Alteration of Gut Microbiota in Carbapenem-Resistant Enterobacteriaceae Carriers during Fecal Microbiota Transplantation According to Decolonization Periods
Abstract
1. Introduction
2. Materials and Methods
2.1. Subject Selection and FMT Procedures
2.2. Culture Assay and Polymerase Chain Reaction (PCR) for CRE Detection in Carriers
2.3. Quantitative Real-Time PCR and MiSeq Sequencing
2.4. Sequencing Data Analysis
2.5. Co-Occurrence Network Analysis
2.6. Statistical Analysis
3. Results
3.1. Clinical Features of CP-CRE Carriers
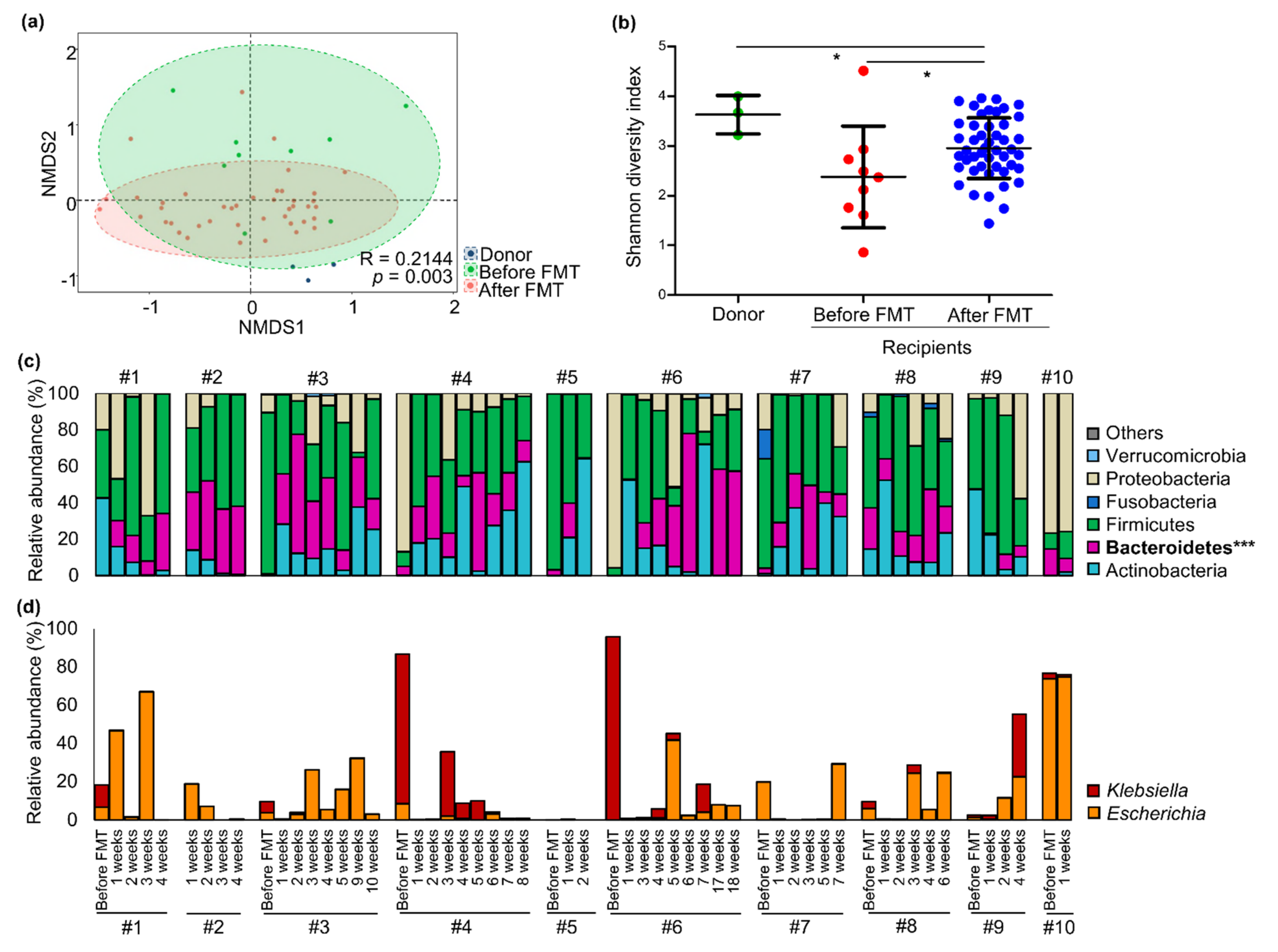
3.2. Changes in Gut Microbiota of CP-CRE Carriers after FMT Treatment
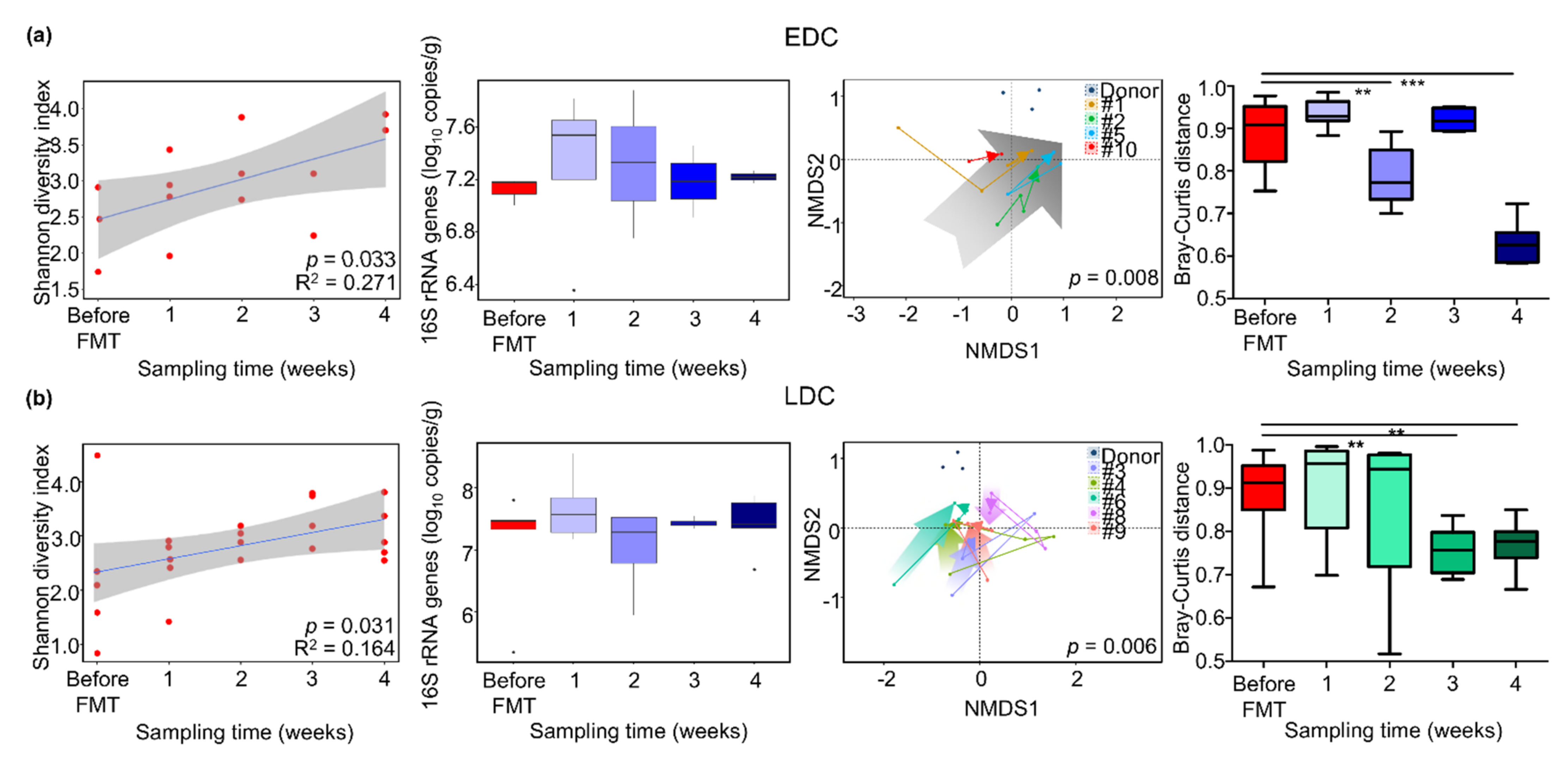
3.3. Comparison of Gut Microbiota between Early Decolonization Carriers and Late Decolonization Carriers
3.4. Different Alteration of Gut Microbiota along Follow-Up Times between EDC and LDC within 4 Weeks after FMT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bonomo, R.A.; Burd, E.M.; Conly, J.; Limbago, B.M.; Poirel, L.; Segre, J.A.; Westblade, L.F. Carbapenemase-Producing Organisms: A Global Scourge. Clin. Infect. Dis. 2018, 66, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Paterson, D.L. Carbapenemase-producing Enterobacteriaceae. Semin. Respir. Crit. Care Med. 2015, 36, 74–84. [Google Scholar] [CrossRef]
- Bar-Yoseph, H.; Hussein, K.; Braun, E.; Paul, M. Natural history and decolonization strategies for ESBL/carbapenem-resistant Enterobacteriaceae carriage: Systematic review and meta-analysis. J. Antimicrob. Chemother. 2016, 71, 2729–2739. [Google Scholar] [CrossRef] [PubMed]
- Campos, A.C.; Albiero, J.; Ecker, A.B.; Kuroda, C.M.; Meirelles, L.E.; Polato, A.; Tognim, M.C.; Wingeter, M.A.; Teixeira, J.J. Outbreak of Klebsiella pneumoniae carbapenemase-producing K pneumoniae: A systematic review. Am. J. Infect. Control 2016, 44, 1374–1380. [Google Scholar] [CrossRef]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Debast, S.B.; Bauer, M.P.; Kuijper, E.J.; European Society of Clinical, M.; Infectious, D. European Society of Clinical Microbiology and Infectious Diseases: Update of the treatment guidance document for Clostridium difficile infection. Clin. Microbiol. Infect. 2014, 20 (Suppl. S2), 1–26. [Google Scholar] [CrossRef]
- Millan, B.; Park, H.; Hotte, N.; Mathieu, O.; Burguiere, P.; Tompkins, T.A.; Kao, D.; Madsen, K.L. Fecal Microbial Transplants Reduce Antibiotic-resistant Genes in Patients with Recurrent Clostridium difficile Infection. Clin. Infect. Dis. 2016, 62, 1479–1486. [Google Scholar] [CrossRef] [PubMed]
- Rossen, N.G.; Fuentes, S.; van der Spek, M.J.; Tijssen, J.G.; Hartman, J.H.; Duflou, A.; Lowenberg, M.; van den Brink, G.R.; Mathus-Vliegen, E.M.; de Vos, W.M.; et al. Findings From a Randomized Controlled Trial of Fecal Transplantation for Patients with Ulcerative Colitis. Gastroenterology 2015, 149, 110–118.e114. [Google Scholar] [CrossRef]
- Suskind, D.L.; Brittnacher, M.J.; Wahbeh, G.; Shaffer, M.L.; Hayden, H.S.; Qin, X.; Singh, N.; Damman, C.J.; Hager, K.R.; Nielson, H.; et al. Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn’s disease. Inflamm. Bowel Dis. 2015, 21, 556–563. [Google Scholar] [CrossRef]
- Saha, S.; Tariq, R.; Tosh, P.K.; Pardi, D.S.; Khanna, S. Faecal microbiota transplantation for eradicating carriage of multidrug-resistant organisms: A systematic review. Clin. Microbiol. Infect. 2019, 25, 958–963. [Google Scholar] [CrossRef]
- Manges, A.R.; Steiner, T.S.; Wright, A.J. Fecal microbiota transplantation for the intestinal decolonization of extensively antimicrobial-resistant opportunistic pathogens: A review. Infect. Dis. 2016, 48, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Korach-Rechtman, H.; Hreish, M.; Fried, C.; Gerassy-Vainberg, S.; Azzam, Z.S.; Kashi, Y.; Berger, G. Intestinal Dysbiosis in Carriers of Carbapenem-Resistant Enterobacteriaceae. mSphere 2020, 5, e00173-20. [Google Scholar] [CrossRef]
- Kim, Y.K.; Song, S.A.; Lee, J.N.; Oh, M.; Jo, K.M.; Kim, H.J.; Lee, J.H.; Park, J.; Jang, H.J.; Kim, H.K.; et al. Clinical factors predicting persistent carriage of Klebsiella pneumoniae carbapenemase-producing carbapenem-resistant Enterobacteriaceae among patients with known carriage. J. Hosp. Infect. 2018, 99, 405–412. [Google Scholar] [CrossRef]
- Cammarota, G.; Ianiro, G.; Tilg, H.; Rajilic-Stojanovic, M.; Kump, P.; Satokari, R.; Sokol, H.; Arkkila, P.; Pintus, C.; Hart, A.; et al. European consensus conference on faecal microbiota transplantation in clinical practice. Gut 2017, 66, 569–580. [Google Scholar] [CrossRef] [PubMed]
- Costello, S.P.; Tucker, E.C.; La Brooy, J.; Schoeman, M.N.; Andrews, J.M. Establishing a Fecal Microbiota Transplant Service for the Treatment of Clostridium difficile Infection. Clin. Infect. Dis. 2016, 62, 908–914. [Google Scholar] [CrossRef]
- Terveer, E.M.; van Beurden, Y.H.; Goorhuis, A.; Seegers, J.; Bauer, M.P.; van Nood, E.; Dijkgraaf, M.G.W.; Mulder, C.J.J.; Vandenbroucke-Grauls, C.; Verspaget, H.W.; et al. How to: Establish and run a stool bank. Clin. Microbiol. Infect. 2017, 23, 924–930. [Google Scholar] [CrossRef]
- Division of Human Blood Safety Surveillance. Transfusion Guideline, 3rd ed.; Korean Centers for Disease Control and Prevention: Cheongju, Korea, 2013.
- Clinical & Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. M100S; CLSI: Wayne, PA, USA, 2016. [Google Scholar]
- Morey, K.E.; Vega, R.; Cassidy, P.M.; Buser, G.L.; Rayar, J.K.; Myers, J.A.; Weissman, S.J.; Beldavs, Z.G.; Pfeiffer, C.D. Evaluation of the Carba NP Test in Oregon, 2013. Antimicrob. Agents Chemother. 2017, 61, e3005-15. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.H.; Kim, H.S.; Kim, J.S.; Shin, D.H.; Kim, H.S.; Park, M.J.; Shin, S.; Hong, J.S.; Lee, S.S.; Song, W. Prevalence and Molecular Characteristics of Carbapenemase-Producing Enterobacteriaceae from Five Hospitals in Korea. Ann. Lab. Med. 2016, 36, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.E.; Lee, J.J.; Lee, M.J.; Kim, B.S. Analysis of microbiome in raw chicken meat from butcher shops and packaged products in South Korea to detect the potential risk of foodborne illness. Food Res. Int. 2019, 122, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Park, J.U.; Oh, B.; Lee, J.P.; Choi, M.H.; Lee, M.J.; Kim, B.S. Influence of Microbiota on Diabetic Foot Wound in Comparison with Adjacent Normal Skin Based on the Clinical Features. BioMed Res. Int. 2019, 2019, 7459236. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Faust, K.; Raes, J. CoNet app: Inference of biological association networks using Cytoscape. F1000Research 2016, 5, 1519. [Google Scholar] [CrossRef] [PubMed]
- Staley, C.; Kaiser, T.; Vaughn, B.P.; Graiziger, C.T.; Hamilton, M.J.; Rehman, T.U.; Song, K.; Khoruts, A.; Sadowsky, M.J. Predicting recurrence of Clostridium difficile infection following encapsulated fecal microbiota transplantation. Microbiome 2018, 6, 166. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Taur, Y.; Jenq, R.R.; Equinda, M.J.; Son, T.; Samstein, M.; Viale, A.; Socci, N.D.; van den Brink, M.R.; Kamboj, M.; et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Investig. 2010, 120, 4332–4341. [Google Scholar] [CrossRef]
- Seekatz, A.M.; Aas, J.; Gessert, C.E.; Rubin, T.A.; Saman, D.M.; Bakken, J.S.; Young, V.B. Recovery of the gut microbiome following fecal microbiota transplantation. mBio 2014, 5, e00893-14. [Google Scholar] [CrossRef]
- Weingarden, A.; Gonzalez, A.; Vazquez-Baeza, Y.; Weiss, S.; Humphry, G.; Berg-Lyons, D.; Knights, D.; Unno, T.; Bobr, A.; Kang, J.; et al. Dynamic changes in short- and long-term bacterial composition following fecal microbiota transplantation for recurrent Clostridium difficile infection. Microbiome 2015, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Wilson, B.C.; Vatanen, T.; Cutfield, W.S.; O’Sullivan, J.M. The Super-Donor Phenomenon in Fecal Microbiota Transplantation. Front. Cell. Infect. Microbiol. 2019, 9, 2. [Google Scholar] [CrossRef]
- Liu, X.; Zheng, H.; Lu, R.; Huang, H.; Zhu, H.; Yin, C.; Mo, Y.; Wu, J.; Liu, X.; Deng, M.; et al. Intervening Effects of Total Alkaloids of Corydalis saxicola Bunting on Rats with Antibiotic-Induced Gut Microbiota Dysbiosis Based on 16S rRNA Gene Sequencing and Untargeted Metabolomics Analyses. Front. Microbiol. 2019, 10, 1151. [Google Scholar] [CrossRef]
- Dragomirescu, C.C.; Lixandru, B.E.; Coldea, I.L.; Corneli, O.N.; Pana, M.; Palade, A.M.; Cristea, V.C.; Suciu, I.; Suciu, G.; Manolescu, L.S.C.; et al. Antimicrobial Susceptibility Testing for Corynebacterium Species Isolated from Clinical Samples in Romania. Antibiotics 2020, 9, 31. [Google Scholar] [CrossRef] [PubMed]
- Pettigrew, M.M.; Gent, J.F.; Kong, Y.; Halpin, A.L.; Pineles, L.; Harris, A.D.; Johnson, J.K. Gastrointestinal Microbiota Disruption and Risk of Colonization with Carbapenem-resistant Pseudomonas aeruginosa in Intensive Care Unit Patients. Clin. Infect. Dis. 2019, 69, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Agudelo-Ochoa, G.M.; Valdes-Duque, B.E.; Giraldo-Giraldo, N.A.; Jaillier-Ramirez, A.M.; Giraldo-Villa, A.; Acevedo-Castano, I.; Yepes-Molina, M.A.; Barbosa-Barbosa, J.; Benitez-Paez, A. Gut microbiota profiles in critically ill patients, potential biomarkers and risk variables for sepsis. Gut Microbes 2020, 12, 1707610. [Google Scholar] [CrossRef]
- Zhu, L.; Liao, R.; Tu, W.; Lu, Y.; Cai, X. Pyrodextrin enhances intestinal function through changing the intestinal microbiota composition and metabolism in early weaned piglets. Appl. Microbiol. Biotechnol. 2020, 104, 4141–4154. [Google Scholar] [CrossRef]
- Engels, C.; Ruscheweyh, H.J.; Beerenwinkel, N.; Lacroix, C.; Schwab, C. The Common Gut Microbe Eubacterium hallii also Contributes to Intestinal Propionate Formation. Front. Microbiol. 2016, 7, 713. [Google Scholar] [CrossRef]
- Trosvik, P.; de Muinck, E.J. Ecology of bacteria in the human gastrointestinal tract--identification of keystone and foundation taxa. Microbiome 2015, 3, 44. [Google Scholar] [CrossRef]
- Comstock, L.E. Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell Host Microbe 2009, 5, 522–526. [Google Scholar] [CrossRef]
- Sequeira, R.P.; McDonald, J.A.K.; Marchesi, J.R.; Clarke, T.B. Commensal Bacteroidetes protect against Klebsiella pneumoniae colonization and transmission through IL-36 signalling. Nat. Microbiol. 2020, 5, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Goloshchapov, O.V.; Olekhnovich, E.I.; Sidorenko, S.V.; Moiseev, I.S.; Kucher, M.A.; Fedorov, D.E.; Pavlenko, A.V.; Manolov, A.I.; Gostev, V.V.; Veselovsky, V.A.; et al. Long-term impact of fecal transplantation in healthy volunteers. BMC Microbiol. 2019, 19, 312. [Google Scholar] [CrossRef]
- Iljazovic, A.; Roy, U.; Galvez, E.J.C.; Lesker, T.R.; Zhao, B.; Gronow, A.; Amend, L.; Will, S.E.; Hofmann, J.D.; Pils, M.C.; et al. Perturbation of the gut microbiome by Prevotella spp. enhances host susceptibility to mucosal inflammation. Mucosal. Immunol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.; Grinspan, A. Fecal Microbiota Transplantation for Inflammatory Bowel Disease. Gastroenterol. Hepatol. 2016, 12, 374–379. [Google Scholar]
- Leszczyszyn, J.J.; Radomski, M.; Leszczyszyn, A.M. Intestinal microbiota transplant—Current state of knowledge. Reumatologia 2016, 54, 24–28. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Umu, O.C.; Bauerl, C.; Oostindjer, M.; Pope, P.B.; Hernandez, P.E.; Perez-Martinez, G.; Diep, D.B. The Potential of Class II Bacteriocins to Modify Gut Microbiota to Improve Host Health. PLoS ONE 2016, 11, e0164036. [Google Scholar] [CrossRef] [PubMed]
- Andoh, A.; Tsujikawa, T.; Hata, K.; Araki, Y.; Kitoh, K.; Sasaki, M.; Yoshida, T.; Fujiyama, Y. Elevated circulating platelet-derived microparticles in patients with active inflammatory bowel disease. Am. J. Gastroenterol. 2005, 100, 2042–2048. [Google Scholar] [CrossRef]
- Leonetti, D.; Reimund, J.M.; Tesse, A.; Viennot, S.; Martinez, M.C.; Bretagne, A.L.; Andriantsitohaina, R. Circulating microparticles from Crohn’s disease patients cause endothelial and vascular dysfunctions. PLoS ONE 2013, 8, e73088. [Google Scholar] [CrossRef]
- Sweere, J.M.; Van Belleghem, J.D.; Ishak, H.; Bach, M.S.; Popescu, M.; Sunkari, V.; Kaber, G.; Manasherob, R.; Suh, G.A.; Cao, X.; et al. Bacteriophage trigger antiviral immunity and prevent clearance of bacterial infection. Science 2019, 363. [Google Scholar] [CrossRef]
- Rasmussen, T.S.; Mentzel, C.M.J.; Kot, W.; Castro-Mejia, J.L.; Zuffa, S.; Swann, J.R.; Hansen, L.H.; Vogensen, F.K.; Hansen, A.K.; Nielsen, D.S. Faecal virome transplantation decreases symptoms of type 2 diabetes and obesity in a murine model. Gut 2020, 69, 2122–2130. [Google Scholar] [CrossRef]
- Duan, Y.; Llorente, C.; Lang, S.; Brandl, K.; Chu, H.; Jiang, L.; White, R.C.; Clarke, T.H.; Nguyen, K.; Torralba, M.; et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 2019, 575, 505–511. [Google Scholar] [CrossRef]
- Wong, W.F.; Santiago, M. Microbial approaches for targeting antibiotic-resistant bacteria. Microb. Biotechnol. 2017, 10, 1047–1053. [Google Scholar] [CrossRef]
- Jouhten, H.; Mattila, E.; Arkkila, P.; Satokari, R. Reduction of Antibiotic Resistance Genes in Intestinal Microbiota of Patients with Recurrent Clostridium difficile Infection after Fecal Microbiota Transplantation. Clin. Infect. Dis. 2016, 63, 710–711. [Google Scholar] [CrossRef] [PubMed]
- Bilinski, J.; Grzesiowski, P.; Sorensen, N.; Madry, K.; Muszynski, J.; Robak, K.; Wroblewska, M.; Dzieciatkowski, T.; Dulny, G.; Dwilewicz-Trojaczek, J.; et al. Fecal Microbiota Transplantation in Patients with Blood Disorders Inhibits Gut Colonization with Antibiotic-Resistant Bacteria: Results of a Prospective, Single-Center Study. Clin. Infect. Dis. 2017, 65, 364–370. [Google Scholar] [CrossRef] [PubMed]


| Carrier No. | Age | Sex | CRE Type | Duration of CRE Carriage before FMT | Carbapenem Use ≥ 3 days after CRE Carriage | CRE (+) in Clinical Specimen after CRE Carriage | Concurrent C. difficile Infection after CRE Carriage | Prolonged Hospitalization ≥2 Months after CRE Carriage | FMT Material | FMT Procedure | Time to Decolonization of CRE from FMT |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 78 | Female | KPC-CRE | 7 months | + | + | - | + | Unrelated donor, frozen stool | Terminal ileum and ascending colon via colonoscopy | 25 days |
| 2 | 68 | Male | KPC-CRE | 4 months | + | + | - | + | Unrelated donor, frozen stool | Duodenum via EGD and ascending colon via colonoscopy | 26 days |
| 3 | 80 | Female | KPC-CRE | 7 months | + | + | - | + | Unrelated donor, frozen stool | Duodenum via EGD (1st and 2nd FMT) | 15 days after 2nd FMT (106 days after 1st FMT) |
| 4 | 79 | Male | KPC-CRE | 6 months | + | + | - | + | Unrelated donor, frozen stool | Terminal ileum via colonoscopy | 51 days |
| 5 | 75 | Female | KPC-CRE (& VRE) | 5 months | + | + | + | + | Unrelated donor, frozen stool | Ascending colon via colonoscopy | 15 days |
| 6 | 75 | Female | KPC-CRE (& VRE) | 4 months | + | + | - | + | Unrelated donor, frozen stool | Terminal ileum via colonoscopy (1st FMT), duodenum via EGD (2nd FMT) | 34 days after 2nd FMT (117 days after 1st FMT) |
| 7 | 57 | Male | KPC-CRE | 7 months | + | - | - | + | Unrelated donor, frozen stool | Terminal ileum and ascending colon via colonoscopy (1st and 2nd FMT) | not decolonized (followed until 138 days after 1st FMT) |
| 8 | 81 | Female | KPC-CRE | 10 months | + | + | + | + | Unrelated donor, frozen and capsulized stool | Duodenum via EGD (1st and 2nd FMT), 20 capsules daily for 2 days (3rd FMT) | 16 days after 3rd FMT (137 days after 1st FMT) |
| 9 | 65 | Female | KPC-CRE | 3 months | + | - | - | + | Unrelated donor, frozen stool | Terminal ileum via colonoscopy (1st FMT) duodenum via EGD (2nd FMT) | 16 days after 2nd FMT (92 days after 1st FMT) |
| 10 | 69 | Female | KPC-CRE | 4 months | + | + | - | + | Unrelated donor, frozen stool | Terminal ileum and ascending colon via colonoscopy | 18 days |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.-J.; Yong, D.; Suk, K.T.; Kim, D.J.; Woo, H.-J.; Lee, S.S.; Kim, B.-S. Alteration of Gut Microbiota in Carbapenem-Resistant Enterobacteriaceae Carriers during Fecal Microbiota Transplantation According to Decolonization Periods. Microorganisms 2021, 9, 352. https://doi.org/10.3390/microorganisms9020352
Lee J-J, Yong D, Suk KT, Kim DJ, Woo H-J, Lee SS, Kim B-S. Alteration of Gut Microbiota in Carbapenem-Resistant Enterobacteriaceae Carriers during Fecal Microbiota Transplantation According to Decolonization Periods. Microorganisms. 2021; 9(2):352. https://doi.org/10.3390/microorganisms9020352
Chicago/Turabian StyleLee, Jin-Jae, Dongeun Yong, Ki Tae Suk, Dong Joon Kim, Heung-Jeong Woo, Seung Soon Lee, and Bong-Soo Kim. 2021. "Alteration of Gut Microbiota in Carbapenem-Resistant Enterobacteriaceae Carriers during Fecal Microbiota Transplantation According to Decolonization Periods" Microorganisms 9, no. 2: 352. https://doi.org/10.3390/microorganisms9020352
APA StyleLee, J.-J., Yong, D., Suk, K. T., Kim, D. J., Woo, H.-J., Lee, S. S., & Kim, B.-S. (2021). Alteration of Gut Microbiota in Carbapenem-Resistant Enterobacteriaceae Carriers during Fecal Microbiota Transplantation According to Decolonization Periods. Microorganisms, 9(2), 352. https://doi.org/10.3390/microorganisms9020352








