Grazing Affects Bacterial and Fungal Diversities and Communities in the Rhizosphere and Endosphere Compartments of Leymus chinensis through Regulating Nutrient and Ion Distribution
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Sampling
2.3. Plant Physiology and Soil Properties
2.4. DNA Extraction, Amplification, and Sequencing
2.5. Data Analysis
3. Results
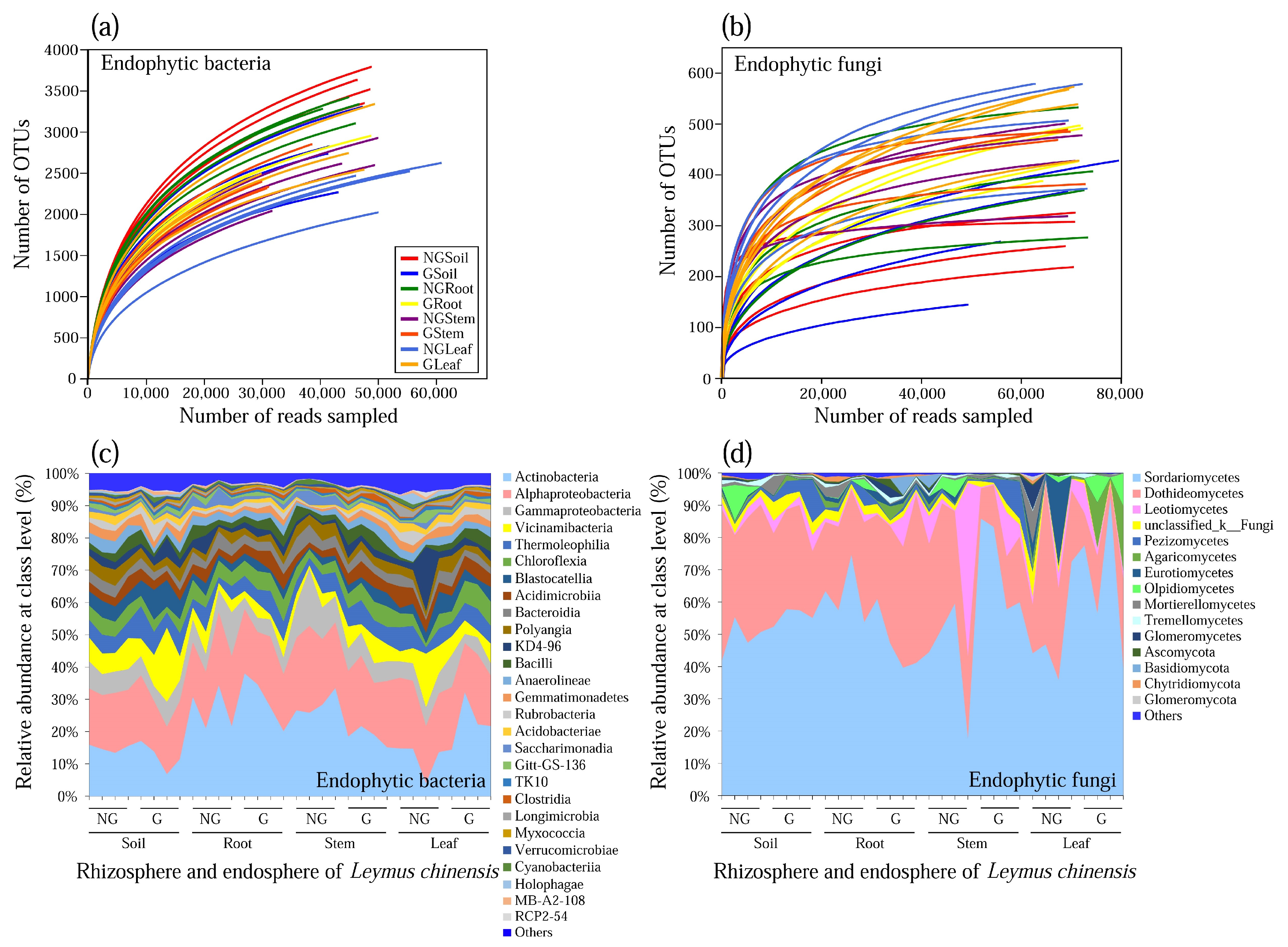
3.1. Community Composition of Endophytic Bacteria and Fungi
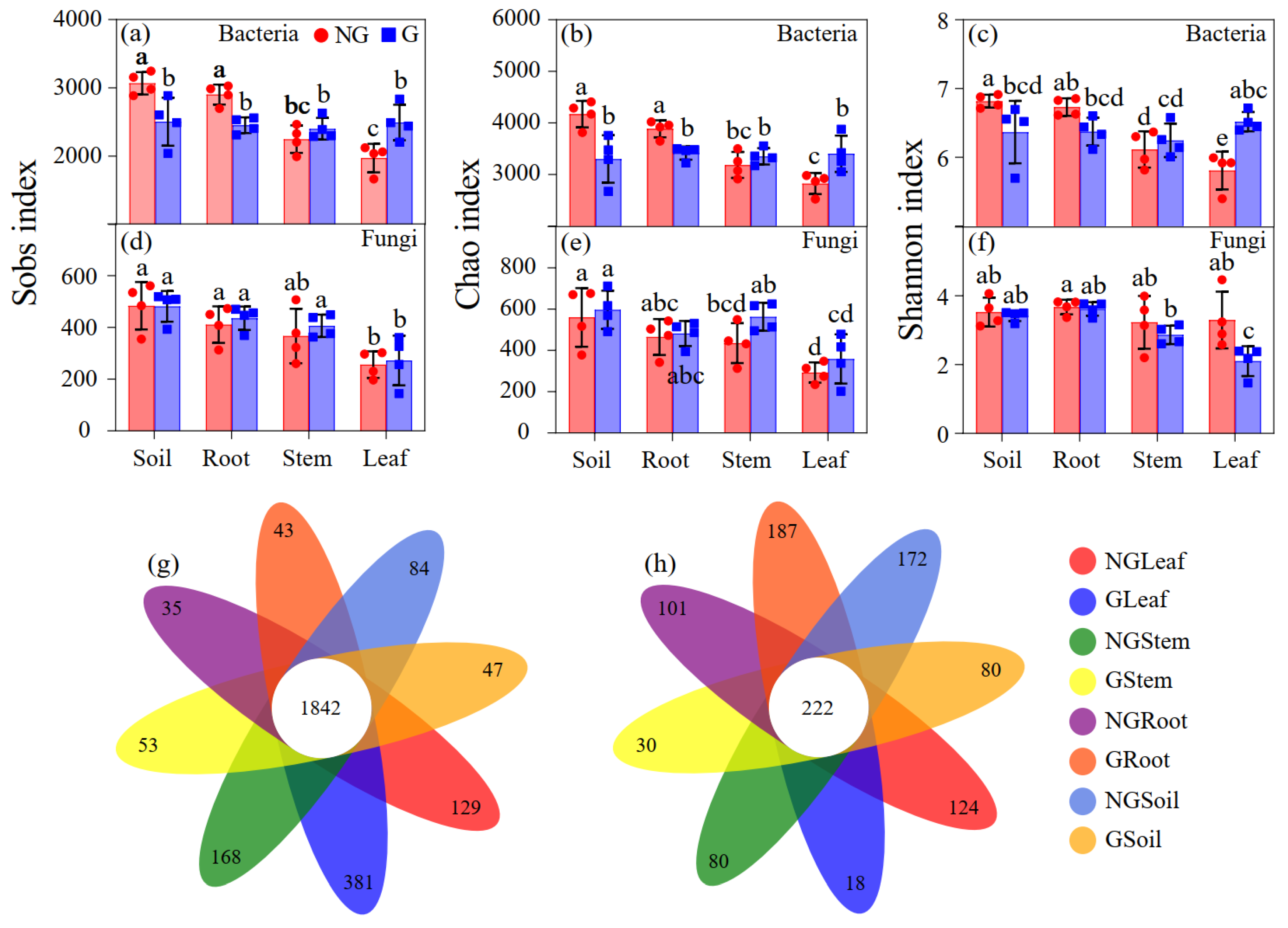
3.2. Alpha Diversity of Endophytic Bacterial and Fungal Communities
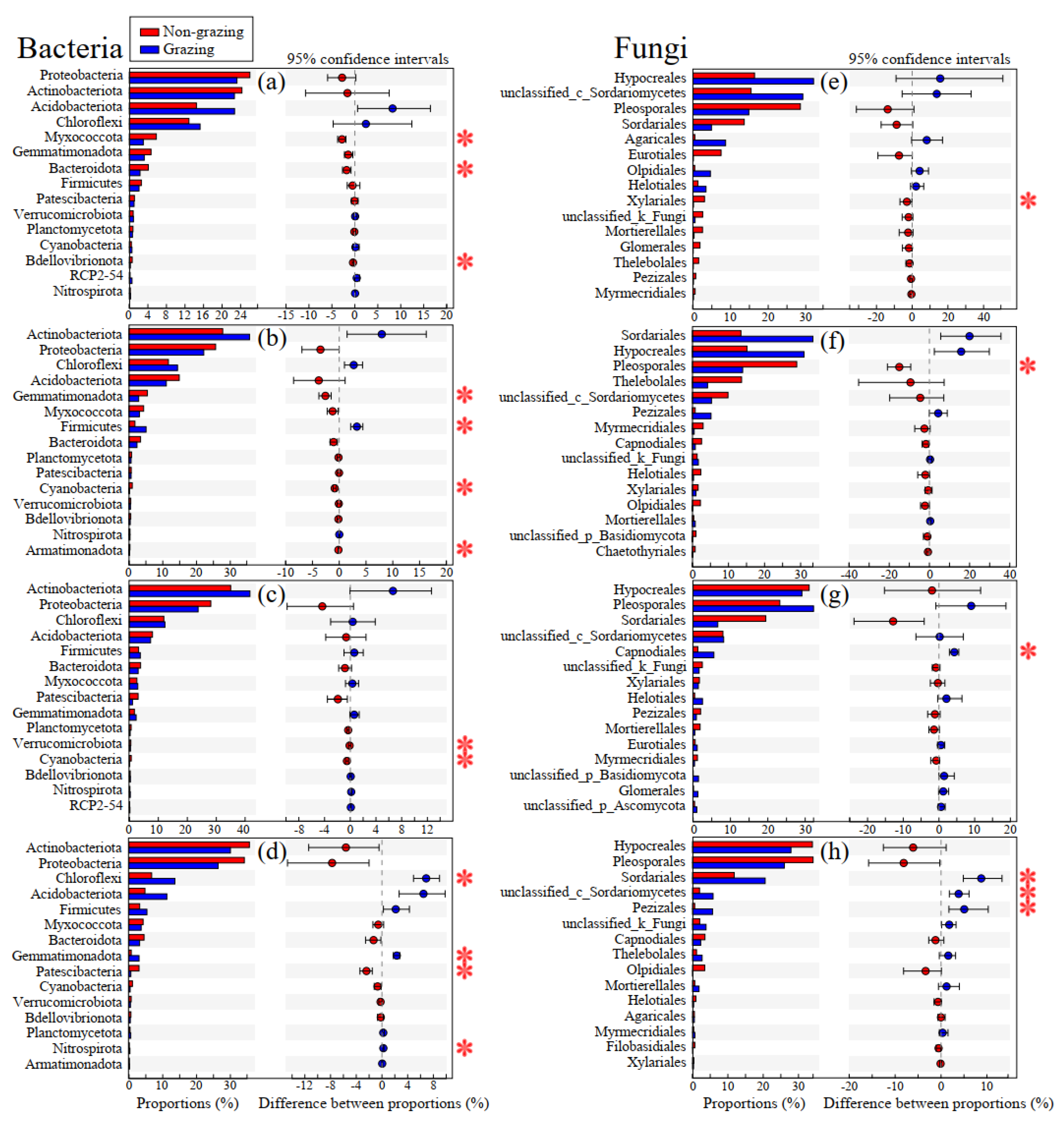
3.3. Difference in Endophytic Bacterial Phylum and Fungal Order among Plant Compartments
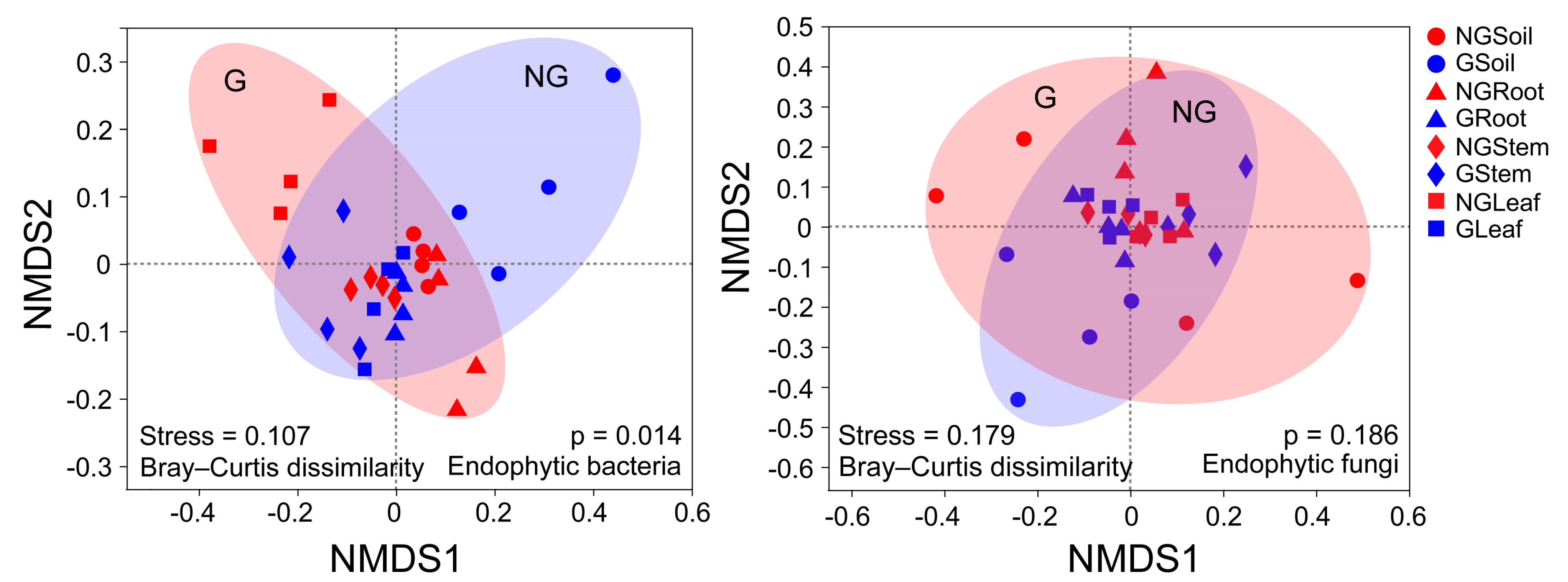
3.4. Beta Diversity of Endophytic Bacterial and Fungal Communities
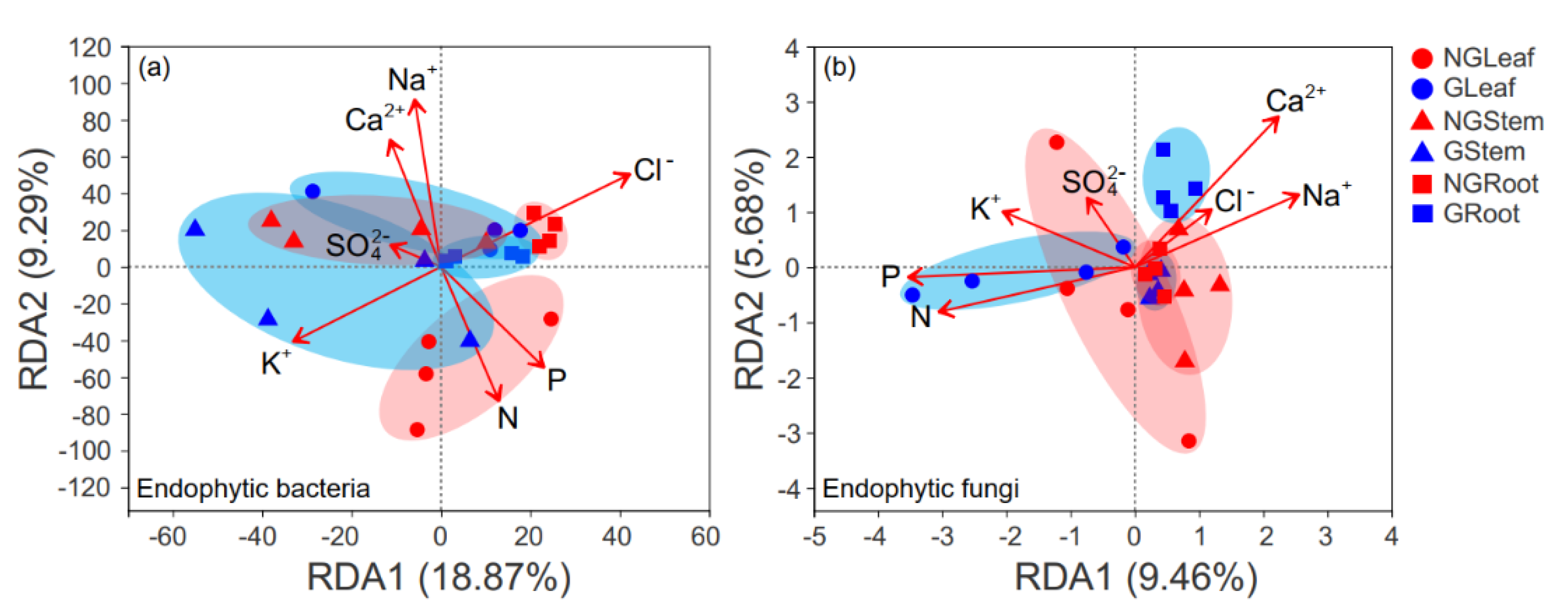
3.5. Relationship between the Endophytic Microbial Community and Environmental Variables
3.6. Co-Occurrence Network Analysis on Endophytic Bacterial and Fungal Communities
4. Discussion
4.1. Grazing-Induced Changes in Soil Properties and Plant Characteristics
4.2. Grazing-Induced Changes in Endophytic Microbial Diversity and Community
4.3. How Grazing Influence Endophytic Microbial Diversity and Community Composition?
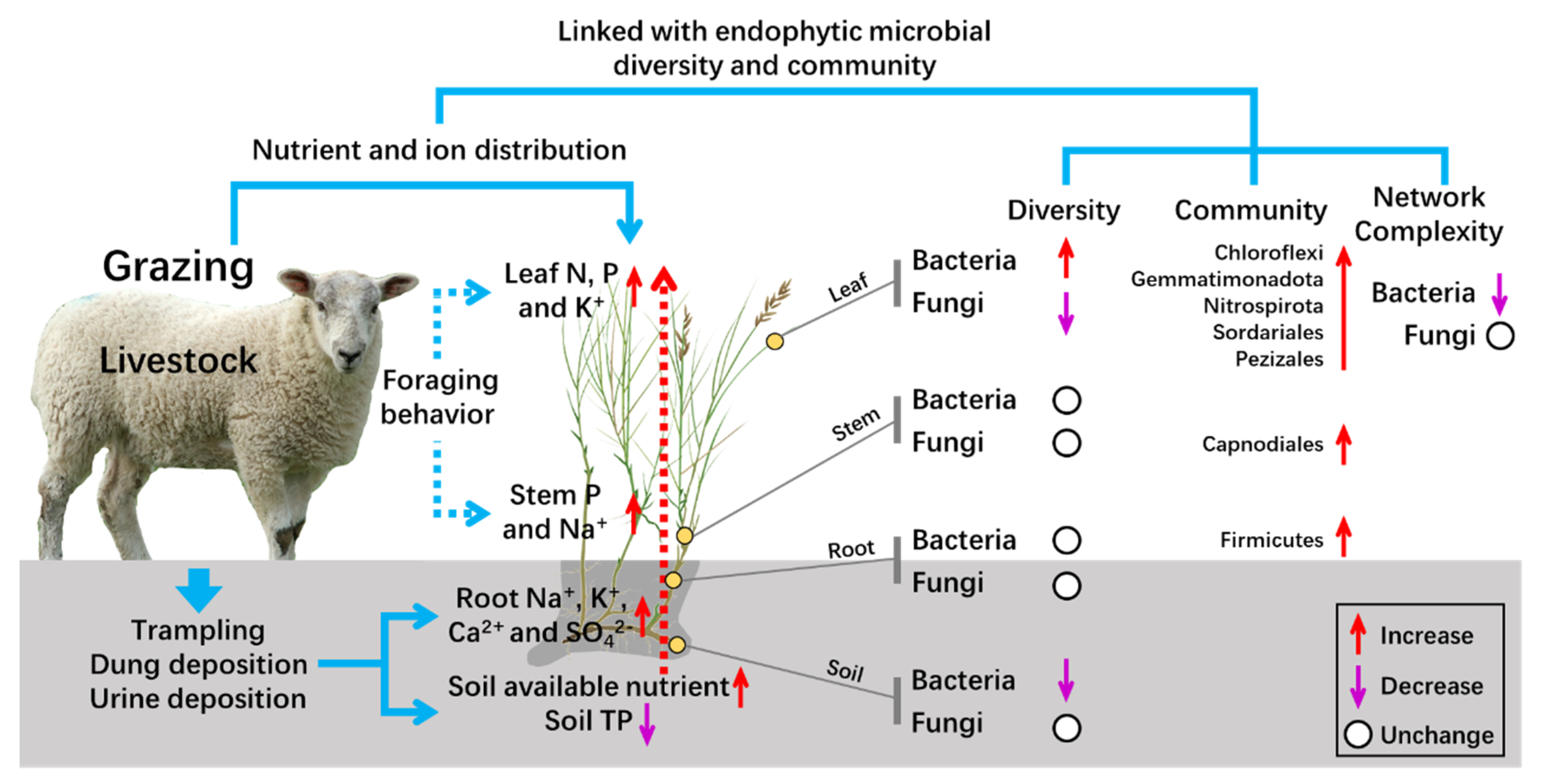
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Endophytic microbes and their potential applications in crop management. Pest. Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef] [PubMed]
- Manganyi, M.C.; Ateba, C.N. Untapped potentials of endophytic fungi: A review of novel bioactive compounds with biological applications. Microorganisms 2020, 8, 1934. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Ryan, R.P.; Germaine, K.; Franks, A.; Ryan, D.J.; Dowling, D.N. Bacterial endophytes: Recent developments and applications. FEMS Microbiol. Lett. 2008, 278, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Amaresan, N.; Jayakumar, V.; Kumar, K.; Thajuddin, N. Endophytic bacteria from tomato and chilli, their diversity and antagonistic potential against Ralstonia solanacearum. Arch. Phytopathol. Pflanzenschutz. 2012, 45, 344–355. [Google Scholar] [CrossRef]
- Nicoletti, R.; Vaio, C.D.; Cirillo, C. Endophytic fungi of olive tree. Microorganisms 2020, 8, 1321. [Google Scholar] [CrossRef] [PubMed]
- Clay, K.; Holah, J. Fungal endophyte symbiosis and plant diversity in successional fields. Science 1999, 285, 1742–1744. [Google Scholar] [CrossRef] [PubMed]
- Griffin, E.A.; Carson, W.P. Tree endophytes: Cryptic drivers of tropical forest diversity. In Endophytes of Forest Trees; Springer: Cham, Switzerland, 2018; pp. 63–103. [Google Scholar]
- Rodriguez, R.J.; White, J.F.; Arnold, A.E.; Redman, R.S. Fungal endophytes: Diversity and functional roles. New Phytol. 2009, 182, 314–330. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Al-Hosni, K.; Kang, S.M.; Seo, C.W.; Lee, I.J. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 2017, 68, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Germaine, K.; Keogh, E.; Garcia-Cabellos, G.; Borremans, B.; Lelie, D.; Barac, T.; Oeyen, L.; Vangronsveld, J.; Moore, F.P.; Moore, E.R.B.; et al. Colonisation of poplar trees by gfp expressing bacterial endophytes. FEMS Microbiol. Ecol. 2004, 48, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Khan Chowdhury, M.D.E.; Jeon, J.; Ok Rim, S.; Park, Y.H.; Kyu Lee, S.; Bae, H. Composition, diversity and bioactivity of culturable bacterial endophytes in mountain-cultivated ginseng in Korea. Sci. Rep. 2017, 7, 10098. [Google Scholar] [CrossRef] [PubMed]
- Rocha, P.D.S.D.; Paula, V.M.B.; Olinto, S.C.F.; Santos, E.L.D.; Souza, K.D.P.; Estevinho, L.M. Diversity, chemical constituents and biological activities of endophytic fungi isolated from Schinus terebinthifolius Raddi. Microorganisms 2020, 8, 859. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.M.; Wang, Z.; Mei, Y.; Wang, L.Q.; Wang, X.; Xu, Q.S.; Peng, S.; Zhou, Y.; Wei, C.L. Isolation, diversity, and growth-promoting activities of endophytic bacteria from tea cultivars of Zijuan and Yunkang-10. Front. Microbiol. 2018, 9, 1848. [Google Scholar] [CrossRef] [PubMed]
- Lodewyckx, C.; Vangronsveld, J.; Porteous, F.; Moore, E.R.; Taghavi, S.; Mezgeay, M.; der Lelie, D.V. Endophytic bacteria and their potential application. Crit. Rev. Plant Sci. 2002, 21, 583–606. [Google Scholar] [CrossRef]
- Ferreira, A.; Quecine, M.C.; Lacava, P.T.; Oda, S.; Azevedo, J.L.; Araújo, W.L. Diversity of endophytic bacteria from eucalyptus species seeds and colonization of seedlings by Pantoea agglomerans. FEMS Microbiol. Lett. 2010, 287, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, P.; Chauhan, P.S.; Saravanan, V.S.; Anandham, R.; Sa, T. Isolation and characterization of plant growth promoting endophytic bacterial isolates from root nodule of Lespedeza sp. Biol. Fert. Soils 2010, 46, 807–816. [Google Scholar] [CrossRef]
- Edwards, J.; Johnson, C.; Santos-Medellín, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, 911–920. [Google Scholar] [CrossRef] [PubMed]
- Robinson, R.J.; Fraaije, B.A.; Clark, I.M.; Jackson, R.W.; Hirsch, P.R.; Mauchline, T.H. Endophytic bacterial community composition in wheat (Triticum aestivum) is determined by plant tissue type, developmental stage and soil nutrient availability. Plant Soil 2015, 405, 1–16. [Google Scholar] [CrossRef]
- Thongsandee, W.; Matsuda, Y.; Ito, S. Temporal variations in endophytic fungal assenmblages of Ginkgo biloba L. J. Forest Res. 2012, 17, 213. [Google Scholar] [CrossRef]
- Franzluebbers, A.J. Short-term responses of soil C and N fractions to tall fescue endophyte infection. Plant Soil 2006, 282, 153–164. [Google Scholar] [CrossRef]
- Ren, F.; Dong, W.; Yan, D.H. Endophytic bacterial communities of Jingbai Pear trees in north China analyzed with Illumina sequencing of 16S rDNA. Arch. Microbiol. 2019, 201, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Kohler, F.; Hamelin, J.; Gillet, F.; Gobat, J.M.; Buttler, A. Soil microbial community changes in wooded mountain pastures due to simulated effects of cattle grazing. Plant Soil 2005, 278, 327–340. [Google Scholar] [CrossRef]
- Wang, D.; Ba, L. Ecology of meadow steppe in northeast china. Rangel. J. 2008, 30, 247–254. [Google Scholar] [CrossRef]
- Wang, L.J.; Li, X.F.; Chen, S.Y.; Liu, G.S. Enhanced drought tolerance in transgenic Leymus chinensis plants with constitutively expressed wheat TaLEA3. Biotechnol. Lett. 2009, 31, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, D.; Xing, F.; Liu, J.; Wang, L. Combined effects of resource heterogeneity and simulated herbivory on plasticity of clonal integration in a rhizomatous perennial herb. Plant Biol. 2014, 16, 774–782. [Google Scholar] [CrossRef]
- Wang, Y.; Zhai, X.; Sun, H.; Chang, H.; Rong, L. Performance of different sampling methods in estimating species richness in semiarid grassland. Ecol. Eng. 2019, 136, 10–16. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Sun, S.; Xing, F.; Mu, X.; Bai, Y. Nitrogen addition interacted with salinity-alkalinity to modify plant diversity, microbial PLFAs and soil coupled elements: A 5-year experiment. Appl. Soil Ecol. 2019, 137, 78–86. [Google Scholar] [CrossRef]
- Yang, C.; Zheng, S.; Huang, H.; Liu, Z.; Zheng, W.; Liu, B.; Shi, D. Comparison of osmotic adjustment and ion balance strategies in nineteen alkali-tolerant halophyte species during adaptation to salt-alkalinized habitats in northeast China. Aust. J. Crop Sci. 2012, 6, 141–148. [Google Scholar]
- Ahmed, M.Z.; Shimazaki, T.; Gulzar, S.; Kikuchi, A.; Gul, B.; Khan, M.A.; Koyro, H.W.; Huchzermeyer, B.; Watanabe, K.N. The influence of genes regulating transmembrane transport of Na+ on the salt resistance of Aeluropus lagopoides. Funct. Plant Biol. 2013, 40, 860–871. [Google Scholar] [CrossRef] [PubMed]
- Yin, R.; Deng, H.; Wang, H.L.; Zhang, B. Vegetation type affects soil enzyme activities and microbial functional diversity following re-vegetation of a severely eroded red soil in sub-tropical China. Catena 2014, 115, 96–103. [Google Scholar] [CrossRef]
- Li, X.; Ding, C.; Bu, H.; Han, L.; Ma, P.; Su, D. Effects of submergence frequency on soil C:N:P ecological stoichiometry in riparian zones of Hulunbuir steppe. J. Soil. Sediment. 2012, 20, 1–14. [Google Scholar] [CrossRef]
- Anderson, J.M.; Ingram, J.S.I. Tropical soil biology and fertility. Soil Sci. 1994, 157, 265. [Google Scholar] [CrossRef]
- Masoud, W.; Takamiya, M.; Vogensen, F.K.; Lillevang, S.; Al-Soud, W.A.; Sørensen, S.J.; Jakobsen, M. Characterization of bacterial populations in Danish raw milk cheeses made with different starter cultures by denaturating gradient gel electrophoresis and pyrosequencing. Int. Dairy J. 2011, 21, 142–148. [Google Scholar] [CrossRef]
- Terhonen, E.; Keriö, S.; Sun, H.; Asiegbu, F.O. Endophytic fungi of Norway spruce roots in boreal pristine mire, drained peatland and mineral soil and their inhibitory effect on Heterobasidion parviporum in vitro. Fungal Ecol. 2014, 9, 17–26. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Stevens, M.H.; Oksanen, M.J.; Suggests, M.A. The Vegan Package. Community Ecology Package. 2007. Available online: http://cran.r-project.org/ (accessed on 10 January 2021).
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5–5. 2019. Available online: http://packages.renjin.org/package/org.renjin.cran/vegan (accessed on 10 January 2021).
- Lumley, T.C.; Gignac, L.D.; Currah, R.S. Microfungus communities of white spruce and trembling aspen logs at different stages of decay in disturbed and undisturbed sites in the boreal mixedwood region of Alberta. Can. J. Bot. 2001, 79, 76–92. [Google Scholar]
- Parks, D.H.; Beiko, R.G. Identifying biologically relevant differences between metagenomic communities. Bioinformatics 2010, 26, 715–721. [Google Scholar] [CrossRef] [PubMed]
- Goslee, S.C.; Urban, D.L. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 2007, 22, 1–9. [Google Scholar] [CrossRef]
- Barberán, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Greenwood, K.L.; McKenzie, B.M. Grazing effects on soil physical properties and the consequences for pastures: A review. Aust. J. Exp. Agr. 2001, 41, 1231–1250. [Google Scholar] [CrossRef]
- Su, Y.-Z.; Li, Y.-L.; Cui, J.-Y.; Zhao, W.-Z. Influences of continuous grazing and livestock exclusion on soil properties in a degraded sandy grassland, Inner Mongolia, northern China. Catena 2005, 59, 267–278. [Google Scholar]
- Liu, C.; Li, W.; Xu, J.; Wei, W.; Xue, P.; Yan, H. Response of soil nutrients and stoichiometry to grazing management in alpine grassland on the Qinghai-Tibet Plateau. Soil Till. Res. 2021, 206, 104822. [Google Scholar] [CrossRef]
- Liu, J.; Feng, C.; Wang, D.; Wang, L.; Wilsey, B.J.; Zhong, Z. Impacts of grazing by different large herbivores in grassland depend on plant species diversity. J. Appl. Ecol. 2015, 52, 1053–1062. [Google Scholar] [CrossRef]
- Chen, L.; Saixi, Y.; Yi, R.; Baoyin, T. Characterization of soil microbes associated with a grazing-tolerant grass species, Stipa breviflora, in the Inner Mongolian desert steppe. Ecol. Evol. 2020, 10, 10607–10618. [Google Scholar] [CrossRef]
- Lu, X.; Kelsey, K.C.; Yan, Y.; Sun, J.; Wang, X.; Cheng, G.; Neff, J.C. Effects of grazing on ecosystem structure and function of alpine grasslands in Qinghai-Tibetan Plateau: A synthesis. Ecosphere 2017, 8, e01656. [Google Scholar] [CrossRef]
- Li, C.; Hao, X.; Zhao, M.; Han, G.; Willms, W.D. Influence of historic sheep grazing on vegetation and soil properties of a Desert Steppe in Inner Mongolia. Agr. Ecosyst. Environ. 2008, 128, 109–116. [Google Scholar] [CrossRef]
- Dormaar, J.F.; Smoliak, S.; Willms, W.D. Distribution of nitrogen fractions in grazed and ungrazed fescue grassland Ah horizons. J. Range Manag. 1990, 43, 6–9. [Google Scholar] [CrossRef]
- Blank, R.R.; Svejcar, T.; Riegel, G. Soil attributes in a Sierra Nevada riparian meadow as influenced by grazing. Rangel. Ecol. Manag. 2006, 59, 321–329. [Google Scholar] [CrossRef]
- Ma, W.; Li, J.; Jimoh, S.O.; Zhang, Y.; Guo, F.; Ding, Y.; Li, X.; Hou, X. Stoichiometric ratios support plant adaption to grazing moderated by soil nutrients and root enzymes. PeerJ 2019, 7, e7047. [Google Scholar] [CrossRef]
- McNaughton, S.J.; Banyikwa, F.F.; McNaughton, M.M. Promotion of the cycling of diet-enhancing nutrients by African grazers. Science 1997, 278, 1798–1800. [Google Scholar] [CrossRef] [PubMed]
- Heyburn, J.; McKenzie, P.; Crawley, M.J.; Fornara, D.A. Effects of grassland management on plant C: N: P stoichiometry: Implications for soil element cycling and storage. Ecosphere 2017, 8, e01963. [Google Scholar] [CrossRef]
- Beckers, B.; De Beeck, M.O.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2009, 321, 341–361. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhang, E.; Hong, M.; Yin, X.; Cai, H.; Yuan, L.; Yuan, F.; Li, L.; Zhao, K.; Lan, X. Analysis of endophytic and rhizosphere bacterial diversity and function in the endangered plant Paeonia ludlowii. Arch. Microbiol. 2020, 20, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; Von Mering, C.; Vorholt, J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef]
- Liu, H.; Carvalhais, L.C.; Crawford, M.; Singh, E.; Dennis, P.G.; Pieterse, C.M.; Schenk, P.M. Inner plant values: Diversity, colonization and benefits from endophytic bacteria. Front. Microbiol. 2017, 8, 2552. [Google Scholar] [CrossRef]
- Dong, L.; Cheng, R.; Xiao, L.; Wei, F.; Wei, G.; Xu, J.; Wang, Y.; Guo, X.; Chen, Z.; Chen, S. Diversity and composition of bacterial endophytes among plant parts of Panax notoginseng. Chin. Med. 2018, 13, 41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Shen, Z.; Fang, W.; Gao, G. Composition of bacterial communities in municipal wastewater treatment plant. Sci. Total Environ. 2019, 689, 1181–1191. [Google Scholar] [CrossRef] [PubMed]
- Moulin, L.; Munive, A.; Dreyfus, B.; Boivin-Masson, C. Nodulation of legumes by members of the β-subclass of Proteobacteria. Nature 2001, 411, 948–950. [Google Scholar] [CrossRef] [PubMed]
- Rasolomampianina, R.; Bailly, X.; Fetiarison, R.; Rabevohitra, R.; Béna, G.; Ramaroson, L.; Raherimandimby, M.; Moulin, L.; De Lajudie, P.; Dreyfus, B.; et al. Nitrogen-fixing nodules from rose wood legume trees (Dalbergia spp.) endemic to Madagascar host seven different genera belonging to α-and β-Proteobacteria. Mol. Ecol. 2005, 14, 4135–4146. [Google Scholar] [CrossRef]
- Yandigeri, M.S.; Meena, K.K.; Singh, D.; Malviya, N.; Singh, D.P.; Solanki, M.K.; Yadav, A.K.; Arora, D.K. Drought-tolerant endophytic actinobacteria promote growth of wheat (Triticum aestivum) under water stress conditions. Plant Growth Regul. 2012, 68, 411–420. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Giovanardi, D.; Stefani, E. Plant growth promoting and biocontrol activity of Streptomyces spp. as endophytes. Int. J. Mol. Sci. 2018, 19, 952. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.; Lin, Y.; Luo, J.; Di, H.J.; Lindsey, S.; Liu, D.; Fan, J.; Ding, W. Responses of soil fungal diversity and community composition to long-term fertilization: Field experiment in an acidic Ultisol and literature synthesis. Appl. Soil Ecol. 2020, 145, 103305. [Google Scholar] [CrossRef]
- Cheng, J.; Jing, G.; Wei, L.; Jing, Z. Long-term grazing exclusion effects on vegetation characteristics, soil properties and bacterial communities in the semi-arid grasslands of China. Ecol. Eng. 2016, 97, 170–178. [Google Scholar] [CrossRef]
- Acosta-Martínez, V.; Dowd, S.E.; Sun, Y.; Wester, D.; Allen, V. Pyrosequencing analysis for characterization of soil bacterial populations as affected by an integrated livestock-cotton production system. Appl. Soil Ecol. 2010, 45, 13–25. [Google Scholar] [CrossRef]
- Gómez-Acata, E.S.; Valencia-Becerril, I.; Valenzuela-Encinas, C.; Velásquez-Rodríguez, A.S.; Navarro-Noya, Y.E.; Montoya-Ciriaco, N.; Suárez-Arriaga, M.C.; Rojas-Valdez, A.; Reyes-Reyes, B.G.; Luna-Guido, M.; et al. Deforestation and cultivation with maize (Zea mays L.) has a profound effect on the bacterial community structure in soil. Land Degrad. Dev. 2016, 27, 1122–1130. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Chen, S.; Wu, X.; Syed, S.I.; Syed, I.U.S.; Huang, B.; Guan, P.; Wang, D. Grazing Affects Bacterial and Fungal Diversities and Communities in the Rhizosphere and Endosphere Compartments of Leymus chinensis through Regulating Nutrient and Ion Distribution. Microorganisms 2021, 9, 476. https://doi.org/10.3390/microorganisms9030476
Yang Y, Chen S, Wu X, Syed SI, Syed IUS, Huang B, Guan P, Wang D. Grazing Affects Bacterial and Fungal Diversities and Communities in the Rhizosphere and Endosphere Compartments of Leymus chinensis through Regulating Nutrient and Ion Distribution. Microorganisms. 2021; 9(3):476. https://doi.org/10.3390/microorganisms9030476
Chicago/Turabian StyleYang, Yurong, Siying Chen, Xuefeng Wu, Sajid Iqbal Syed, Irfan Ullah Shah Syed, Beitong Huang, Pingting Guan, and Deli Wang. 2021. "Grazing Affects Bacterial and Fungal Diversities and Communities in the Rhizosphere and Endosphere Compartments of Leymus chinensis through Regulating Nutrient and Ion Distribution" Microorganisms 9, no. 3: 476. https://doi.org/10.3390/microorganisms9030476
APA StyleYang, Y., Chen, S., Wu, X., Syed, S. I., Syed, I. U. S., Huang, B., Guan, P., & Wang, D. (2021). Grazing Affects Bacterial and Fungal Diversities and Communities in the Rhizosphere and Endosphere Compartments of Leymus chinensis through Regulating Nutrient and Ion Distribution. Microorganisms, 9(3), 476. https://doi.org/10.3390/microorganisms9030476





