Disproportionate CH4 Sink Strength from an Endemic, Sub-Alpine Australian Soil Microbial Community
Abstract
1. Introduction
2. Materials and Methods
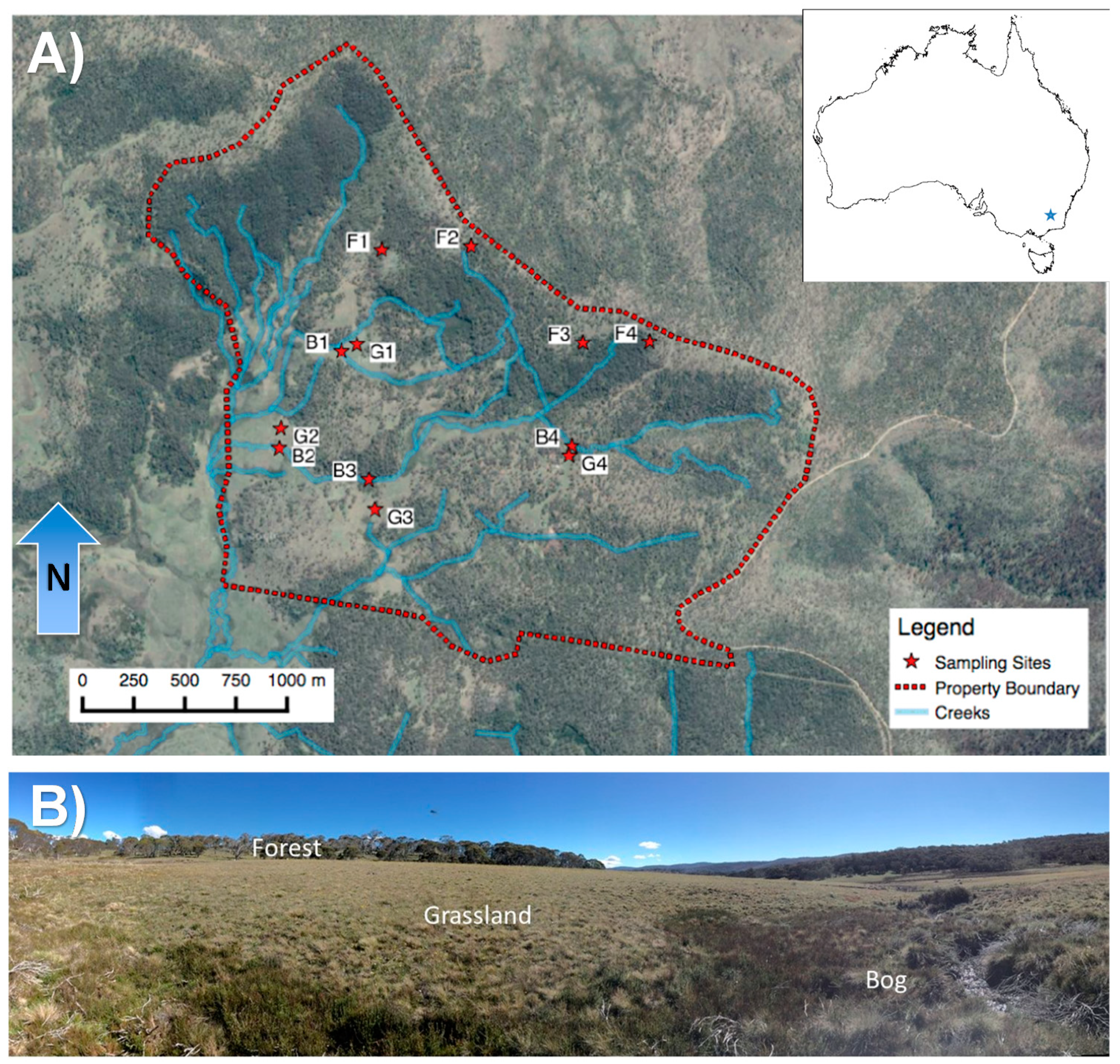
2.1. Study Site Characteristics
2.2. Experimental Design, Soil Sampling, and Gas Sampling
2.3. Soil Processing and Analyses
2.4. DNA Extraction and Quantitative PCR
2.5. Illumina Amplicon Library Preparation and Sequencing
2.6. Bioinformatics, Data Processing, GIS Modeling, and Statistical Analyses
3. Results
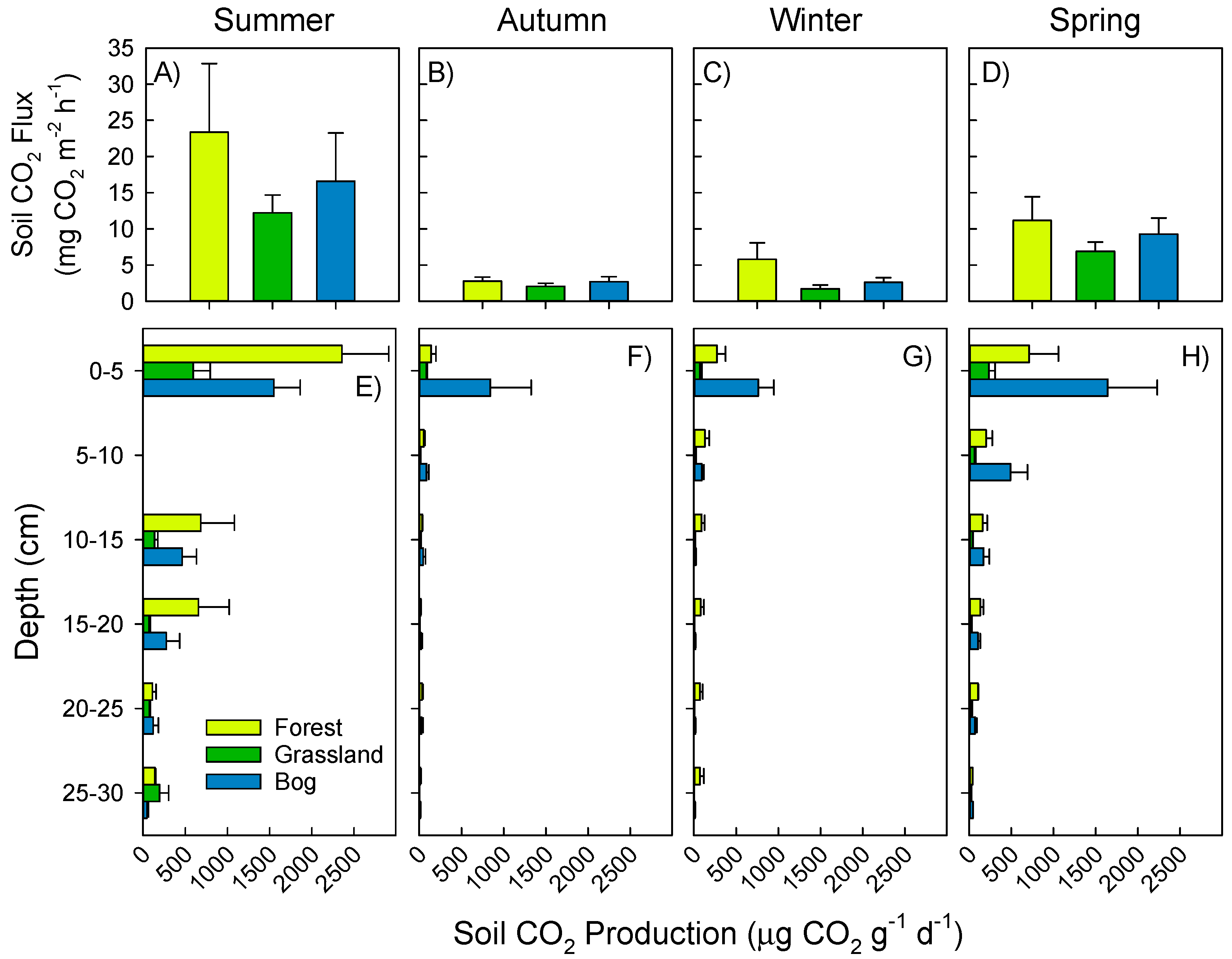
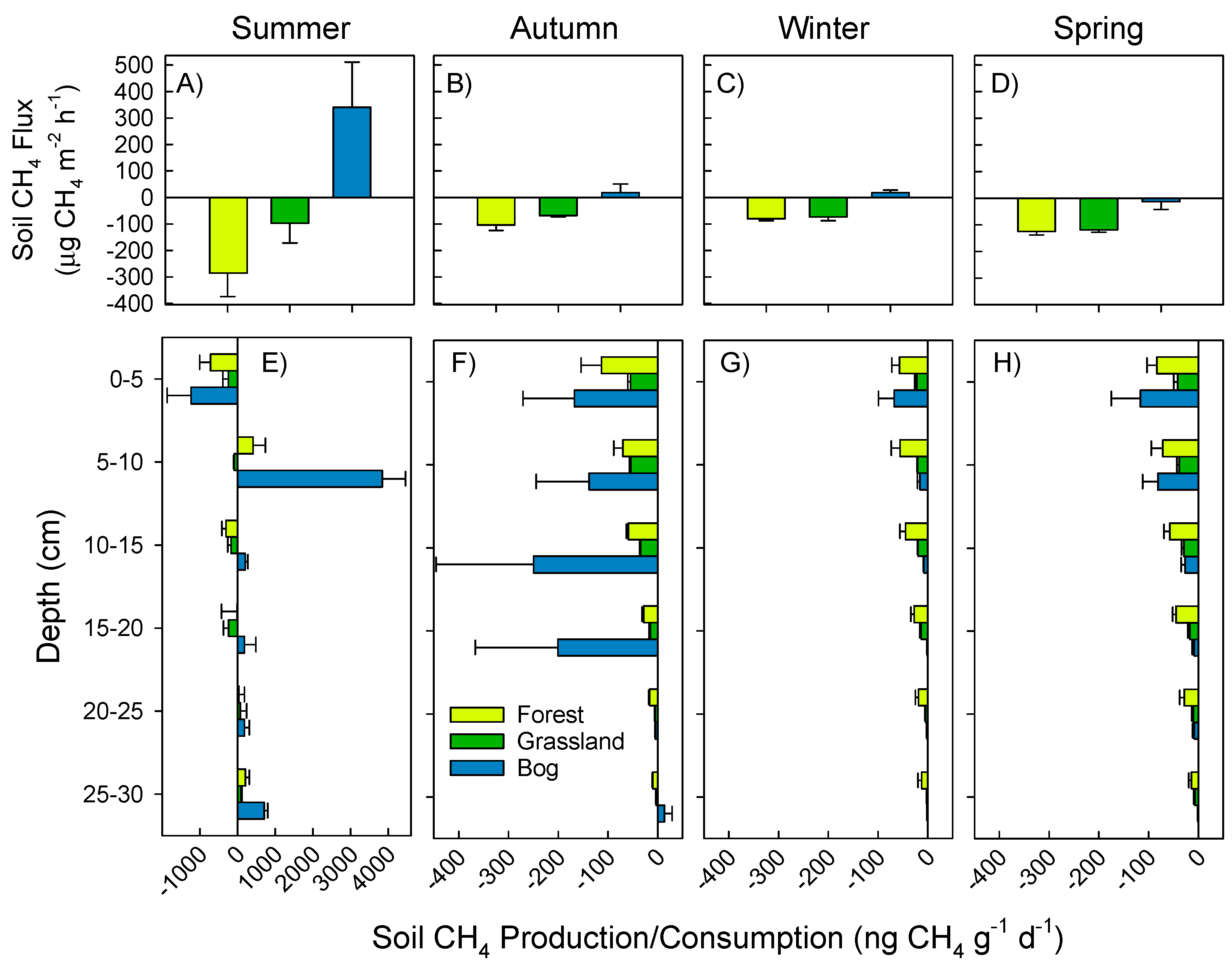
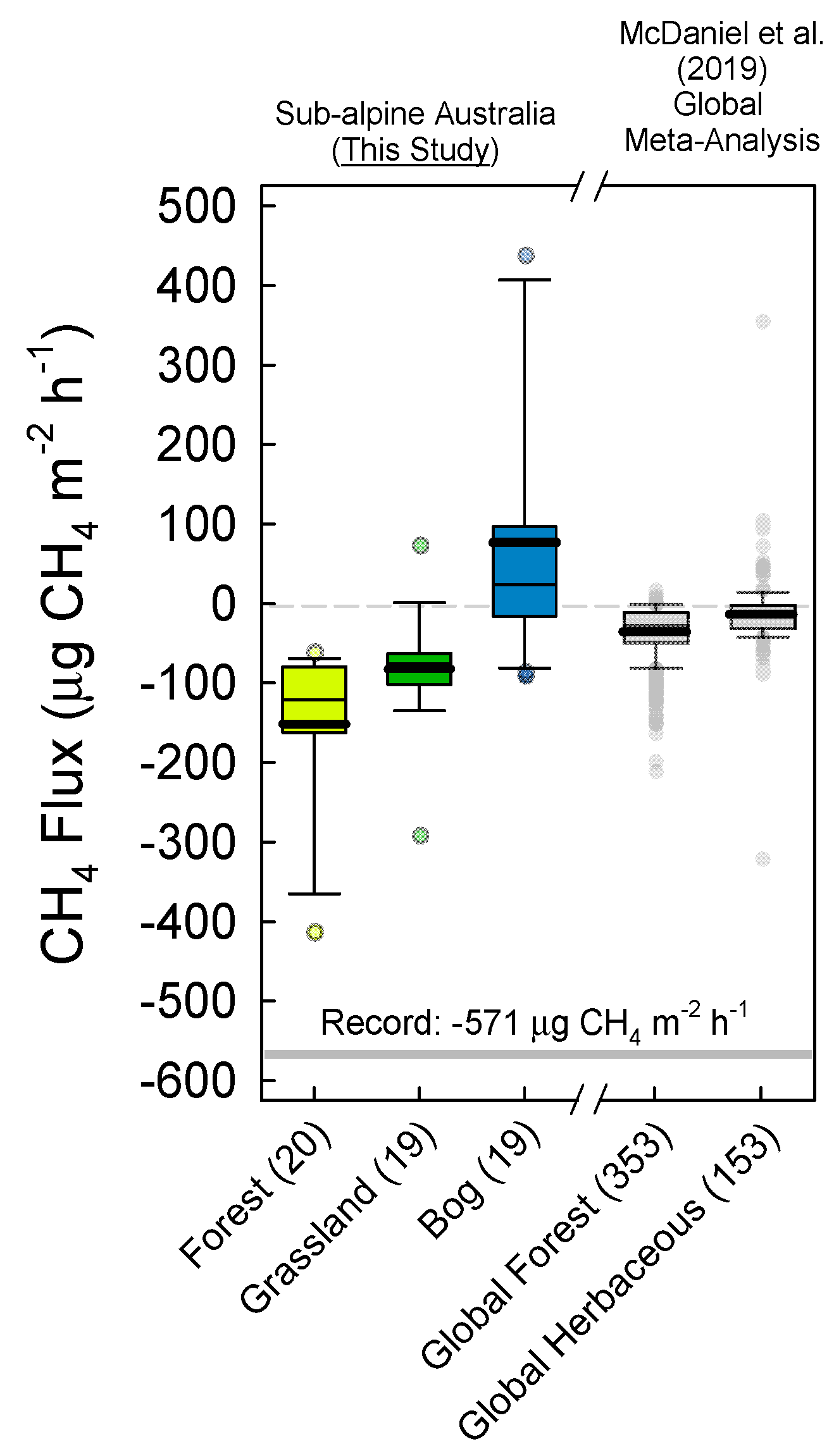
3.1. Soil Greenhouse Gas Fluxes and Production at Depth
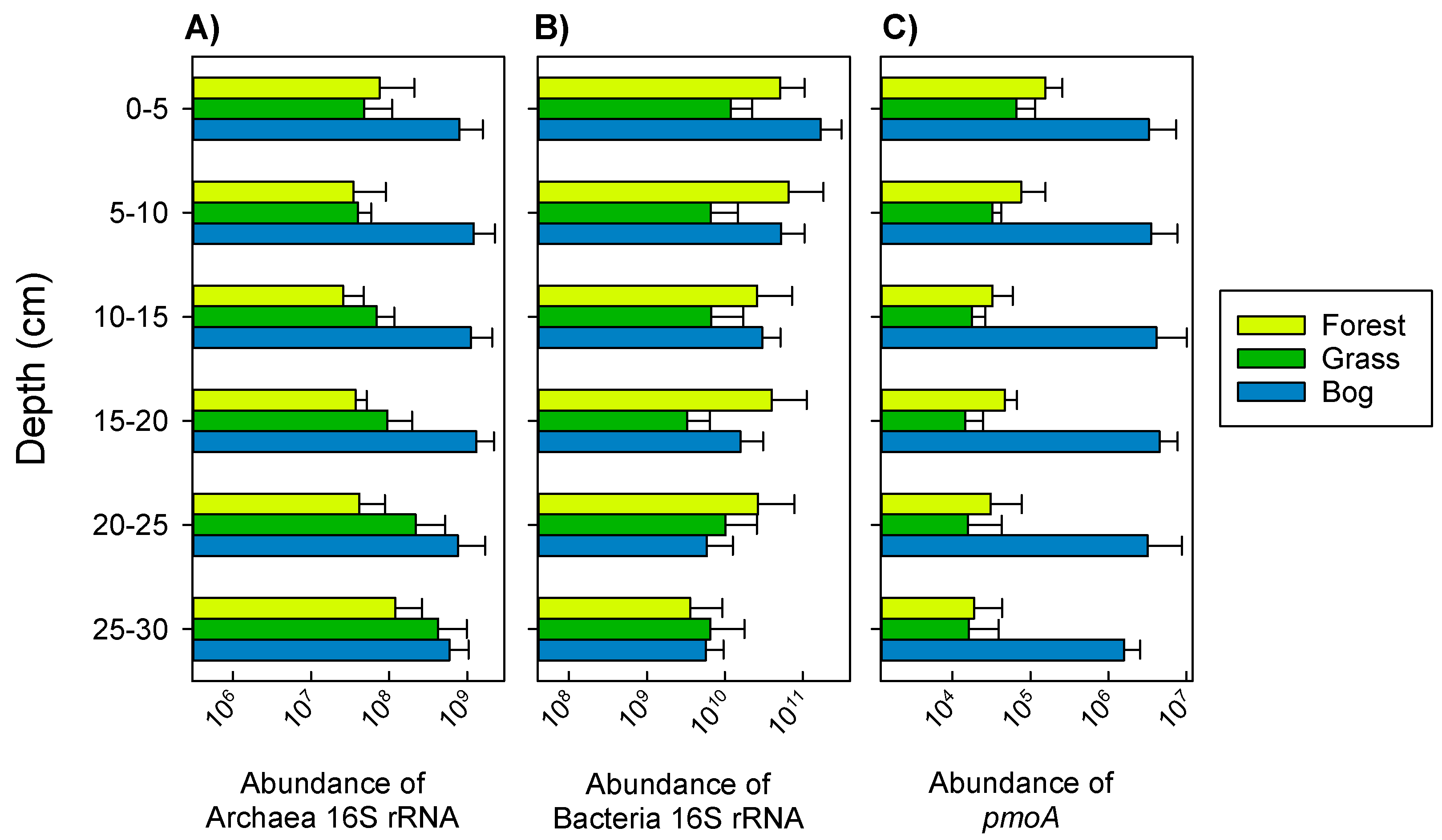
3.2. Archaeal and Bacterial Gene Abundance (qPCR) during Summer (17 February 2015)
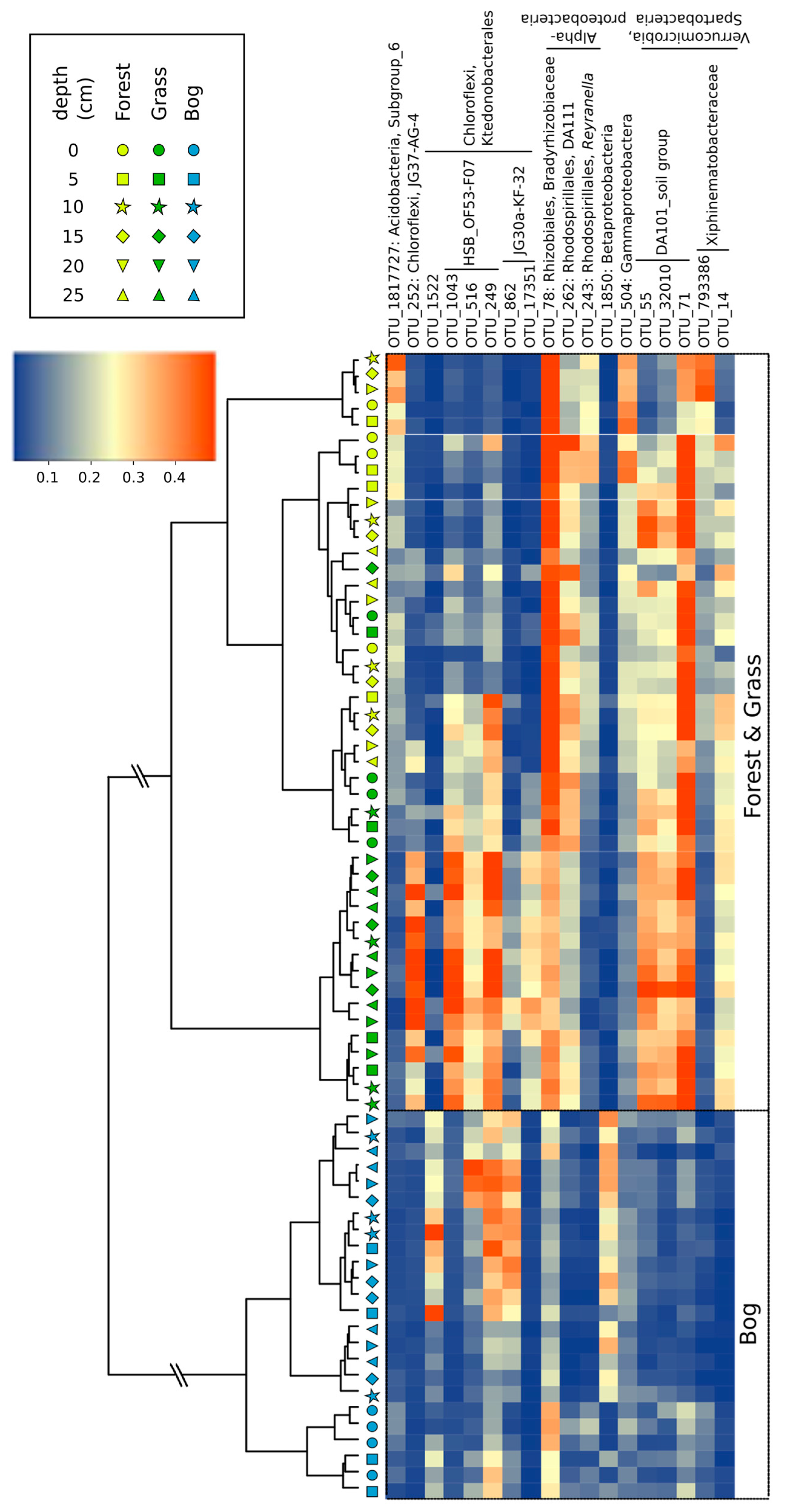
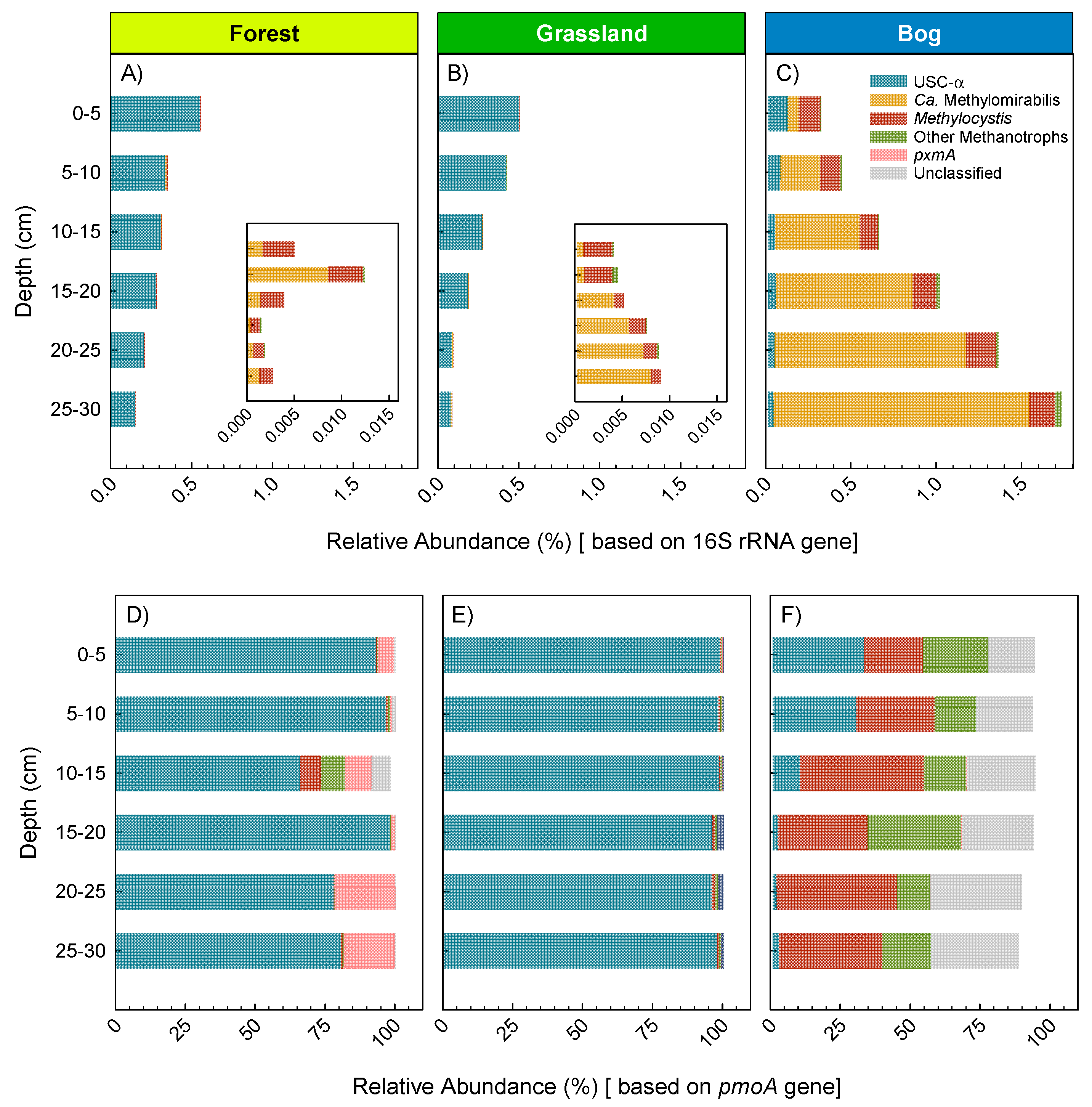
3.3. Microbial Community Composition and Diversity during Summer (17 February 2015)
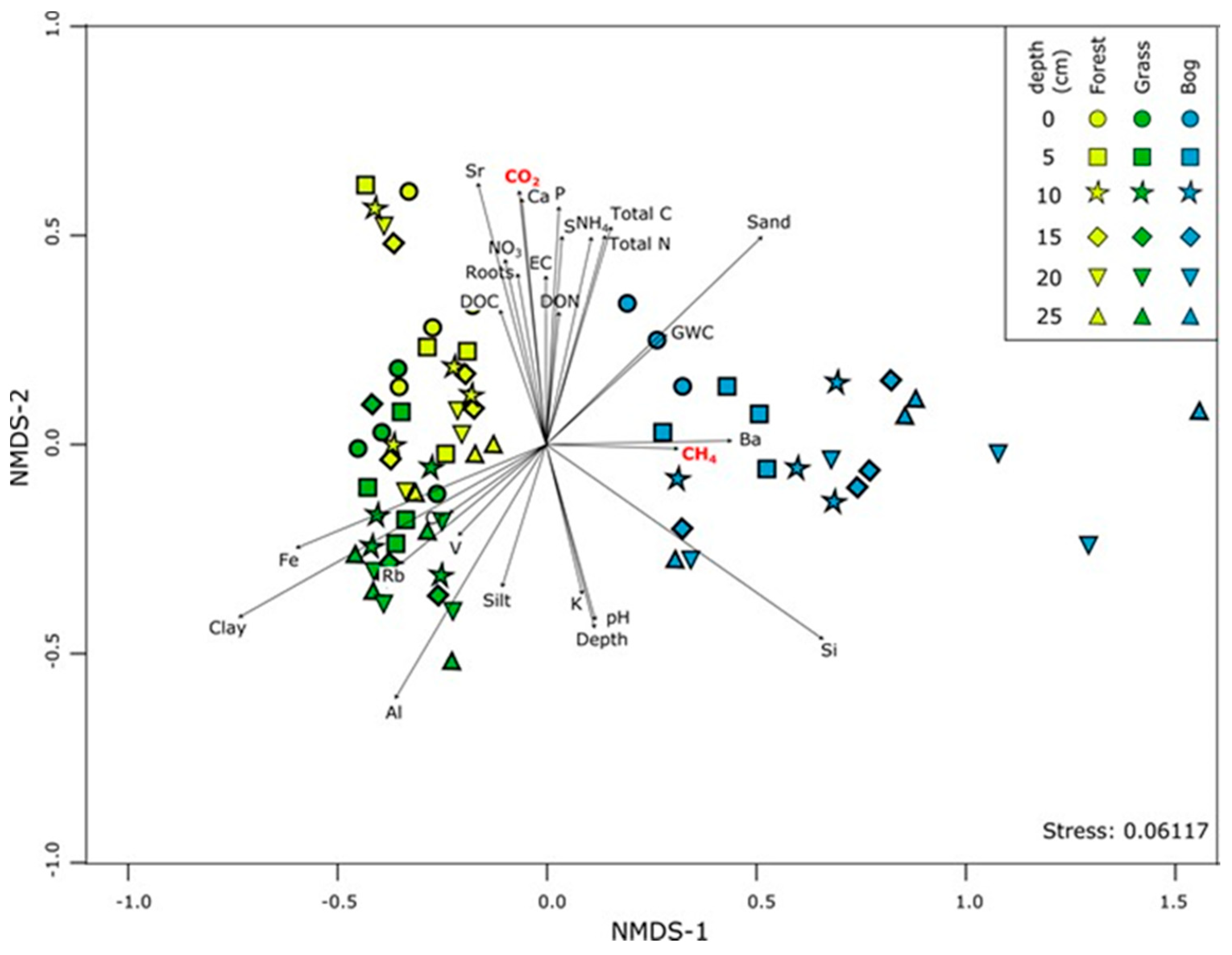
3.4. Patterns in Community Composition and Relationships with Soil Properties
4. Discussion & Conclusions
4.1. Soil Microbial Community Composition and Links to Greenhouse Gas Fluxes
4.2. The Australian Alps: Unique Soils with Disproportionate CH4 Sink Strength
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bender, M.; Conrad, R. Methane oxidation activity in various soils and freshwater sediments: Occurrence, characteristics, vertical profiles, and distribution on grain size fractions. J. Geophys. Res. Atmos. 1994, 99, 16531–16540. [Google Scholar] [CrossRef]
- Conrad, R.; Rothfuss, F. Methane oxidation in the soil surface layer of a flooded rice field and the effect of ammonium. Biol. Fertil. Soils 1991, 12, 28–32. [Google Scholar] [CrossRef]
- He, R.; Wooller, M.J.; Pohlman, J.W.; Quensen, J.; Tiedje, J.M.; Leigh, M.B. Diversity of active aerobic methanotrophs along depth profiles of arctic and subarctic lake water column and sediments. ISME J. 2012, 6, 1937–1948. [Google Scholar] [CrossRef]
- Sexstone, A.J.; Revsbech, N.P.; Parkin, T.B.; Tiedje, J.M. Direct measurement of oxygen profiles and denitrification rates in soil aggregates. Soil Sci. Soc. Am. J. 1985, 49, 645–651. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Or, D. Microbial community dynamics in soil aggregates shape biogeochemical gas fluxes from soil profiles–upscaling an aggregate biophysical model. Glob. Chang. Biol. 2016, 22, 3141–3156. [Google Scholar] [CrossRef] [PubMed]
- Karbin, S.; Hagedorn, F.; Hiltbrunner, D.; Zimmermann, S.; Niklaus, P.A. Spatial micro-distribution of methanotrophic activity along a 120-year afforestation chronosequence. Plant. Soil 2016, 415, 1–11. [Google Scholar] [CrossRef]
- Yuan, H.; Ge, T.; Chen, C.; O’Donnell, A.G.; Wu, J. Significant role for microbial autotrophy in the sequestration of soil carbon. Appl. Environ. Microbiol. 2012, 78, 2328–2336. [Google Scholar] [CrossRef]
- Hernández, M.; Conrad, R.; Klose, M.; Ma, K.; Lu, Y. Structure and function of methanogenic microbial communities in soils from flooded rice and upland soybean fields from Sanjiang plain, NE China. Soil Biol. Biochem. 2017, 105, 81–91. [Google Scholar] [CrossRef]
- Harriss, R.C.; Sebacher, D.I.; Day, F.P. Methane flux in the Great Dismal swamp. Nature 1982, 297, 673–674. [Google Scholar] [CrossRef]
- McDaniel, M.D.; Kaye, J.P.; Kaye, M.W. Do “hot moments” become hotter under climate change? Soil nitrogen dynamics from a climate manipulation experiment in a post-harvest forest. Biogeochemistry 2014, 121, 339–354. [Google Scholar] [CrossRef]
- Velthof, G.L.; Jarvis, S.C.; Stein, A.; Allen, A.G.; Oenema, O. Spatial variability of nitrous oxide fluxes in mown and grazed grasslands on a poorly drained clay soil. Soil Biol. Biochem. 1996, 28, 1215–1225. [Google Scholar] [CrossRef]
- McDaniel, M.D.; Simpson, R.R.; Malone, B.P.; McBratney, A.B.; Minasny, B.; Adams, M.A. Quantifying and predicting spatio-temporal variability of soil CH4 and N2O fluxes from a seemingly homogeneous Australian agricultural field. Agric. Ecosyst. Environ. 2017, 240, 182–193. [Google Scholar] [CrossRef]
- Priemé, A.; Christensen, S.; Dobbie, K.E.; Smith, K.A. Slow increase in rate of methane oxidation in soils with time following land use change from arable agriculture to woodland. Soil Biol. Biochem. 1997, 29, 1269–1273. [Google Scholar] [CrossRef]
- IPCC. Climate change 2013: The physical science basis. In Contribution of Working Group I to the Fifth Assessment Report of the Intergovern-Mental Panel on Climate Change; Stocker, T.F., Qin, D., Plattner, G.K., Tignor, M., Allen, S.K., Boschung, J., Nauels, A., Xia, Y., Bex, V., Mi, P.M., Eds.; Cambridge University Press: Cambridge, UK, 2013; p. 1535. [Google Scholar]
- Nazaries, L.; Murrell, J.C.; Millard, P.; Baggs, L.; Singh, B.K. Methane, microbes and models: Fundamental understanding of the soil methane cycle for future predictions. Environ. Microbiol. 2013, 15, 2395–2417. [Google Scholar] [CrossRef] [PubMed]
- Murrell, J.C.; Jetten, M.S.M. The microbial methane cycle. Environ. Microbiol. Rep. 2009, 1, 279–284. [Google Scholar] [CrossRef]
- Wu, X.; Yao, Z.; Brüggemann, N.; Shen, Z.Y.; Wolf, B.; Dannenmann, M.; Zheng, X.; Butterbach-Bahl, K. Effects of soil moisture and temperature on CO2 and CH4 soil–atmosphere exchange of various land use/cover types in a semi-arid grassland in Inner Mongolia, China. Soil Biol. Biochem. 2010, 42, 773–787. [Google Scholar] [CrossRef]
- Sullivan, B.W.; Kolb, T.E.; Hart, S.C.; Kaye, J.P.; Hungate, B.A.; Dore, S.; Montes-Helu, M. Wildfire reduces carbon dioxide efflux and increases methane uptake in ponderosa pine forest soils of the southwestern USA. Biogeochemistry 2010, 104, 251–265. [Google Scholar] [CrossRef]
- Wolf, K.; Flessa, H.; Veldkamp, E. Atmospheric methane uptake by tropical montane forest soils and the contribution of organic layers. Biogeochemistry 2012, 111, 469–483. [Google Scholar] [CrossRef]
- Knief, C. Diversity and habitat preferences of cultivated and uncultivated aerobic methanotrophic bacteria evaluated based on pmoA as molecular marker. Front. Microbiol. 2015, 6, 1346. [Google Scholar] [CrossRef]
- Khadem, A.F.; Wieczorek, A.S.; Pol, A.; Vuilleumier, S.; Harhangi, H.R.; Dunfield, P.F.; Kalyuzhnaya, M.G.; Murrell, J.C.; Francoijs, K.J.; Stunnenberg, H.G.; et al. Draft genome sequence of the volcano-inhabiting thermoacidophilic methanotroph Methylacidiphilum fumariolicum strain SolV. J. Bacteriol. 2012, 194, 3729. [Google Scholar] [CrossRef]
- Ettwig, K.F.; Butler, M.K.; Le Paslier, D.; Pelletier, E.; Mangenot, S.; Kuypers, M.M.M.; Schreiber, F.; Dutilh, B.E.; Zedelius, J.; de Beer, D.; et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature 2010, 464, 543–548. [Google Scholar] [CrossRef]
- Pratscher, J.; Vollmers, J.; Wiegand, S.; Dumont, M.G.; Kaster, A. Unravelling the identity, metabolic potential and global biogeography of the atmospheric methane-oxidizing Upland Soil Cluster α. Environ. Microbiol. 2018, 20, 1016–1029. [Google Scholar] [CrossRef] [PubMed]
- Singleton, C.M.; McCalley, C.K.; Woodcroft, B.J.; Boyd, J.A.; Evans, P.N.; Hodgkins, S.B.; Chanton, J.P.; Frolking, S.; Crill, P.M.; Saleska, S.R. Methanotrophy across a natural permafrost thaw environment. ISME J. 2018, 12, 2544–2558. [Google Scholar] [CrossRef]
- Tveit, A.T.; Hestnes, A.G.; Robinson, S.L.; Schintlmeister, A.; Dedysh, S.N.; Jehmlich, N.; von Bergen, M.; Herbold, C.; Wagner, M.; Richter, A.; et al. Widespread soil bacterium that oxidizes atmospheric methane. Proc. Natl. Acad. Sci. USA 2019, 116, 8515. [Google Scholar] [CrossRef]
- Thauer, R.K.; Kaster, A.-K.; Seedorf, H.; Buckel, W.; Hedderich, R. Methanogenic archaea: Ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 2008, 6, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Frenzel, P.; Rothfuss, F.; Conrad, R. Oxygen profiles and methane turnover in a flooded rice microcosm. Biol. Fertil. Soils 1992, 14, 84–89. [Google Scholar] [CrossRef]
- Sass, R.; Denmead, O.T.; Conrad, R.; Freney, J.; Klug, M.; Minami, K.; Mosier, A.; Neue, H.U.; Rennenberg, H.; Su, W.H. Exchange of methane and other trace gases in rice cultivation. Ecol. Bull. 1992, 199–206. [Google Scholar]
- Oremland, R.S.; Culbertson, C.W. Evaluation of methyl fluoride and dimethyl ether as inhibitors of aerobic methane oxidation. Appl. Environ. Microbiol. 1992, 58, 2983–2992. [Google Scholar] [CrossRef]
- Christiansen, J.R.; Levy-Booth, D.; Prescott, C.E.; Grayston, S.J. Microbial and environmental controls of methane fluxes along a soil moisture gradient in a Pacific coastal temperate rainforest. Ecosystems 2016, 19, 1255–1270. [Google Scholar] [CrossRef]
- Freitag, T.E.; Toet, S.; Ineson, P.; Prosser, J.I. Links between methane flux and transcriptional activities of methanogens and methane oxidizers in a blanket peat bog. FEMS Microbiol. Ecol. 2010, 73, 157–165. [Google Scholar] [CrossRef]
- Krause, S.; Meima-Franke, M.; Hefting, M.M.; Bodelier, P.L.E. Spatial patterns of methanotrophic communities along a hydrological gradient in a riparian wetland. FEMS Microbiol. Ecol. 2013, 86, 59–70. [Google Scholar] [CrossRef] [PubMed][Green Version]
- SILO. Queensland Government. Available online: https://silo.longpaddock.qld.gov.au/ (accessed on 21 March 2018).
- Jenkins, M.; Adams, M.A. Vegetation type determines heterotrophic respiration in subalpine Australian ecosystems. Glob. Chang. Biol. 2010, 16, 209–219. [Google Scholar] [CrossRef]
- Costin, A.B.; Hallsworth, E.G.; Woof, M. Studies in pedogenesis in New South Wales. J. Soil Sci. 1952, 3, 190–218. [Google Scholar] [CrossRef]
- Gee, G.W.; Bauder, J.W. Particle-size analysis. In Methods of Soil Analysis: Part 1—Physical and Mineralogical Methods; Soil Science Society of America: Madison, WI, USA, 1986; pp. 383–411. [Google Scholar]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef] [PubMed]
- Horz, H.P.; Rich, V.; Avrahami, S.; Bohannan, B.J.M. Methane-oxidizing bacteria in a California upland grassland soil: Diversity and response to simulated global change. Appl. Environ. Microbiol. 2005, 71, 2642–2652. [Google Scholar] [CrossRef] [PubMed]
- Kolb, S.; Knief, C.; Stubner, S.; Conrad, R. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 2003, 69, 2423–2429. [Google Scholar] [CrossRef] [PubMed]
- Bates, S.T.; Cropsey, G.W.G.; Caporaso, J.G.; Knight, R.; Fierer, N. Bacterial communities associated with the lichen symbiosis. Appl. Environ. Microbiol. 2011, 77, 1309–1314. [Google Scholar] [CrossRef]
- Hernández, M.; Dumont, M.G.; Yuan, Q.; Conrad, R. Different bacterial populations associated with the roots and rhizosphere of rice incorporate plant-derived carbon. Appl. Environ. Microbiol. 2015, 81, 2244–2253. [Google Scholar] [CrossRef]
- Holmes, A.J.; Costello, A.; Lidstrom, M.E.; Murrell, J.C. Evidence that participate methane monooxygenase and ammonia monooxygenase may be evolutionarily related. FEMS Microbiol. Lett. 1995, 132, 203–208. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Reim, A.; Hernández, M.; Klose, M.; Chidthaisong, A.; Yuttitham, M.; Conrad, R. Response of methanogenic microbial communities to desiccation stress in flooded and rain-fed paddy soil from Thailand. Front. Microbiol. 2017, 8, 785. [Google Scholar] [CrossRef] [PubMed]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Dumont, M.G.; Lüke, C.; Deng, Y.; Frenzel, P. Classification of pmoA amplicon pyrosequences using BLAST and the lowest common ancestor method in MEGAN. Front. Microbiol. 2014, 5, 1–11. [Google Scholar] [CrossRef]
- Zuur, A.F.; Ieno, E.N.; Elphick, C.S. A protocol for data exploration to avoid common statistical problems. Methods Ecol. Evol. 2010, 1, 3–14. [Google Scholar] [CrossRef]
- RC Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Vegan: Community ecology package. Agric. Sci. 2016, 7, 1–48. [Google Scholar]
- Warnes, G.; Bolker, B.; Lumley, T. gplots: Various R programming tools for plotting data. R Package Vers 2015, 2, 1. [Google Scholar]
- BOM Bureau of Meteorology. Maps. Recent and Past Conditions. Available online: http://www.bom.gov.au/climate/maps/ (accessed on 5 October 2020).
- Frost, A.J.; Ramchurn, A.; Smith, A. The Australian Landscape Water Balance Model; Bureau of Meteorology: Melbourne, Australia, 2018.
- IBRA7. Interim Bioregionalisation for Australia, version 7. Available online: https://environment.gov.au/land/nrs/science/ibra#ibra (accessed on 5 April 2020).
- Department of Energy and Environment. Woody Cover National Forest and Sparse Woody Vegetation Data (Version 3). Available online: https://data.gov.au/data/dataset/d734c65e-0e7b-4190-9aa5-ddbb5844e86d (accessed on 6 December 2019).
- Tarboton, D.G. Terrain Analysis using Digital Elevation Models (TauDEM); Utah State University: Logan, UT, USA, 2005. [Google Scholar]
- Bastida, F.; Eldridge, D.J.; García, C.; Png, G.K.; Bardgett, R.D.; Delgado-Baquerizo, M. Soil microbial diversity–biomass relationships are driven by soil carbon content across global biomes. ISME J. 2021, 1–11. [Google Scholar] [CrossRef]
- Zhou, J.; Xia, B.; Treves, D.S.; Wu, L.-Y.; Marsh, T.L.; O’Neill, R.V.; Palumbo, A.V.; Tiedje, J.M. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 2002, 68, 326–334. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, R.B. The diversity and biogeography of soil bacterial communities. Proc. Natl. Acad. Sci. USA 2006, 103, 626–631. [Google Scholar] [CrossRef]
- Hu, B.; Shen, L.; Lian, X.; Zhu, Q.; Liu, S.; Huang, Q.; He, Z.; Geng, S.; Cheng, D.; Lou, L.; et al. Evidence for nitrite-dependent anaerobic methane oxidation as a previously overlooked microbial methane sink in wetlands. Proc. Natl. Acad. Sci. USA 2014, 111, 4495–4500. [Google Scholar] [CrossRef] [PubMed]
- Luesken, F.A.; Wu, M.L.; Op den Camp, H.J.M.; Keltjens, J.T.; Stunnenberg, H.; Francoijs, K.; Strous, M.; Jetten, M.S.M. Effect of oxygen on the anaerobic methanotroph ‘Candidatus Methylomirabilis oxyfera’: Kinetic and transcriptional analysis. Environ. Microbiol. 2012, 14, 1024–1034. [Google Scholar] [CrossRef]
- Le Mer, J.; Roger, P. Production, oxidation, emission and consumption of methane by soils: A review. Eur. J. Soil Biol. 2001, 37, 25–50. [Google Scholar] [CrossRef]
- Yang, W.H.; Silver, W.L. Net soil-atmosphere fluxes mask patterns in gross production and consumption of nitrous oxide and methane in a managed ecosystem. Biogeosciences 2016, 13, 1705. [Google Scholar] [CrossRef]
- Bender, M.; Conrad, R. Kinetics of CH4 oxidation in oxic soils exposed to ambient air or high CH4 mixing ratios. FEMS Microbiol. Lett. 1992, 101, 261–269. [Google Scholar] [CrossRef]
- Von Fischer, J.C.; Hedin, L.O. Separating methane production and consumption with a field-based isotope pool dilution technique. Glob. Biogeochem. Cycles 2002, 16, 1–13. [Google Scholar] [CrossRef]
- Conrad, R. Control of microbial methane production in wetland rice fields. Nutr. Cycl. Agroecosyst. 2002, 64, 59–69. [Google Scholar] [CrossRef]
- Angel, R.; Claus, P.; Conrad, R. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 2012, 6, 847–862. [Google Scholar] [CrossRef]
- Kolb, S.; Knief, C.; Dunfield, P.F.; Conrad, R. Abundance and activity of uncultured methanotrophic bacteria involved in the consumption of atmospheric methane in two forest soils. Environ. Microbiol. 2005, 7, 1150–1161. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Dumont, M.G.; Cébron, A.; Murrell, J.C. Identification of active methanotrophs in a landfill cover soil through detection of expression of 16S rRNA and functional genes. Environ. Microbiol. 2007, 9, 2855–2869. [Google Scholar] [CrossRef] [PubMed]
- Bengtson, P.; Basiliko, N.; Dumont, M.G.; Hills, M.; Murrell, J.C.; Roy, R.; Grayston, S.J. Links between methanotroph community composition and CH4 oxidation in a pine forest soil. FEMS Microbiol. Ecol. 2009, 70, 356–366. [Google Scholar] [CrossRef]
- Kolb, S. The quest for atmospheric methane oxidizers in forest soils. Environ. Microbiol. Rep. 2009, 1, 336–346. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, P.M.; Kammann, C.; Lenhart, K.; Dam, B.; Liesack, W. Linking activity, composition and seasonal dynamics of atmospheric methane oxidizers in a meadow soil. ISME J. 2012, 6, 1115–1126. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Yang, S.; Rozova, O.N.; Smalley, N.E.; Clubb, J.; Lamb, A.; Gowda, G.A.N.; Raftery, D.; Fu, Y.; Bringel, F.; et al. Highly efficient methane biocatalysis revealed in a methanotrophic bacterium. Nat. Commun. 2013, 4, 2785. [Google Scholar] [CrossRef]
- Kits, K.D.; Campbell, D.J.; Rosana, A.R.; Stein, L.Y. Diverse electron sources support denitrification under hypoxia in the obligate methanotroph Methylomicrobium album strain BG8. Front. Microbiol. 2015, 6, 1072. [Google Scholar] [CrossRef] [PubMed]
- Wartiainen, I.; Hestnes, A.G.; McDonald, I.R.; Svenning, M.M. Methylocystis rosea sp. nov., a novel methanotrophic bacterium from Arctic wetland soil, Svalbard, Norway (78° N). Int. J. Syst. Evol. Micr. 2006, 56, 541–547. [Google Scholar] [CrossRef]
- Leng, L.; Chang, J.; Geng, K.; Lu, Y.; Ma, K. Uncultivated Methylocystis species in paddy soil include facultative methanotrophs that utilize acetate. Micr. Ecol. 2015, 70, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Zheng, Y.; Bodelier, P.L.E.; Conrad, R.; Jia, Z. Conventional methanotrophs are responsible for atmospheric methane oxidation in paddy soils. Nat. Commun. 2016, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.A.; Dobbie, K.E.; Ball, B.C.; Bakken, L.R.; Sitaula, B.K.; Hansen, S.; Brumme, R.; Borken, W.; Christensen, S.; Priemé, A.; et al. Oxidation of atmospheric methane in Northern European soils, comparison with other ecosystems, and uncertainties in the global terrestrial sink. Glob. Chang. Biol. 2000, 6, 791–803. [Google Scholar] [CrossRef]
- Shukla, P.N.; Pandey, K.D.; Mishra, V.K. Environmental determinants of soil methane oxidation and methanotrophs. Crit. Rev. Environ. Sci. Technol. 2013, 43, 1945–2011. [Google Scholar] [CrossRef]
- Malghani, S.; Reim, A.; von Fischer, J.; Conrad, R.; Kuebler, K.; Trumbore, S.E. Soil methanotroph abundance and community composition are not influenced by substrate availability in laboratory incubations. Soil Biol. Biochem. 2016, 101, 184–194. [Google Scholar] [CrossRef]
- Knief, C.; Lipski, A.; Dunfield, P.F. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 2003, 69, 6703–6714. [Google Scholar] [CrossRef] [PubMed]
- Nazaries, L.; Tate, K.R.; Ross, D.J.; Singh, J.; Dando, J.; Saggar, S.; Baggs, E.M.; Millard, P.; Murrell, J.C.; Singh, B.K. Response of methanotrophic communities to afforestation and reforestation in New Zealand. ISME J. 2011, 5, 1832–1836. [Google Scholar] [CrossRef] [PubMed]
- Nazaries, L.; Pan, Y.; Bodrossy, L.; Baggs, E.M.; Millard, P.; Murrell, J.C.; Singh, B.K. Evidence of microbial regulation of biogeochemical cycles from a study on methane flux and land use change. Appl. Environ. Microbiol. 2013, 79, 4031–4040. [Google Scholar] [CrossRef]
- Saari, A.; Martikainen, P.J.; Ferm, A.; Ruuskanen, J.; De Boer, W.; Troelstra, S.R.; Laanbroek, H.J. Methane oxidation in soil profiles of Dutch and Finnish coniferous forests with different soil texture and atmospheric nitrogen deposition. Soil Biol. Biochem. 1997, 29, 1625–1632. [Google Scholar] [CrossRef]
- Fest, B.; Wardlaw, T.; Livesley, S.J.; Duff, T.J.; Arndt, S.K. Changes in soil moisture drive soil methane uptake along a fire regeneration chronosequence in a eucalypt forest landscape. Glob. Chang. Biol. 2015, 21, 4250–4264. [Google Scholar] [CrossRef] [PubMed]
- D’imperio, L.; Nielsen, C.S.; Westergaard-Nielsen, A.; Michelsen, A.; Elberling, B. Methane oxidation in contrasting soil types: Responses to experimental warming with implication for landscape-integrated CH4 budget. Glob. Chang. Biol. 2017, 23, 966–976. [Google Scholar] [CrossRef]
- Pratscher, J.; Dumont, M.G.; Conrad, R. Assimilation of acetate by the putative atmospheric methane oxidizers belonging to the USCα clade. Environ. Microbiol. 2011, 13, 2692–2701. [Google Scholar] [CrossRef]
- Sullivan, B.W.; Selmants, P.C.; Hart, S.C. Does dissolved organic carbon regulate biological methane oxidation in semiarid soils? Glob. Chang. Biol. 2013, 19, 2149–2157. [Google Scholar] [CrossRef]
- McDaniel, M.D.; Saha, D.; Dumont, M.G.; Hernández, M.; Adams, M.A. The effect of land-use change on soil CH4 and N2O fluxes: A global meta-analysis. Ecosystems 2019, 22, 1424–1443. [Google Scholar] [CrossRef]
- Singh, J.S.; Singh, S.; Raghubanshi, A.S.; Singh, S.; Kashyap, A.K.; Reddy, V.S. Effect of soil nitrogen, carbon and moisture on methane uptake by dry tropical forest soils. Plant Soil 1997, 196, 115–121. [Google Scholar] [CrossRef]
- Reeburgh, W.S. Global methane biogeochemistry. Treatise Geochem. 2003, 4, 347. [Google Scholar]
- Dutaur, L.; Verchot, L.V. A global inventory of the soil CH4 sink. Glob. Biogeochem. Cycles 2007, 21, 1–9. [Google Scholar] [CrossRef]
- Kirschke, S.; Bousquet, P.; Ciais, P.; Saunois, M.; Canadell, J.G.; Dlugokencky, E.J.; Bergamaschi, P.; Bergmann, D.; Blake, D.R.; Bruhwiler, L. Three decades of global methane sources and sinks. Nat. Geosci. 2013, 6, 813–823. [Google Scholar] [CrossRef]
- Saunois, M.; Bousquet, P.; Poulter, B.; Peregon, A.; Ciais, P.; Canadell, J.G.; Dlugokencky, E.J.; Etiope, G.; Bastviken, D.; Houweling, S. The global methane budget 2000–2012. Earth Syst. Sci. Data 2016, 8, 697–751. [Google Scholar] [CrossRef]
- Gatica, G.; Fernández, M.E.; Juliarena, M.P.; Gyenge, J. Environmental and anthropogenic drivers of soil methane fluxes in forests: Global patterns and among-biomes differences. Glob. Chang. Biol. 2020, 26, 6604–6615. [Google Scholar] [CrossRef] [PubMed]
- Koschorreck, M.; Conrad, R. Oxidation of atmospheric methane in soil: Measurements in the field, in soil cores and in soil samples. Glob. Biogeochem. Cycles 1993, 7, 109–121. [Google Scholar] [CrossRef]
- Hütsch, B.W. Tillage and land use effects on methane oxidation rates and their vertical profiles in soil. Biol. Fertil. Soils 1998, 27, 284–292. [Google Scholar] [CrossRef]
- Priemé, A.; Christensen, S. Seasonal and spatial variation of methane oxidation in a Danish spruce forest. Soil Biol. Biochem. 1997, 29, 1165–1172. [Google Scholar] [CrossRef]
- Hütsch, B.W.; Webster, C.P.; Powlson, D.S. Methane oxidation in soil as affected by land use, soil pH and N fertilization. Soil Biol. Biochem. 1994, 26, 1613–1622. [Google Scholar] [CrossRef]
- Schnell, S.; King, G.M. Mechanistic analysis of ammonium inhibition of atmospheric methane consumption in forest soils. Appl. Environ. Microbiol. 1994, 60, 3514–3521. [Google Scholar] [CrossRef] [PubMed]
- Ni, X.; Groffman, P.M. Declines in methane uptake in forest soils. Proc. Natl. Acad. Sci.USA 2018, 115, 8587–8590. [Google Scholar] [CrossRef]
- Adams, M.A.; Cunningham, S.C.; Taranto, M.T. A critical review of the science underpinning fire management in the high altitude ecosystems of south-eastern Australia. For. Ecol. Manag. 2013, 294, 225–237. [Google Scholar] [CrossRef]
- Adams, M.A. Mega-fires, tipping points and ecosystem services: Managing forests and woodlands in an uncertain future. For. Ecol. Manag. 2013, 294, 250–261. [Google Scholar] [CrossRef]
- Jónsson, J.Ö.G.; Davíðsdóttir, B. Classification and valuation of soil ecosystem services. Agric. Syst. 2016, 145, 24–38. [Google Scholar] [CrossRef]
| Ecosystem | % of 548 ha Watershed | Net Ecosystem Flux | |
|---|---|---|---|
| CO2 | CH4 | ||
| CO2-equivalents g m−2 y−1 | |||
| Forest | 8–75 | 911 ± 165 | −506 ± 22 |
| Grassland | 25–91 | 484 ± 50 | −259 ± 38 |
| Bog | 1 | 646 ± 109 | 256 ± 82 |
| CO2-equivalents kg ha−1 y−1 | |||
| Total Watershed | 5207 to 8031 | −2762 to −4391 | |
| Measurement | Values | ||
|---|---|---|---|
| Ecosystems Areal Coverage | |||
| Global forest + grassland ecosystems (M ha) | 5100 | ||
| Areal coverage of Australian Alps—Figure S5 (M ha) | 1.23 | ||
| Fraction of Australian Alps to global forest + grassland (%) | 0.024 | ||
| Soil CH4 Sink Estimates | Low Estimate | Best Estimate | High Estimate |
| Global CH4 soil sink † (Tg y−1) | −9 | −30 | −100 |
| Mean annual Australian Alps CH4 sink ‡ (kg ha−1 y−1) | −4.2 | −19.2 | −33.2 |
| Australian Alps Contribution to Global CH4 Sink (%) | Low Estimate | Best Estimate | High Estimate |
| Low estimate (−4.2 kg ha−1 y−1) | 69 | 21 | 6 |
| Best estimate, or our projection (−19.2 kg ha−1 y−1) | 213 | 64 | 19 |
| High estimate (−33.2 kg ha−1 y−1) | 359 | 108 | 32 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
McDaniel, M.D.; Hernández, M.; Dumont, M.G.; Ingram, L.J.; Adams, M.A. Disproportionate CH4 Sink Strength from an Endemic, Sub-Alpine Australian Soil Microbial Community. Microorganisms 2021, 9, 606. https://doi.org/10.3390/microorganisms9030606
McDaniel MD, Hernández M, Dumont MG, Ingram LJ, Adams MA. Disproportionate CH4 Sink Strength from an Endemic, Sub-Alpine Australian Soil Microbial Community. Microorganisms. 2021; 9(3):606. https://doi.org/10.3390/microorganisms9030606
Chicago/Turabian StyleMcDaniel, Marshall D., Marcela Hernández, Marc G. Dumont, Lachlan J. Ingram, and Mark A. Adams. 2021. "Disproportionate CH4 Sink Strength from an Endemic, Sub-Alpine Australian Soil Microbial Community" Microorganisms 9, no. 3: 606. https://doi.org/10.3390/microorganisms9030606
APA StyleMcDaniel, M. D., Hernández, M., Dumont, M. G., Ingram, L. J., & Adams, M. A. (2021). Disproportionate CH4 Sink Strength from an Endemic, Sub-Alpine Australian Soil Microbial Community. Microorganisms, 9(3), 606. https://doi.org/10.3390/microorganisms9030606







