Novel Template Plasmids pCyaA’-Kan and pCyaA’-Cam for Generation of Unmarked Chromosomal cyaA’ Translational Fusion to T3SS Effectors in Salmonella
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Standard DNA Techniques
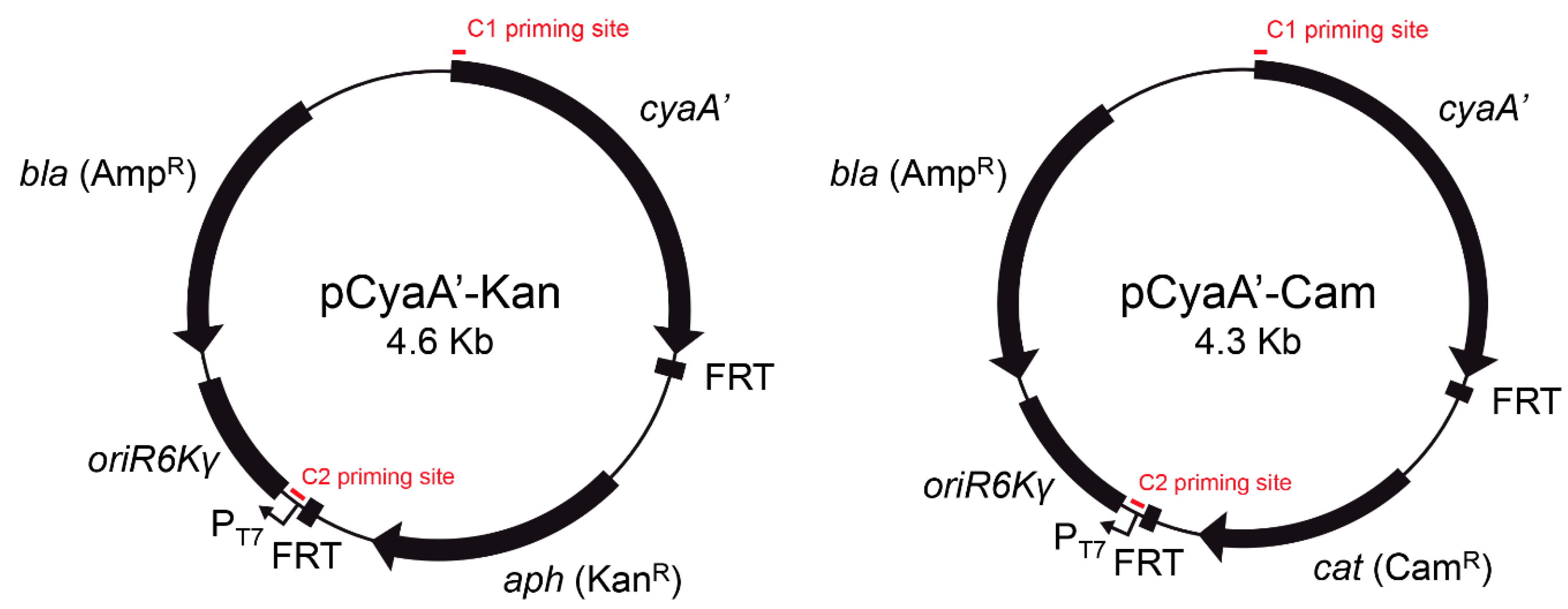
2.3. Construction of Plasmids pCyaA’-Kan and pCyaA’-Cam
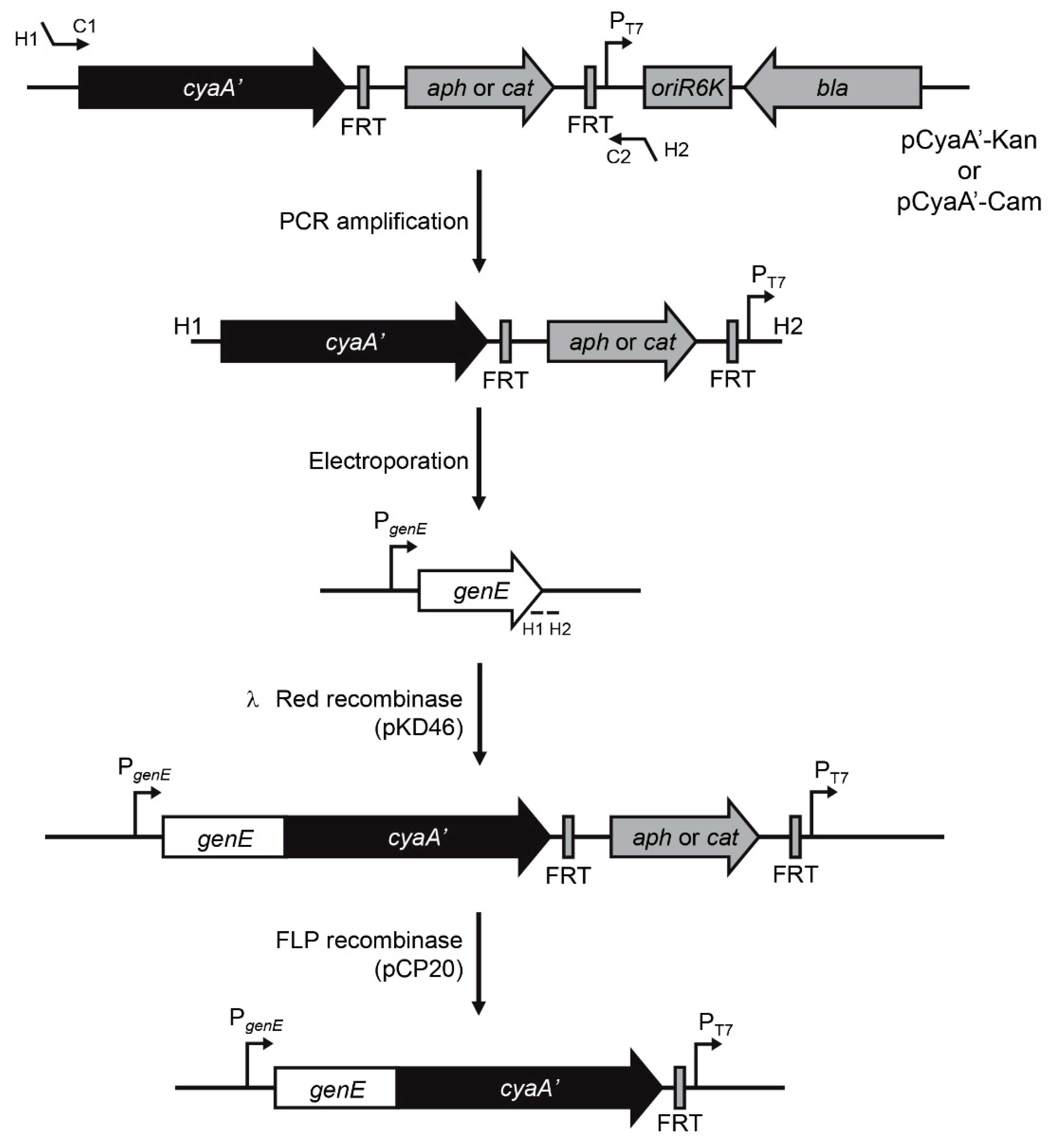
2.4. Generation of cyaA’ Translational Fusions
2.5. Western Blot Analyses
2.6. Mammalian Cell Culture and Infection Assays
2.7. Intracellular cAMP Measurement
3. Results
3.1. Rationale and Design
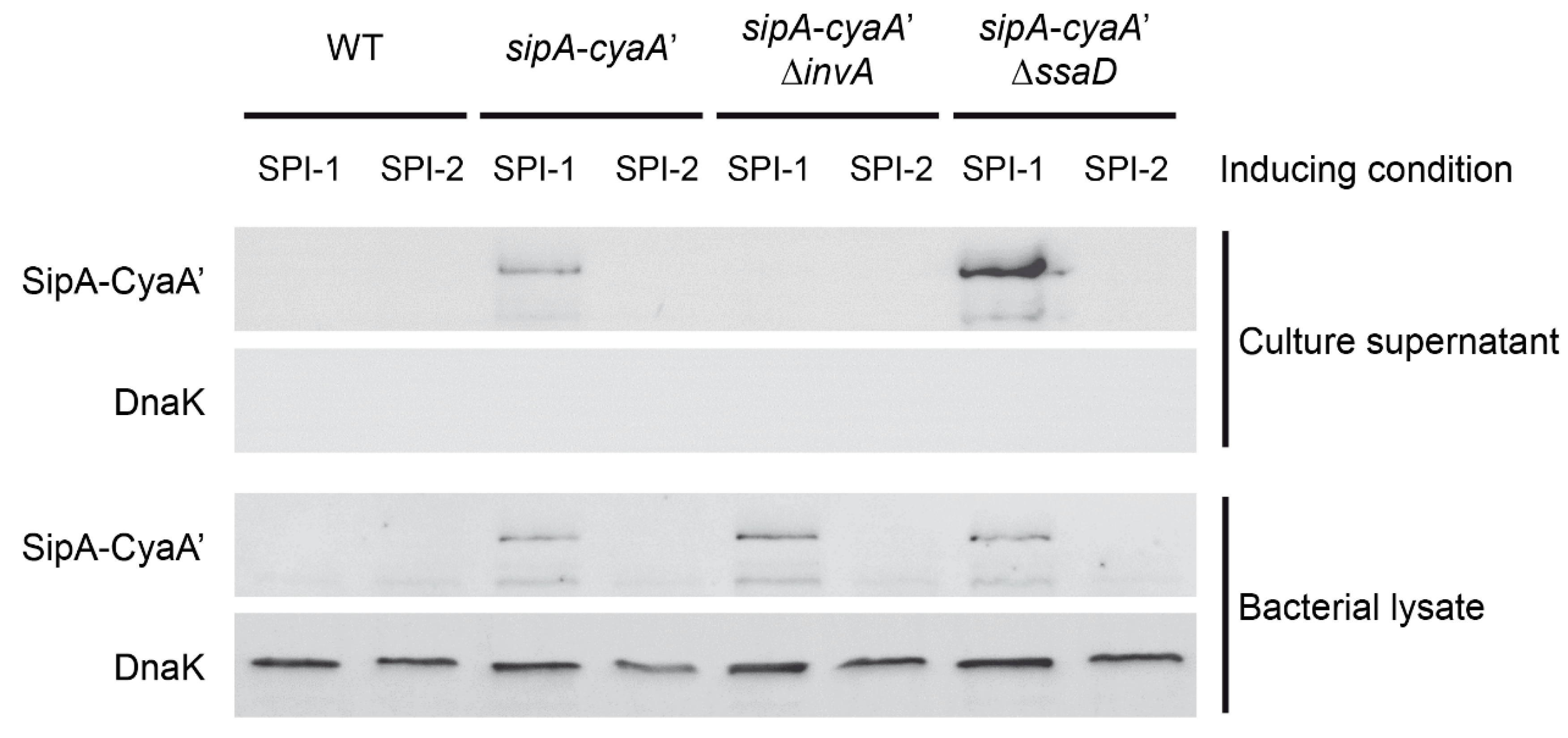
3.2. Generation and Immunodetection of CyaA’ Translational Fusions to S. Typhimurium T3SS Effectors
3.3. Secretion of a Selected CyaA’ Fusion Protein by S. Typhimurium during in Vitro Growth
3.4. Translocation of a Selected CyaA’ Fusion Protein during S. Typhimurium Infection of HeLa Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brenner, F.W.; Villar, R.G.; Angulo, F.J.; Tauxe, R.; Swaminathan, B. Salmonella Nomenclature. J. Clin. Microbiol. 2000, 38, 2465–2467. [Google Scholar] [CrossRef] [PubMed]
- Haraga, A.; Ohlson, M.B.; Miller, S.I. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 2008, 6, 53–66. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Morales, F. Impact of Salmonella enterica type III secretion system effectors on the eukaryotic host cell. ISRN Cell Biol. 2012, 2012, 787934. [Google Scholar] [CrossRef]
- Dean, P. Functional domains and motifs of bacterial type III effector proteins and their roles in infection. FEMS Microbiol. Rev. 2011, 35, 1100–1125. [Google Scholar] [CrossRef] [PubMed]
- Sory, M.P.; Boland, A.; Lambermont, I.; Cornelis, G.R. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. USA 1995, 92, 11998–12002. [Google Scholar] [CrossRef] [PubMed]
- Ramos-Morales, F.; Cardenal-Muñoz, E.; Cordero-Alba, M.; Baisón-Olmo, F. Generation and use of site-directed chromosomal cyaA’ translational fusions in Salmonella enterica. Methods Mol. Biol. 2015, 1225, 93–104. [Google Scholar] [CrossRef]
- Datsenko, K.A.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef]
- Fields, P.I.; Swanson, R.V.; Haidaris, C.G.; Heffron, F. Mutants of Salmonella typhimurium that cannot survive within the macrophage are avirulent. Proc. Natl. Acad. Sci. USA 1986, 83, 5189–5193. [Google Scholar] [CrossRef]
- Jarvik, T.; Smillie, C.; Groisman, E.A.; Ochman, H. Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J. Bacteriol. 2010, 192, 560–567. [Google Scholar] [CrossRef]
- Nelson, D.L.; Kennedy, E.P. Magnesium transport in Escherichia coli. Inhibition by cobaltous ion. J. Biol. Chem. 1971, 246, 3042–3049. [Google Scholar] [CrossRef]
- Santiviago, C.A.; Reynolds, M.M.; Porwollik, S.; Choi, S.H.; Long, F.; Andrews-Polymenis, H.L.; McClelland, M. Analysis of pools of targeted Salmonella deletion mutants identifies novel genes affecting fitness during competitive infection in mice. PLoS Pathog. 2009, 5, e1000477. [Google Scholar] [CrossRef]
- Cherepanov, P.P.; Wackernagel, W. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 1995, 158, 9–14. [Google Scholar] [CrossRef]
- Wessel, D.; Flügge, U.I. A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal Biochem. 1984, 138, 141–143. [Google Scholar] [CrossRef]
- Hansen-Wester, I.; Stecher, B.; Hensel, M. Type III secretion of Salmonella enterica serovar Typhimurium translocated effectors and SseFG. Infect. Immun. 2002, 70, 1403–1409. [Google Scholar] [CrossRef]
- Geddes, K.; Worley, M.; Niemann, G.; Heffron, F. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect. Immun. 2005, 73, 6260–6271. [Google Scholar] [CrossRef]
- Uzzau, S.; Figueroa-Bossi, N.; Rubino, S.; Bossi, L. Epitope tagging of chromosomal genes in Salmonella. Proc. Natl. Acad. Sci. USA 2001, 98, 15264–15269. [Google Scholar] [CrossRef] [PubMed]
- Gerlach, R.G.; Hölzer, S.U.; Jäckel, D.; Hensel, M. Rapid engineering of bacterial reporter gene fusions by using Red recombination. Appl. Environ. Microbiol. 2007, 73, 4234–4242. [Google Scholar] [CrossRef] [PubMed]
- Baisón-Olmo, F.; Cardenal-Muñoz, E.; Ramos-Morales, F. PipB2 is a substrate of the Salmonella pathogenicity island 1-encoded type III secretion system. Biochem. Biophys. Res. Commun. 2012, 423, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Baisón-Olmo, F.; Galindo-Moreno, M.; Ramos-Morales, F. Host cell type-dependent translocation and PhoP-mediated positive regulation of the effector SseK1 of Salmonella enterica. Front. Microbiol. 2015, 6, 396. [Google Scholar] [CrossRef] [PubMed]
- Wolff, C.; Nisan, I.; Hanski, E.; Frankel, G.; Rosenshine, I. Protein translocation into host epithelial cells by infecting enteropathogenic Escherichia coli. Mol. Microbiol. 1998, 28, 143–155. [Google Scholar] [CrossRef]
- Neal-McKinney, J.M.; Konkel, M.E. The Campylobacter jejuni CiaC virulence protein is secreted from the flagellum and delivered to the cytosol of host cells. Front. Cell. Infect. Microbiol. 2012, 2, 31. [Google Scholar] [CrossRef] [PubMed]
- Dubytska, L.P.; Rogge, M.L.; Thune, R.L. Identification and characterization of putative translocated effector proteins of the Edwardsiella ictaluri Type III Secretion System. mSphere 2016, 1, e00039-16. [Google Scholar] [CrossRef]
- Scherer, C.A.; Cooper, E.; Miller, S.I. The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 2000, 37, 1133–1145. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Ansong, C.; Adkins, J.N.; Heffron, F. Discovery of Salmonella virulence factors translocated via outer membrane vesicles to murine macrophages. Infect. Immun. 2011, 79, 2182–2192. [Google Scholar] [CrossRef]
- Bauler, L.D.; Hackstadt, T. Expression and targeting of secreted proteins from Chlamydia trachomatis. J. Bacteriol. 2014, 196, 1325–1334. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, P.A.; Zabner, M.; Ortega, J.; Morgado, C.; Amaya, F.; Vera, G.; Rubilar, C.; Salas, B.; Cuevas, V.; Valenzuela, C.; et al. Novel Template Plasmids pCyaA’-Kan and pCyaA’-Cam for Generation of Unmarked Chromosomal cyaA’ Translational Fusion to T3SS Effectors in Salmonella. Microorganisms 2021, 9, 475. https://doi.org/10.3390/microorganisms9030475
Fernández PA, Zabner M, Ortega J, Morgado C, Amaya F, Vera G, Rubilar C, Salas B, Cuevas V, Valenzuela C, et al. Novel Template Plasmids pCyaA’-Kan and pCyaA’-Cam for Generation of Unmarked Chromosomal cyaA’ Translational Fusion to T3SS Effectors in Salmonella. Microorganisms. 2021; 9(3):475. https://doi.org/10.3390/microorganisms9030475
Chicago/Turabian StyleFernández, Paulina A., Marcela Zabner, Jaime Ortega, Constanza Morgado, Fernando Amaya, Gabriel Vera, Carolina Rubilar, Beatriz Salas, Víctor Cuevas, Camila Valenzuela, and et al. 2021. "Novel Template Plasmids pCyaA’-Kan and pCyaA’-Cam for Generation of Unmarked Chromosomal cyaA’ Translational Fusion to T3SS Effectors in Salmonella" Microorganisms 9, no. 3: 475. https://doi.org/10.3390/microorganisms9030475
APA StyleFernández, P. A., Zabner, M., Ortega, J., Morgado, C., Amaya, F., Vera, G., Rubilar, C., Salas, B., Cuevas, V., Valenzuela, C., Baisón-Olmo, F., Álvarez, S. A., & Santiviago, C. A. (2021). Novel Template Plasmids pCyaA’-Kan and pCyaA’-Cam for Generation of Unmarked Chromosomal cyaA’ Translational Fusion to T3SS Effectors in Salmonella. Microorganisms, 9(3), 475. https://doi.org/10.3390/microorganisms9030475






