A Novel Moderately Thermophilic Facultative Methylotroph within the Class Alphaproteobacteria
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Procedures and Cultivation Strategy
2.2. Isolation and Purification of Strain LS7-MT
2.3. Morphology, Electron Microscopy and Cellular Fatty Acid Analysis
2.4. Genome Analysis
3. Results and Discussion
3.1. Isolation of a Moderately Thermophilic Methylotroph
3.2. Physiological Properties of Strain LS7-MT
3.3. Identification of Phospholipid Fatty Acids (PLFA)
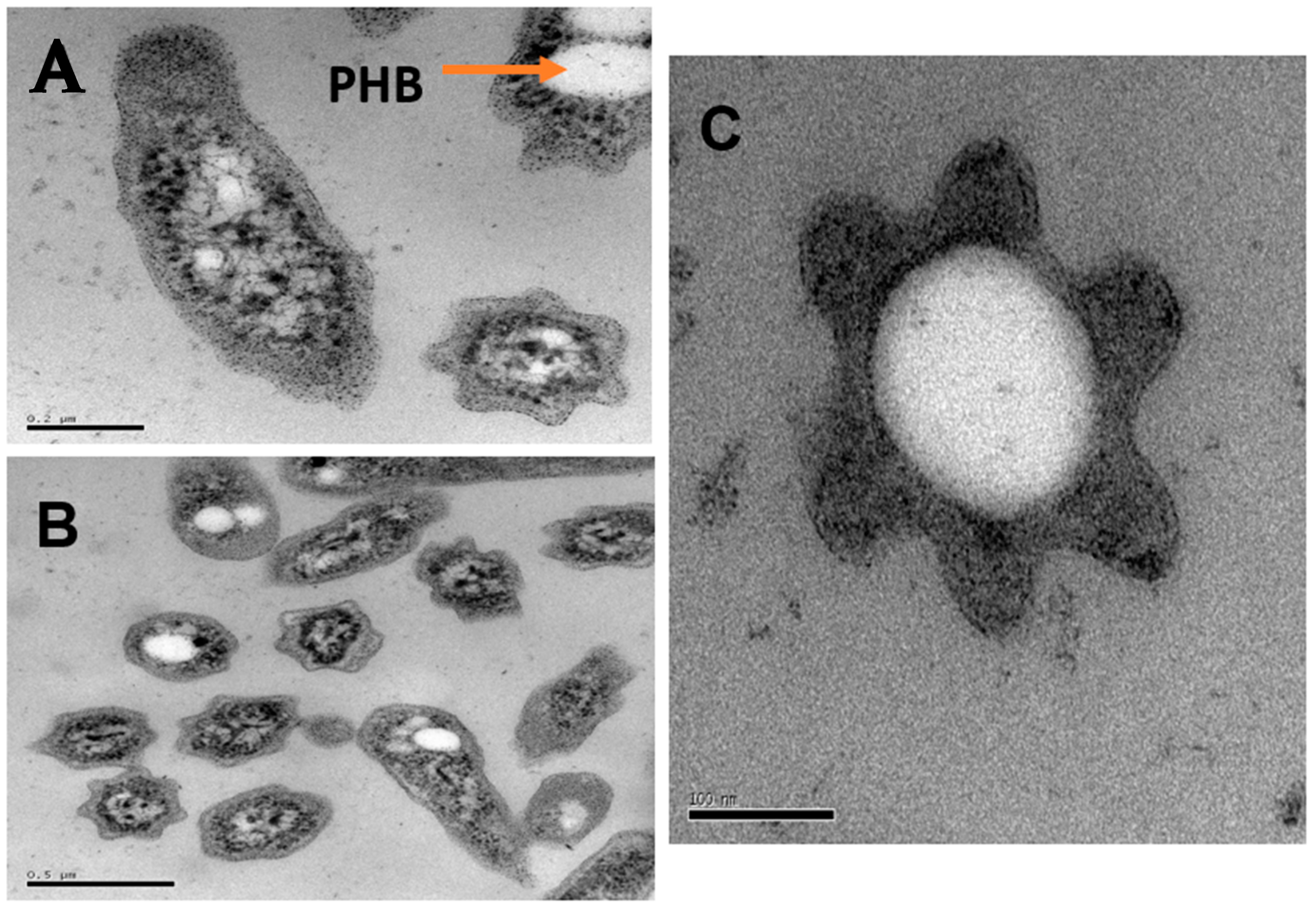
3.4. Microscopic Observations
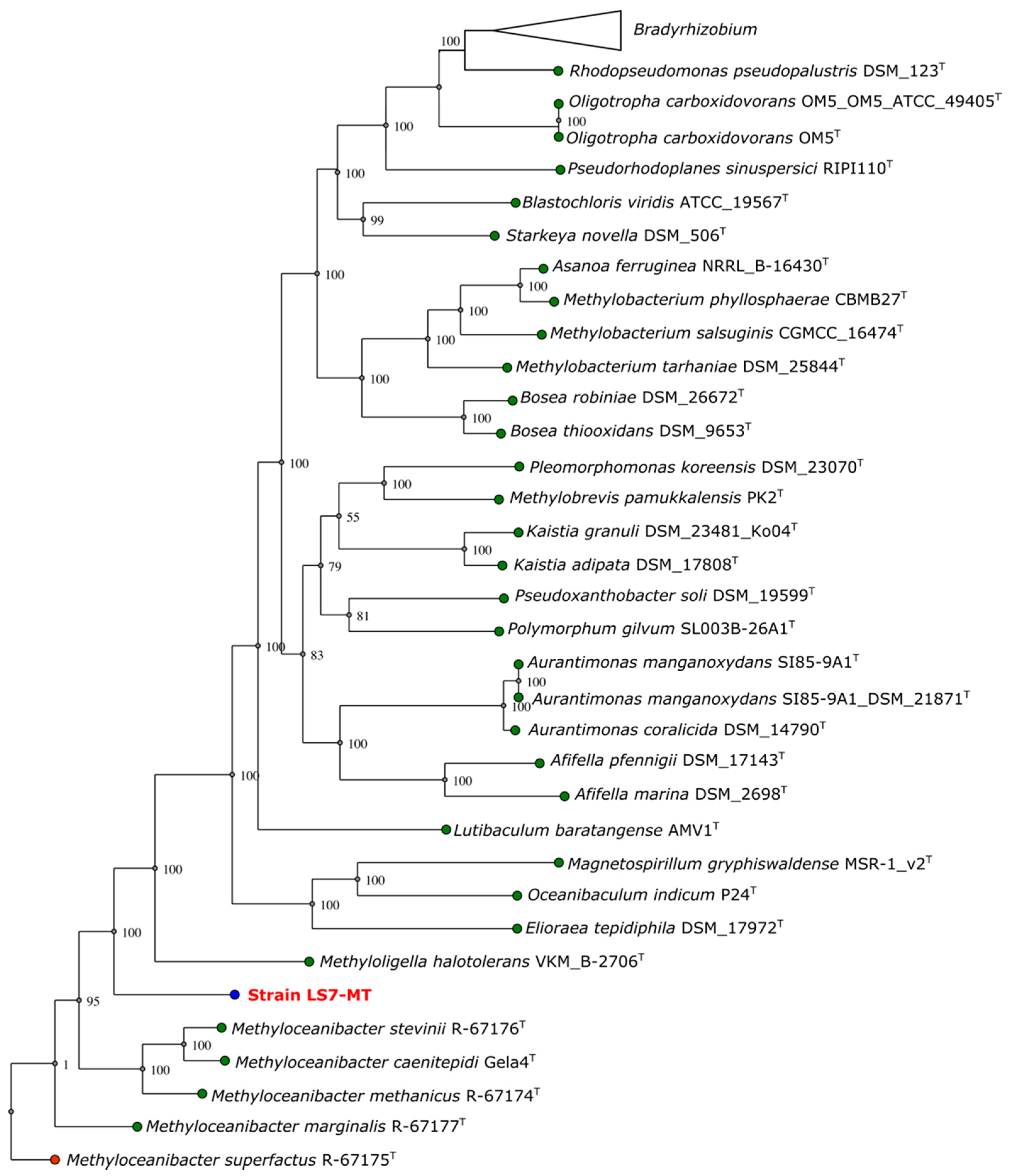
3.5. Phylogeny of Strain LS7-MT
3.6. Genome Assembly and Annotation
3.7. Methanol Metabolism
Description of Methylothermalis aethiopiae Strain LS7-MT gen. nov. sp. nov.
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Houghton, K.M.; Carere, C.R.; Stott, M.B.; McDonald, I.R. Thermophilic methanotrophs: In hot pursuit. FEMS Microbiol. Ecol. 2019, 95, fiz125. [Google Scholar] [CrossRef] [PubMed]
- Islam, T.; Gessesse, A.; Garcia-Moyano, A.; Murrell, J.C.; Øvreås, L. A novel moderately thermophilic Type Ib methanotroph Isolated from an alkaline thermal spring in the Ethiopian Rift Valley. Microorganisms 2020, 8, 250. [Google Scholar] [CrossRef]
- Kolb, S. Aerobic methanol-oxidizing bacteria in soil. FEMS Microbiol. Lett. 2009, 300, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Chistoserdova, L.; Lidstrom, M.E. Aerobic Methylotrophic Prokaryotes. In The Prokaryotes; Rosenberg, E., DeLong, E.F., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 267–285. [Google Scholar]
- Chistoserdova, L.; Kalyuzhnaya, M.G.; Lidstrom, M.E. The expanding world of methylotrophic metabolism. Ann. Rev. Microbiol. 2009, 63, 477–499. [Google Scholar] [CrossRef] [PubMed]
- Anthony, C. The quinoprotein dehydrogenases for methanol and glucose. Arch. Biochem. Biophys. 2004, 428, 2–9. [Google Scholar] [CrossRef]
- Dumont, M.G.; Murrell, J.C. Stable isotope probing—Linking microbial identity to function. Nat. Rev. Microbiol. 2005, 3, 499–504. [Google Scholar] [CrossRef]
- McDonald, I.R.; Bodrossy, L.; Chen, Y.; Murrell, J.C. Molecular ecology techniques for the study of aerobic methanotrophs. Appl. Environ. Microbiol. 2008, 74, 1305–1315. [Google Scholar] [CrossRef]
- Chistoserdova, L. Modularity of methylotrophy revisited. Environ. Microbiol. 2011, 13, 2603–2622. [Google Scholar] [CrossRef]
- Keltjens, J.T.; Pol, A.; Reimann, J.; den Camp, H.J.O. PQQ-dependent methanol dehydrogenases: Rare-earth elements make a difference. Appl. Microbiol. Biotechnol. 2014, 98, 6163–6183. [Google Scholar] [CrossRef]
- McDonald, I.R.; Murrell, J.C. The methanol dehydrogenase structural gene mxaF and its use as a functional gene probe for methanotrophs and methylotrophs. Appl. Environ. Microbiol. 1997, 63, 3218–3224. [Google Scholar] [CrossRef]
- Taubert, M.; Grob, C.; Howat, A.M.; Burns, O.J.; Dixon, J.L.; Chen, Y.; Murrell, J.C. XoxF encoding an alternative methanol dehydrogenase is widespread in coastal marine environments. Environ. Microbiol. 2015, 17, 3937–3948. [Google Scholar] [CrossRef] [PubMed]
- Hördt, A.; López, M.G.; Meier-Kolthoff, J.P.; Schleuning, M.; Weinhold, L.M.; Tindall, B.J.; Göker, M. Analysis of 1000+ type-strain genomes substantially improves taxonomic classification of Alphaproteobacteria. Front. Microbiol. 2020, 11, 468. [Google Scholar] [CrossRef] [PubMed]
- Garrity, G.M.; Bell, J.A.; Lilburn, T. Family VIII. Hyphomicrobiaceae Babudieri. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2005; Volume 2, p. 476. [Google Scholar]
- Oren, A.; Xu, X.W. The Family Hyphomicrobiaceae. In The Prokaryotes; Rosenberg, E., DeLong, E., Lory, S., Stackebrandt, E., Thompson, F., Eds.; Springer: Berlin, Germany, 2014; pp. 247–281. [Google Scholar]
- Hirsch, P.; Hoffmann, B. Dichotomicrobium thermohalophilum gen. nov., spec, nov., budding prosthecate bacteria from the Solar Lake (Sinai) and some related strains. Syst. Appl. Microbiol. 1989, 11, 291–301. [Google Scholar] [CrossRef]
- Gude, H.; Haibel, B.; Muller, H. Development of planktonic bacterial populations in a water column of Lake Constance (Bodensee-Obersee). Arch. Hydrobiol. 1985, 105, 59–77. [Google Scholar]
- Takeuchi, M.; Katayama, T.; Yamagishi, T.; Hanada, S.; Tamaki, H.; Kamagata, Y.; Oshima, K.; Hattori, M.; Marumi, K.; Nedachi, M.; et al. Methyloceanibacter caenitepidi gen. nov., sp. nov., a facultatively methylotrophic bacterium isolated from marine sediments near a hydrothermal vent. Int. J. Syst. Evol. Microbiol. 2014, 64, 462–468. [Google Scholar] [CrossRef]
- Doronina, N.; Poroshina, M.N.; Kaparullina, E.N.; Ezhov, V.A.; Trotsenko, Y.A. Methyloligella halotolerans gen. nov., sp. nov. and Methyloligella solikamskensis sp. nov., two non-pigmented halotolerant obligately methylotrophic bacteria isolated from the Ural saline environments. Syst. Appl. Microbiol. 2013, 36, 148–154. [Google Scholar] [CrossRef]
- McDonald, I.R.; Doronina, N.V.; Trotsenko, Y.A.; McAnulla, C.; Murrell, J.C. Hyphomicrobium chloromethanicum sp. nov. and Methylobacterium chloromethanicum sp. nov., chloromethane-utilizing bacteria isolated from a polluted environment. Int. J. Syst. Evol. Microbiol. 2001, 51, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Denner, E.B.; Kolari, M.; Hoornstra, D.; Tsitko, I.; Kämpfer, P.; Busse, H.J.; Salkinoja-Salonen, M. Rubellimicrobium thermophilum gen. nov., sp. nov., a red-pigmented, moderately thermophilic bacterium isolated from coloured slime deposits in paper machines. Int. J. Syst. Evol. Microbiol. 2006, 56, 1355–1362. [Google Scholar] [CrossRef]
- Panday, D.; Das, S.K. Chelatococcus sambhunathii sp. nov., a moderately thermophilic alphaproteobacterium isolated from hot spring sediment. Int. J. Syst. Evol. Microbiol. 2010, 60, 861–865. [Google Scholar] [CrossRef]
- Albuquerque, L.; Rainey, F.A.; Pena, A.; Tiago, I.; Veríssimo, A.; Nobre, M.F.; da Costa, M.S. Tepidamorphus gemmatus gen. nov., sp. nov., a slightly thermophilic member of the Alphaproteobacteria. Syst. Appl. Microbiol. 2010, 33, 60–66. [Google Scholar] [CrossRef]
- Habib, N.; Khan, I.U.; Xiao, M.; Hejazi, M.S.; Tarhriz, V.; Zhi, X.Y.; Li, W.J. Elioraea thermophila sp. nov., a thermophilic bacterium from hot spring of the class Alphaproteobacteria, emended description of the genus Elioraea and proposal of Elioraeaceae fam. nov. Int. J. Syst. Evol. Microbiol. 2020, 70, 1300–1306. [Google Scholar] [CrossRef]
- Hanada, S.; Kawase, Y.; Hiraishi, A.; Takaichi, S.; Matsuura, K.; Shimada, K.; Nagashima, K.V. Porphyrobacter tepidarius sp. nov., a moderately thermophilic aerobic photosynthetic bacterium isolated from a hot spring. Int. J. Syst. Evol. Microbiol. 1997, 47, 408–413. [Google Scholar] [CrossRef][Green Version]
- Islam, T.; Larsen, Ø.; Torsvik, V.; Øvreås, L.; Panosyan, H.; Murrell, J.C.; Birkeland, N.K.; Bodrossy, L. Novel methanotrophs of the family Methylococcaceae from different geographical regions and habitats. Microorganisms 2015, 3, 484–499. [Google Scholar] [CrossRef]
- Islam, T.; Jensen, S.; Reigstad, L.J.; Larsen, O.; Birkeland, N.K. Methane oxidation at 55 degrees C and pH 2 by a thermoacidophilic bacterium belonging to the Verrucomicrobia phylum. Proc. Natl. Acad. Sci. USA 2008, 105, 300–304. [Google Scholar] [CrossRef]
- Vallenet, D.; Belda, E.; Calteau, A.; Cruveiller, S.; Engelen, S.; Lajus, A.; Medigue, C. MicroScope—An integrated microbial resource for the curation and comparative analysis of genomic and metabolic data. Nucleic Acids. Res. 2013, 41, D636–D647. [Google Scholar] [CrossRef] [PubMed]
- Alanjary, M.; Steinke, K.; Ziemert, N. AutoMLST: An automated web server for generating multi-locus species trees highlighting natural product potential. Nucleic Acids Res. 2019, 47, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular Evolutionary Genetics Analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef]
- Kim, H.S.; Srinivasan, S.; Lee, S.S. Methyloterrigena soli gen. nov., sp. nov., a methanol-utilizing bacterium isolated from chloroethylene-contaminated soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 101–106. [Google Scholar] [CrossRef]
- Bodelier, P.L.; Gillisen, M.J.B.; Hordijk, K.; Damsté, J.S.S.; Rijpstra, W.I.C.; Geenevasen, J.A.; Dunfield, P.F. A reanalysis of phospholipid fatty acids as ecological biomarkers for methanotrophic bacteria. ISME J. 2009, 3, 606–617. [Google Scholar] [CrossRef] [PubMed]
- Goldfine, H.; Hagen, P.O. N-Methyl groups in bacterial lipids III. phospholipids of hyphomicrobia. J. Bacteriol. 1968, 95, 367–375. [Google Scholar] [CrossRef] [PubMed]
- Jones, D.T.; Taylor, W.R.; Thornton, J.M. The rapid generation of mutation data matrices from protein sequences. Comp. Appl. Biosci. 1992, 8, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Pomper, B.K.; Saurel, O.; Milon, A.; Vorholt, J.A. Generation of formate by the formyltransferase/hydrolase complex (Fhc) from Methylobacterium extorquens AM1. FEBS Lett. 2002, 523, 133–137. [Google Scholar] [CrossRef]


| Characteristics | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Ecology (Habitats) | Alkaline thermal spring | Marine sediment | Saline environment | Soil, water, sewage | Contaminated soil | Paper mill (coloured biofilms) | Hot spring sediment | Hot spring water | Hot spring sediment | Brackish Hot spring |
| Methylotrophy (M) | Fac. M. | Fac. M. | Obligately M. | Fac. M. | Fac. M. | Strictly aerobic | Fac. | Fac. | Fac. | Fac. |
| Cell morphology | Rods | Rods | Short rods | Rods | Rods | Rods | Curved rods | Rods | ||
| Cell length (µM) | 0.4–1.5 | 2.8–10.4 | 1.8–2.0 | 0.5–5.0 | 1.0–1.5 | 2.0–4.0 | 0.5–5.0 | 0.5–2.0 | 2.2–3.2 | 0.5–5.0 |
| Colony colour | Shiny white | White | Colourless | Brownish | Cream | Red-pigmented | Brownish | Non-pigmented | Light pink | Orange |
| Flagellation | − | − | − | + | + | − | + | − | − | + |
| Hyphae formation | − | − | − | + | − | − | − | − | − | − |
| Temp. (opt.) °C | 30–60 (55) | 19–43 (35) | 10–40 (29) | 5–45 (28–35) | 20–37 (30) | 25–55 (45–54) | 20–50 (37–42) | 30–53 (45–50) | 45–60 (55) | 30–53 (40–48) |
| Utilization of: | ||||||||||
| methanol | + | + | + | + | + | − | − | − | + a | − |
| Methane | − | − | − | − | − | − | − | − | − | − |
| methylamine | + | + | + | + | nr | + | + | − | nr | + |
| acetate | + | + | − | + | − | + | + | + | + | + |
| pyruvate | + | nr | nr | nr | nr | nr | nr | + | + | − |
| glucose | + | − | − | +/− | + | + | +/− | + | + | + |
| mxaF gene | + | + | + | + | + | − | − | − | nr | − |
| Optimum pH | 7–8 | 6–8 | 7–8 | 6.5–7.5 | 7–8 | 6.5–7.5 | 7.5–8.0 | 7.5–8.5 | 7–7.5 | 6.5–7.5 |
| NaCl tolerance (%) | 0.5 | 9 | 16 | 5.5 | 0–3.5 | 5.5 | 0.0–3.0 | 0–3 | 0.5 | 5.5 |
| G + C content (%) | 65.9 | 63.9 | 60.5 | 59–65 | 60.5 | 70.0 | 59–65 | 66.9 | 70.8 | 65.0 |
| Fatty Acid | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| C14:0 3-OH | 3.99 | 0.2 | 2.0–2.8 | 3.0 | 1.6 | |||||
| C16:0 2-OH | 5.4 | |||||||||
| C16:0 3-OH | 1.8–3.4 | |||||||||
| C16:0 | 2.28 | 1.6 | 0.6 | 1.5–2.5 | 23.6 | 22.34 | 1.2 | 7.4 | 12.6 | nr |
| C17:0 cyclo | 0.48 | |||||||||
| C17:0 | 2.0 | 0.44 | 1.5 | 0.7 | ||||||
| C18:1w9 | ||||||||||
| C18:1w7c 11-Me | 4.8 | 27.9 | 0.92 | 8.3 | ||||||
| C 18:1w7c | 24.20 | 60.2 | 36.7 | 74.4–85.2 | 12.1 | 4.49 | 73.1 | 3.7 | 35.8 | |
| C 18:0 | 40.45 | 36.6 | 17.4 | 3.1–7.6 | 9.4 | 21.62 | 1.7 | 16.2 | ||
| C 18:0 3-OH | 4.68 | 4.5 | 3.4 | 2.3 | 2.5 | |||||
| C 18:0 2-OH | 1.5 | |||||||||
| C 18:1 2-OH | 7.1 | 5.6 | ||||||||
| C 19:0 cyc w8c | 20.77 | 28.7 | 15.3 | 43.91 | 7.8 | 58.2 | 4.2 | |||
| C19:0 | 1.6 | 0.1–2.2 | ||||||||
| C19:1 | 0.67 | |||||||||
| C 20:2 w6,9c | 1.98 | 0.6 | 0.66 | 0.5 | ||||||
| C 20:1 w7c | 0.99 | 0.8 | 0.7 | |||||||
| C 20:1 w9 | 5.2 | |||||||||
| C 20:0 | 1.7 | |||||||||
| C24:0 3-OH | 1.7 | |||||||||
| C 25:0 3-OH | 1.4 | |||||||||
| C 25:0 2-OH | ||||||||||
| C 26:0 3-OH | 1.6 | 1.5 | ||||||||
| C 10:0 3-OH | 1.28 | |||||||||
| Summed feature 2 | 1.2 | 1.8 | ||||||||
| Summed feature 7 | 30.1 | |||||||||
| Summed feature 8 Growth (°C) | 55 | 35 | 29 | 30 | 45 | 30 | 45 | 50 |
| Characteristics | Number | % of Total |
|---|---|---|
| Genome size (total number of bases) | 2,954,375 | 100 |
| DNA coding number of bases | 2,669,286 | 90.35 |
| DNA G + C number of bases | 1,947,002 | 65.90 |
| DNA scaffolds | 237 | 100 |
| Total number of genes | 3152 | 100 |
| Protein coding genes | 3098 | 98.29 |
| RNA genes | 54 | 1.71 |
| Number of rRNA genes (16S, 23S, and 5S) | 3 | 0.10 |
| Number of tRNA genes | 46 | 1.46 |
| Protein coding genes with function prediction | 2355 | 74.71 |
| Protein coding genes without function prediction | 743 | 23.57 |
| Protein coding genes with enzymes | 752 | 23.86 |
| Protein coding genes connected to Transporter Classification | 315 | 9.99 |
| Genes in Biosynthetic Clusters | 94 | 2.98 |
| Protein coding genes coding signal peptides | 367 | 11.64 |
| Protein coding genes coding transmembrane proteins | 834 | 26.46 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Islam, T.; Hernández, M.; Gessesse, A.; Murrell, J.C.; Øvreås, L. A Novel Moderately Thermophilic Facultative Methylotroph within the Class Alphaproteobacteria. Microorganisms 2021, 9, 477. https://doi.org/10.3390/microorganisms9030477
Islam T, Hernández M, Gessesse A, Murrell JC, Øvreås L. A Novel Moderately Thermophilic Facultative Methylotroph within the Class Alphaproteobacteria. Microorganisms. 2021; 9(3):477. https://doi.org/10.3390/microorganisms9030477
Chicago/Turabian StyleIslam, Tajul, Marcela Hernández, Amare Gessesse, J. Colin Murrell, and Lise Øvreås. 2021. "A Novel Moderately Thermophilic Facultative Methylotroph within the Class Alphaproteobacteria" Microorganisms 9, no. 3: 477. https://doi.org/10.3390/microorganisms9030477
APA StyleIslam, T., Hernández, M., Gessesse, A., Murrell, J. C., & Øvreås, L. (2021). A Novel Moderately Thermophilic Facultative Methylotroph within the Class Alphaproteobacteria. Microorganisms, 9(3), 477. https://doi.org/10.3390/microorganisms9030477






