67.5 | 2.39 | 645 | 111.60 | 0.864% | 354.7 |
| Sample | No. of Clones | No. of OTUs | Coverage | Simpson (1/D) | Shannon (H) |
|---|---|---|---|---|---|
| QZM-4 | 25 | 2 | 100.0% | 0.50 | 0.69 |
| QZM-5 | 31 | 7 | 90.3% | 0.75 | 1.60 |
| QZM-6 | 35 | 9 | 91.4% | 0.83 | 1.94 |
| QZM-7 | 42 | 12 | 90.5% | 0.86 | 2.17 |
| QZM-9 | 65 | 12 | 92.3% | 0.82 | 2.03 |
| QZM-10 | 24 | 5 | 91.7% | 0.63 | 1.20 |
| QZM-11 | 25 | 4 | 96.0% | 0.67 | 1.19 |
| QZM-12 | 33 | 8 | 90.9% | 0.82 | 1.84 |
| QZM-13 | 19 | 4 | 100.0% | 0.60 | 1.14 |
| QZM-14 | 35 | 8 | 91.4% | 0.69 | 1.55 |
| DG-1 | 58 | 4 | 96.6% | 0.38 | 0.68 |
| DG-4 | 23 | 2 | 100.0% | 0.16 | 0.30 |
| DG-5 | 24 | 2 | 100.0% | 0.49 | 0.68 |
| DG-14 | 37 | 6 | 94.6% | 0.72 | 1.42 |
| DG-16 | 21 | 7 | 90.5% | 0.75 | 1.66 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, L.; She, W.; Wu, G.; Yang, J.; Phurbu, D.; Jiang, H. Influence of Temperature and Sulfate Concentration on the Sulfate/Sulfite Reduction Prokaryotic Communities in the Tibetan Hot Springs. Microorganisms 2021, 9, 583. https://doi.org/10.3390/microorganisms9030583
Ma L, She W, Wu G, Yang J, Phurbu D, Jiang H. Influence of Temperature and Sulfate Concentration on the Sulfate/Sulfite Reduction Prokaryotic Communities in the Tibetan Hot Springs. Microorganisms. 2021; 9(3):583. https://doi.org/10.3390/microorganisms9030583
Chicago/Turabian StyleMa, Li, Weiyu She, Geng Wu, Jian Yang, Dorji Phurbu, and Hongchen Jiang. 2021. "Influence of Temperature and Sulfate Concentration on the Sulfate/Sulfite Reduction Prokaryotic Communities in the Tibetan Hot Springs" Microorganisms 9, no. 3: 583. https://doi.org/10.3390/microorganisms9030583
APA StyleMa, L., She, W., Wu, G., Yang, J., Phurbu, D., & Jiang, H. (2021). Influence of Temperature and Sulfate Concentration on the Sulfate/Sulfite Reduction Prokaryotic Communities in the Tibetan Hot Springs. Microorganisms, 9(3), 583. https://doi.org/10.3390/microorganisms9030583








