Biogeographical Distribution and Community Assembly of Active Protistan Assemblages along an Estuary to a Basin Transect of the Northern South China Sea
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. Enumeration of Virus and Picoplankton
2.3. High Throughput Sequencing
2.4. Sequence Processing and Statistical Analyses
3. Results
3.1. Environmental Factors
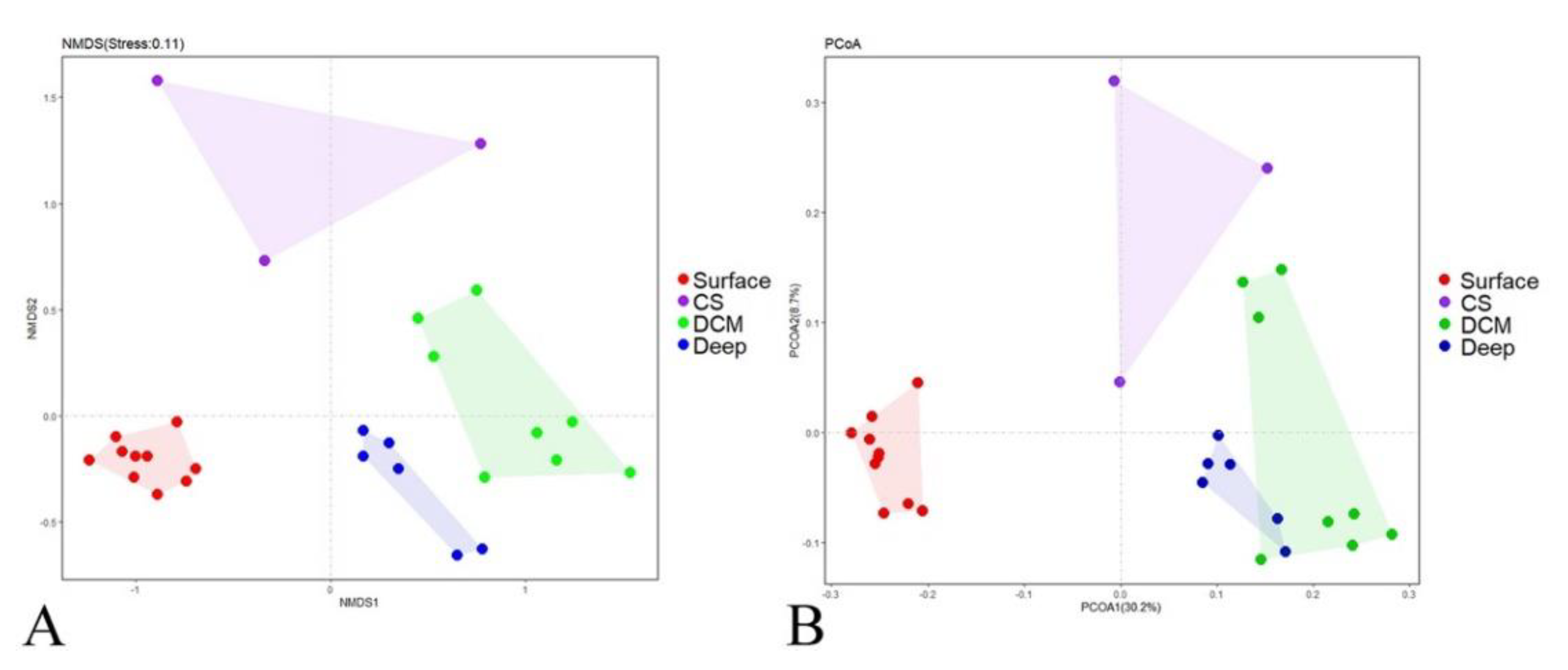
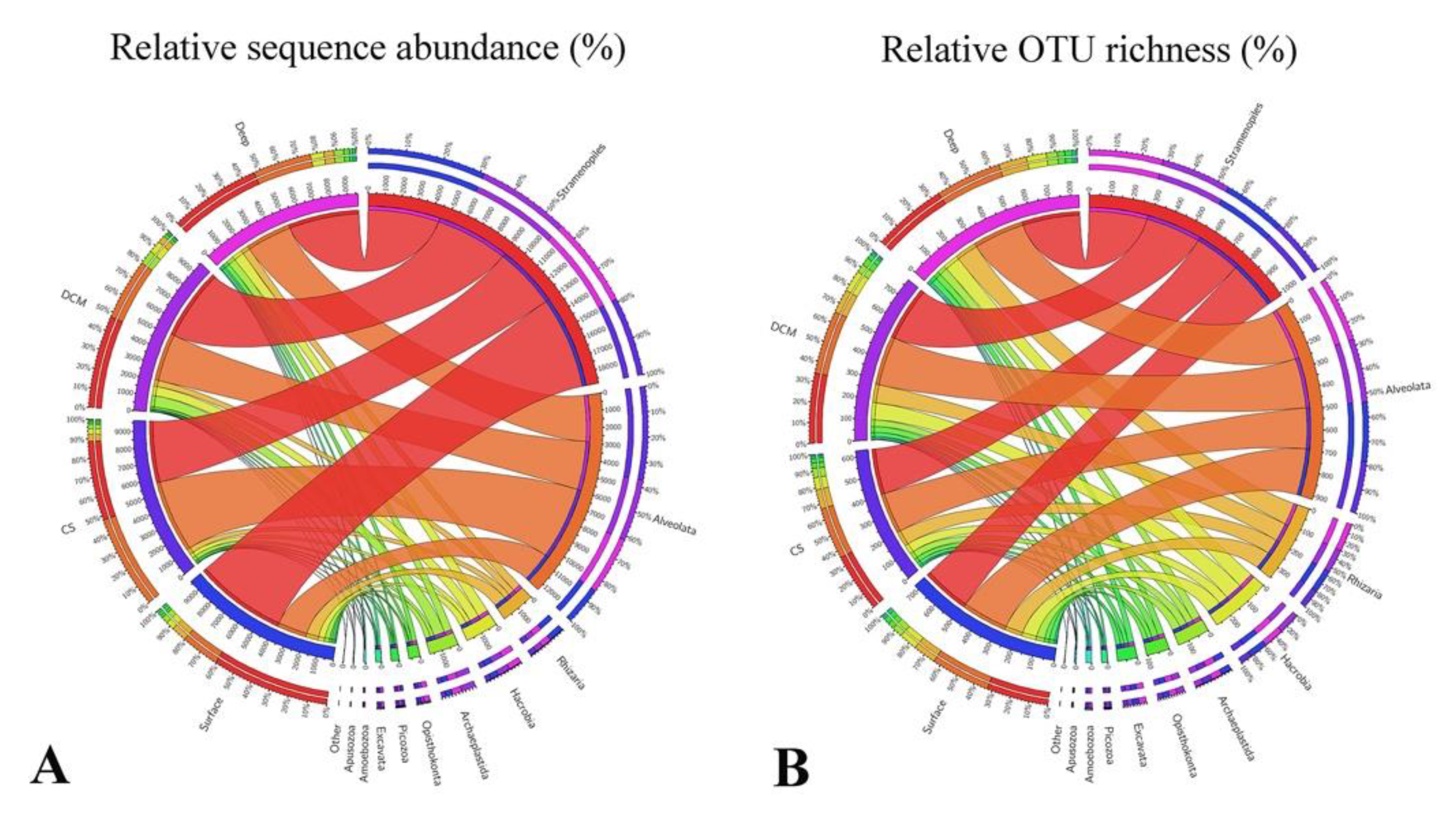
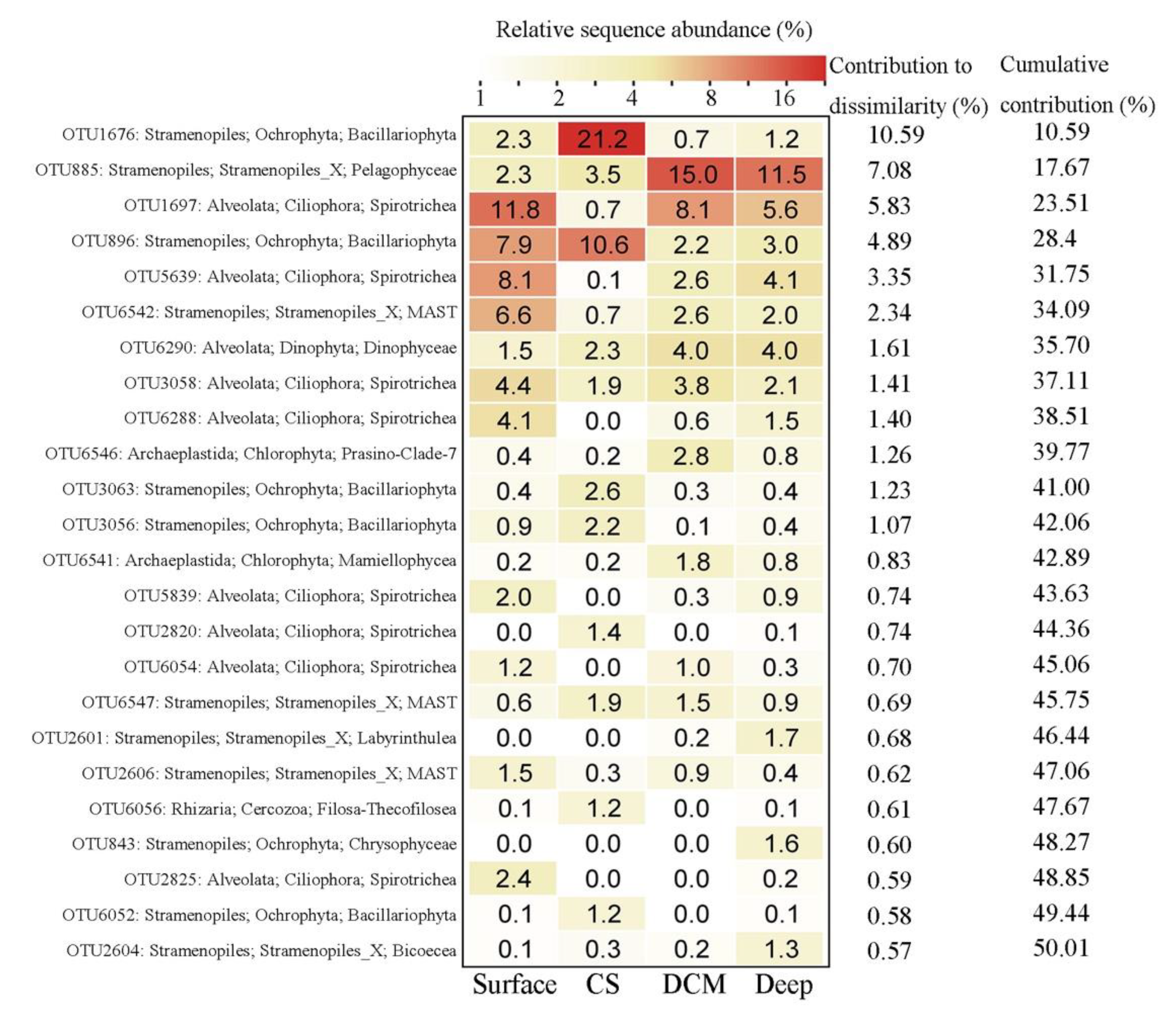
3.2. Beta Diversity and Community Composition
3.3. Alpha Diversity and Its Driving Factors
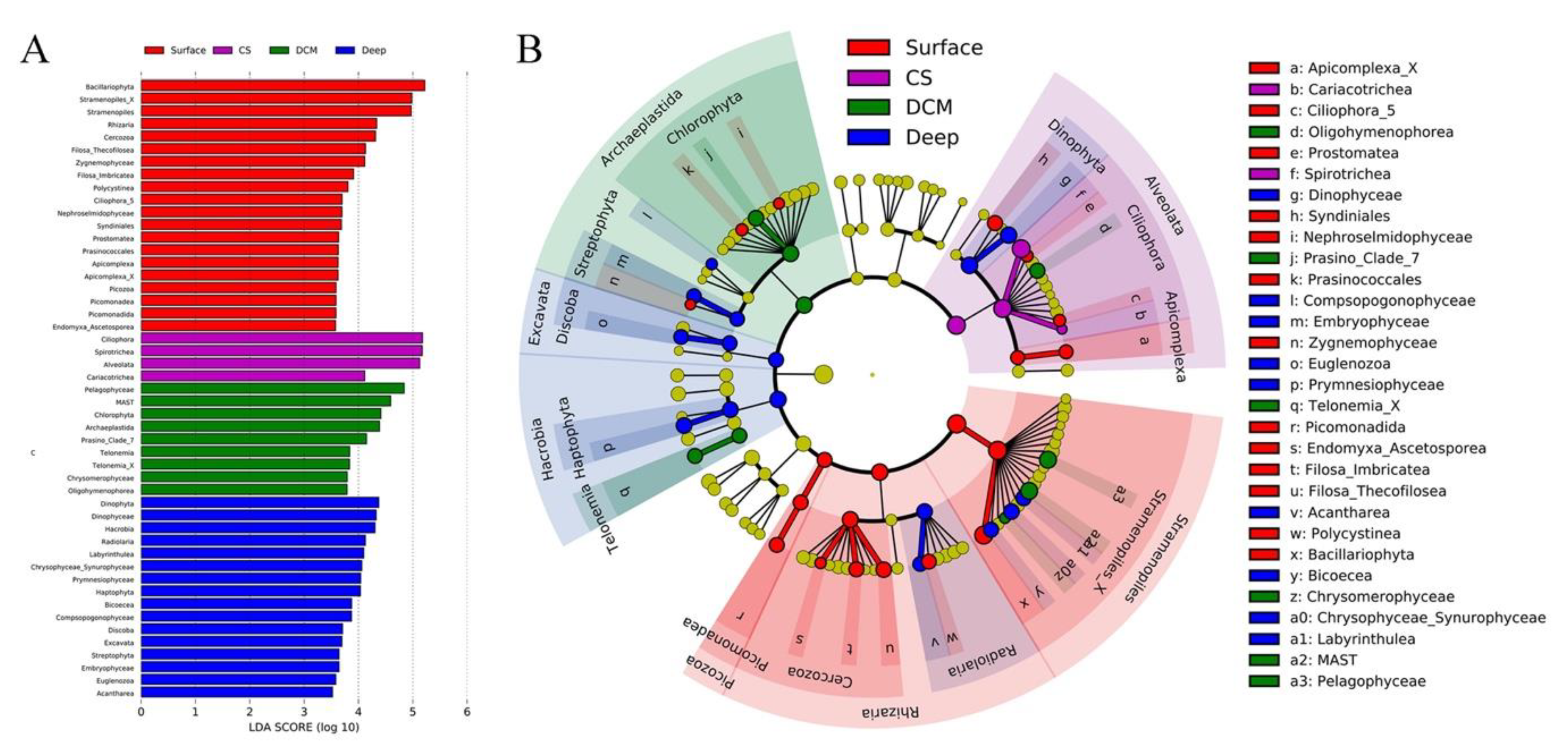
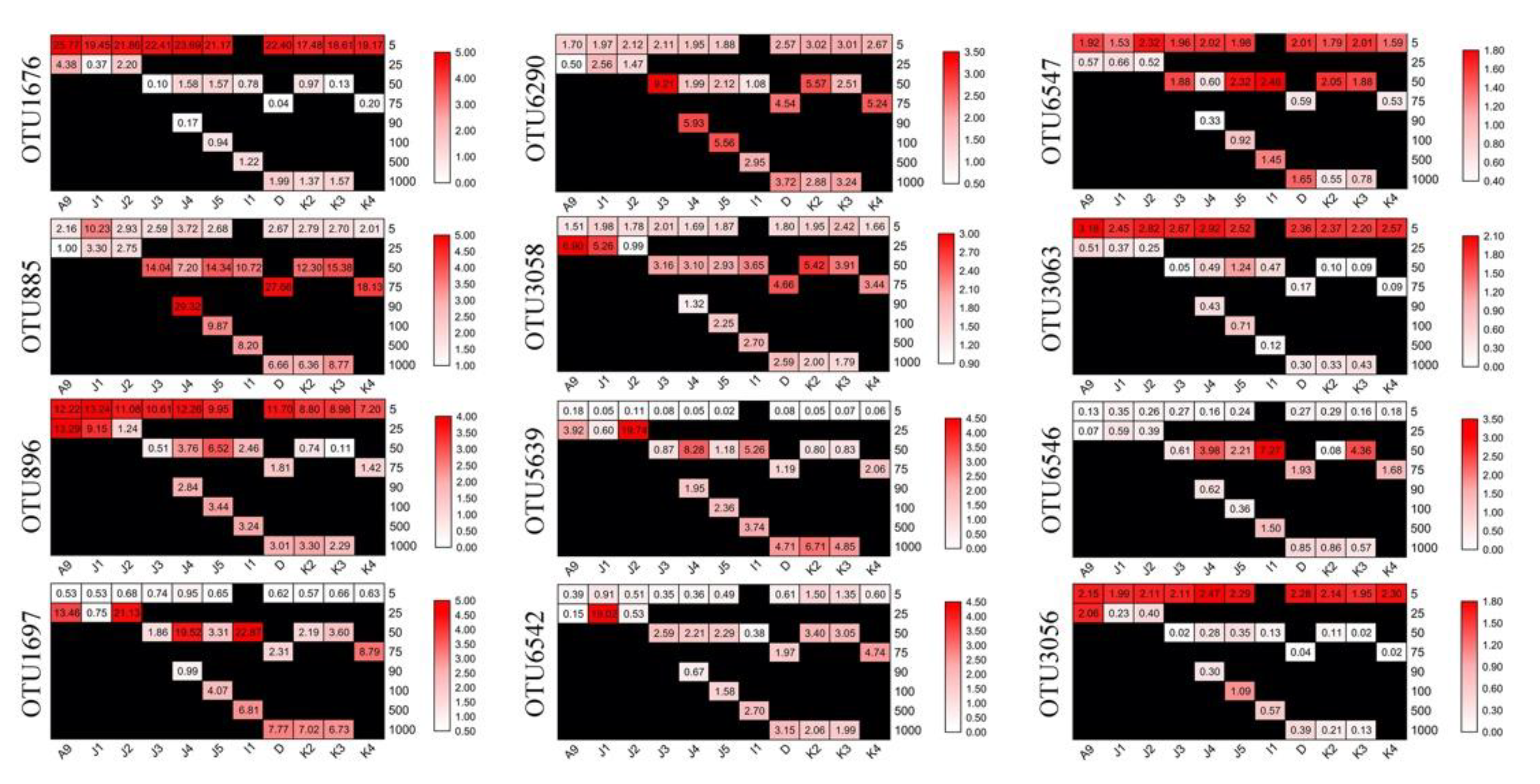
3.4. Abundant and Rare Groups
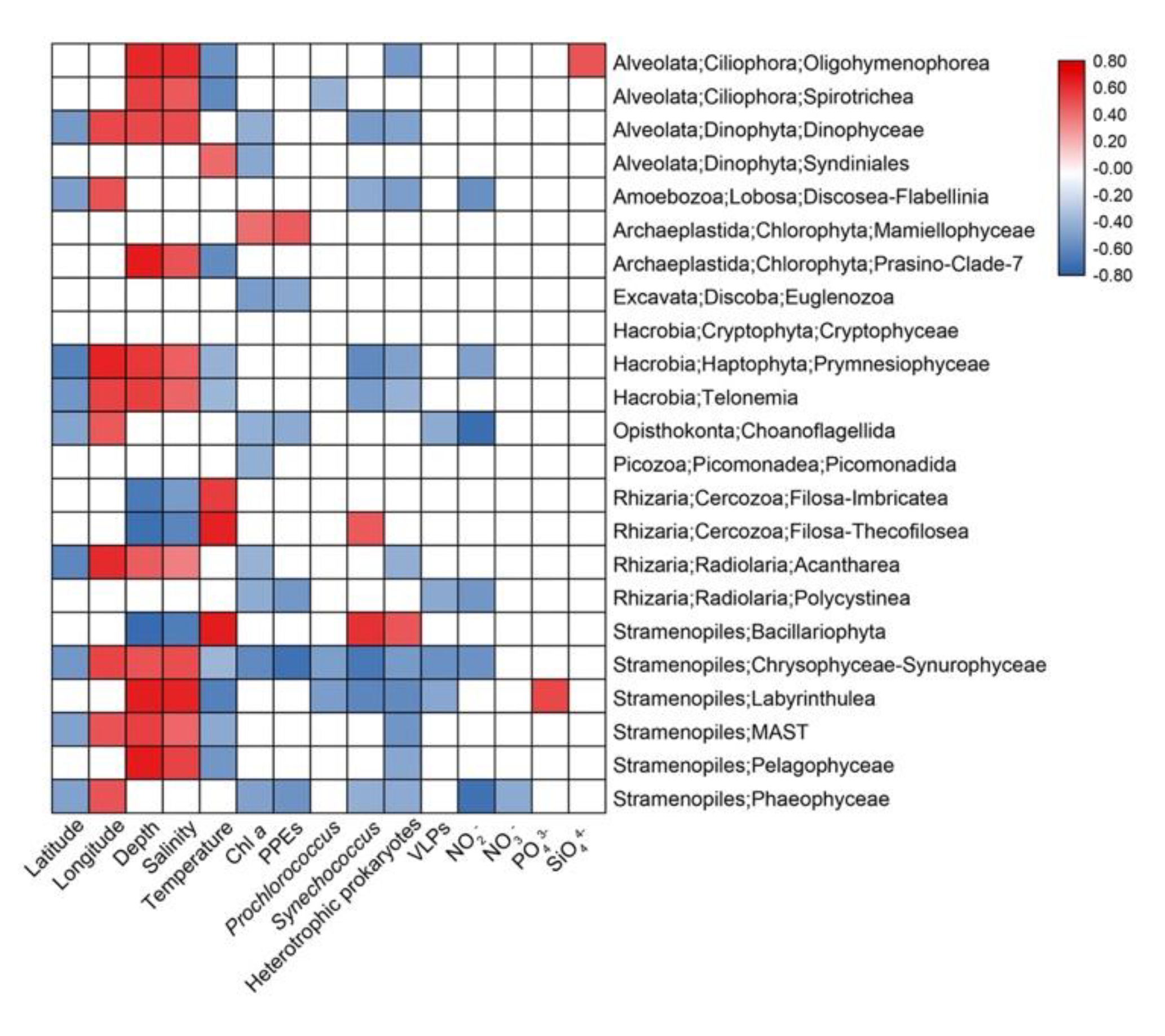
3.5. Effects of Environmental Parameters on the Total (TG), Abundant (AG) and Rare (RG) Groups
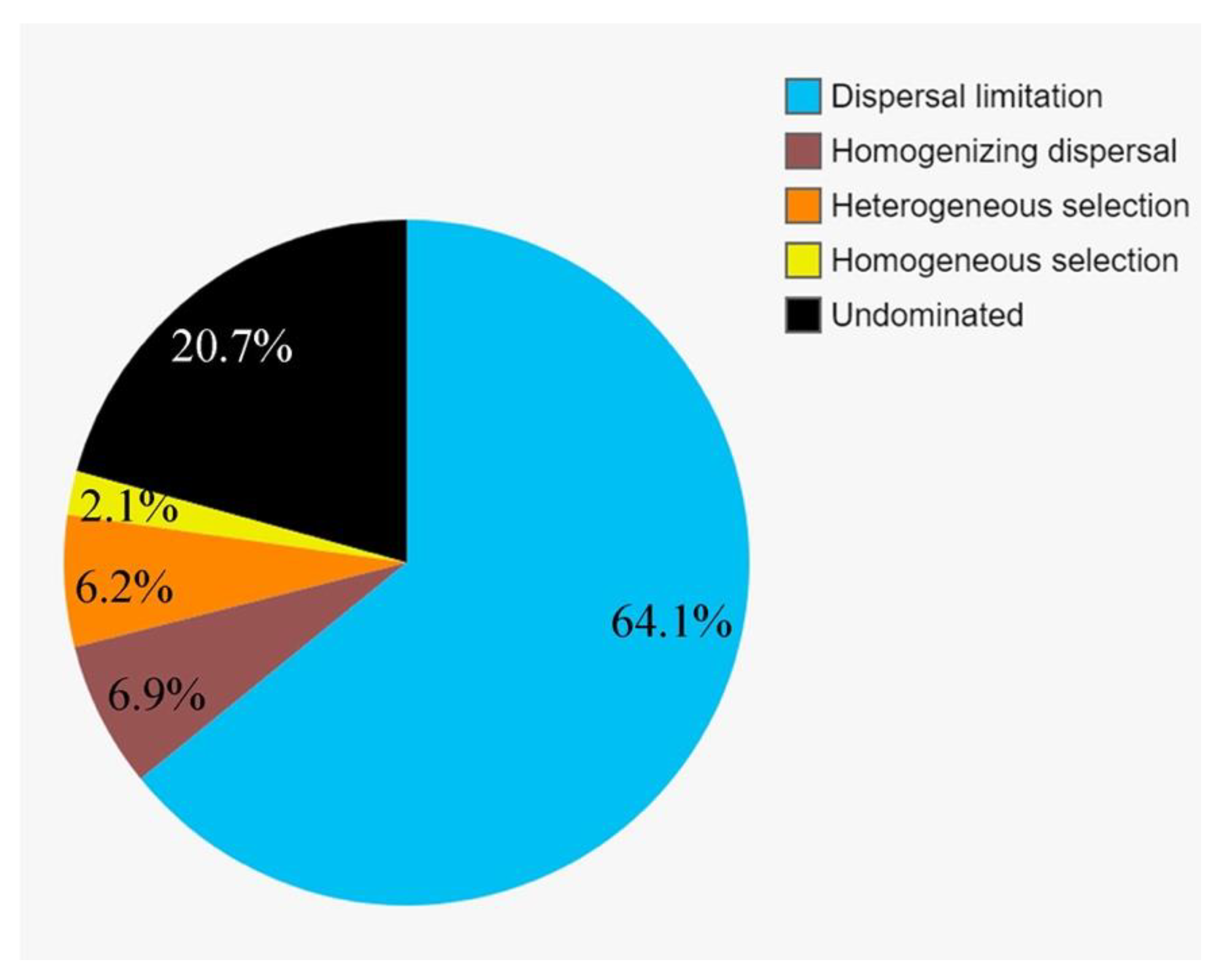
3.6. The Community Assembly of Protists
4. Discussion
4.1. Environmental Parameters
4.2. Variations of Major Protistan Assemblages along the Transect
4.3. Driving Factors of the Total, Abundant, and Rare Protistan Groups
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sunagawa, S.; Coelho, L.P.; Chaffron, S.; Kultima, J.R.; Labadie, K.; Salazar, G.; Djahanschiri, B.; Zeller, G.; Mende, D.R.; Alberti, A.; et al. Structure and function of the global ocean microbiome. Science 2015, 348, 1261359. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, J.A. Microbial community structure and its functional implications. Nat. Cell Biol. 2009, 459, 193–199. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.; Artigas, L.F.; Baross, J.; Bharathi, L.; Boetius, A.; Chandramohan, D.; Herndl, G.; Kogure, K.; Neal, P.; Pedrós-Alió, C.; et al. A Global Census of Marine Microbes. In Life in the World’s Oceans: Diversity, Distribution, and Abundance; Wiley-Blackwell: Hoboken, NJ, USA, 2010; ISBN 9781405192972. [Google Scholar]
- Caron, D.A.; Alexander, H.; Allen, A.E.; Archibald, J.M.; Armbrust, E.V.; Bachy, C.; Bell, C.J.; Bharti, A.; Dyhrman, S.T.; Guida, S.M.; et al. Probing the evolution, ecology and physiology of marine protists using transcriptomics. Nat. Rev. Genet. 2016, 15, 6–20. [Google Scholar] [CrossRef]
- Pace, N.R. A molecular view of microbial diversity and the biosphere. Science 1997, 276, 734–740. [Google Scholar] [CrossRef] [PubMed]
- Levinsen, H.; Nielsen, T.G. The trophic role of marine pelagic ciliates and heterotrophic dinoflagellates in arctic and temperate coastal ecosystems: A cross-latitude comparison. Limnol. Oceanogr. 2002, 47, 427–439. [Google Scholar] [CrossRef]
- Follows, M.J.; Dutkiewicz, S.; Grant, S.; Chisholm, S.W. Emergent biogeography of microbial communities in a model ocean. Science 2007, 315, 1843–1846. [Google Scholar] [CrossRef]
- Caron, D.; Countway, P. Hypotheses on the role of the protistan rare biosphere in a changing world. Aquat. Microb. Ecol. 2009, 57, 227–238. [Google Scholar] [CrossRef]
- Logares, R.; Audic, S.; Bass, D.; Bittner, L.; Boutte, C.; Christen, R.; Claverie, J.-M.; Decelle, J.; Dolan, J.R.; Dunthorn, M.; et al. Patterns of rare and abundant marine microbial eukaryotes. Curr. Biol. 2014, 24, 813–821. [Google Scholar] [CrossRef]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Genet. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Pedrós-Alió, C. The Rare Bacterial Biosphere. Annu. Rev. Mar. Sci. 2012, 4, 449–466. [Google Scholar] [CrossRef] [PubMed]
- Jiao, S.; Chen, W.; Weimin, C. Biogeography and ecological diversity patterns of rare and abundant bacteria in oil-contaminated soils. Mol. Ecol. 2017, 26, 5305–5317. [Google Scholar] [CrossRef]
- Wu, W.; Logares, R.; Huang, B.; Hsieh, C.-H. Abundant and rare picoeukaryotic sub-communities present contrasting patterns in the epipelagic waters of marginal seas in the northwestern Pacific Ocean. Environ. Microbiol. 2017, 19, 287–300. [Google Scholar] [CrossRef]
- Jia, X.; Dini-Andreote, F.; Salles, J.F. Community assembly processes of the microbial rare biosphere. Trends Microbiol. 2018, 26, 738–747. [Google Scholar] [CrossRef]
- Jousset, A.; Bienhold, C.; Chatzinotas, A.; Gallien, L.; Gobet, A.; Kurm, V.; Küsel, K.; Rillig, M.C.; Rivett, D.W.; Salles, J.F.; et al. Where less may be more: How the rare biosphere pulls ecosystems strings. ISME J. 2017, 11, 853–862. [Google Scholar] [CrossRef]
- Mallon, C.A.; Poly, F.; Le Roux, X.; Marring, I.; Van Elsas, J.D.; Salles, J.F. Resource pulses can alleviate the biodiversity-invasion relationship in soil microbial communities. Ecology 2015, 96, 915–926. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hatt, J.K.; Tsementzi, D.; Rodriguez-R, L.M.; Ruiz-Pérez, C.A.; Weigand, M.R.; Kizer, H.; Maresca, G.; Krishnan, R.; Poretsky, R.; et al. Quantifying the importance of the rare biosphere for microbial community response to organic pollutants in a freshwater ecosystem. Appl. Environ. Microbiol. 2017, 83, e03321-16. [Google Scholar] [CrossRef]
- Van Elsas, J.D.; Chiurazzi, M.; Mallon, C.A.; Elhottova, D.; Kristufek, V.; Salles, J.F. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc. Natl. Acad. Sci. USA 2012, 109, 1159–1164. [Google Scholar] [CrossRef] [PubMed]
- Delong, E.F.; Karl, D.M. Genomic perspectives in microbial oceanography. Nat. Cell Biol. 2005, 437, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Caron, D.A.; Countway, P.D.; Jones, A.C.; Kim, D.Y.; Schnetzer, A. Marine protistan diversity. Annu. Rev. Mar. Sci. 2012, 4, 467–493. [Google Scholar] [CrossRef]
- De Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; Berney, C.; Le Bescot, N.; Probert, I.; et al. Ocean plankton. Eukaryotic plankton diversity in the sunlit ocean. Science 2015, 348, 1261605. [Google Scholar] [CrossRef]
- Gran-Stadniczeñko, S.; Egge, E.D.; Hostyeva, V.; Logares, R.; Eikrem, W.; Edvardsen, B. Protist diversity and seasonal dynamics in Skagerrak plankton communities as revealed by metabarcoding and microscopy. J. Eukaryot. Microbiol. 2018, 66, 494–513. [Google Scholar] [CrossRef]
- Bork, P.; Bowler, C.; De Vargas, C.; Gorsky, G.; Karsenti, E.; Wincker, P. Tara Oceans studies plankton at planetary scale. Science 2015, 348, 873. [Google Scholar] [CrossRef]
- Xu, D.; Li, R.; Hu, C.; Sun, P.; Jiao, N.; Warren, A. Microbial eukaryote diversity and activity in the water column of the South China Sea based on DNA and RNA high throughput sequencing. Front. Microbiol. 2017, 8, 1121. [Google Scholar] [CrossRef]
- Nolte, V.; Pandey, R.V.; Jost, S.; Medinger, R.; Ottenwälder, B.; Boenigk, J.; Schlötterer, C. Contrasting seasonal niche separation between rare and abundant taxa conceals the extent of protist diversity. Mol. Ecol. 2010, 19, 2908–2915. [Google Scholar] [CrossRef] [PubMed]
- Hugoni, M.; Taib, N.; Debroas, D.; Domaizon, I.; Dufournel, I.J.; Bronner, G.; Salter, I.; Agogué, H.; Mary, I.; Galand, P.E. Structure of the rare archaeal biosphere and seasonal dynamics of active ecotypes in surface coastal waters. Proc. Natl. Acad. Sci. USA 2013, 110, 6004–6009. [Google Scholar] [CrossRef]
- Gobet, A.; Böer, S.; Huse, S.M.; Van Beusekom, J.; Quince, C.; Sogin, M.L.; Boetius, A.; Ramette, A. Diversity and dynamics of rare and of resident bacterial populations in coastal sands. ISME J. 2011, 6, 542–553. [Google Scholar] [CrossRef]
- Galand, P.E.; Casamayor, E.O.; Kirchman, D.L.; Lovejoy, C. Ecology of the rare microbial biosphere of the Arctic Ocean. Proc. Natl. Acad. Sci. USA 2009, 106, 22427–22432. [Google Scholar] [CrossRef]
- Jones, S.E.; Lennon, J.T. Dormancy contributes to the maintenance of microbial diversity. Proc. Natl. Acad. Sci. USA 2010, 107, 5881–5886. [Google Scholar] [CrossRef] [PubMed]
- Aanderud, Z.T.; Jones, S.E.; Efierer, N.; Lennon, J.T. Resuscitation of the rare biosphere contributes to pulses of ecosystem activity. Front. Microbiol. 2015, 6, 24. [Google Scholar] [CrossRef]
- Campbell, B.J.; Yu, L.; Heidelberg, J.F.; Kirchman, D.L. Activity of abundant and rare bacteria in a coastal ocean. Proc. Natl. Acad. Sci. USA 2011, 108, 12776–12781. [Google Scholar] [CrossRef] [PubMed]
- Genitsaris, S.; Monchy, S.; Viscogliosi, E.; Sime-Ngando, T.; Ferreira, S.; Christaki, U. Seasonal variations of marine protist community structure based on taxon-specific traits using the eastern English Channel as a model coastal system. FEMS Microbiol. Ecol. 2015, 91. [Google Scholar] [CrossRef] [PubMed]
- Mangot, J.-F.; Domaizon, I.; Taib, N.; Marouni, N.; Duffaud, E.; Bronner, G.; Debroas, D. Short-term dynamics of diversity patterns: Evidence of continual reassembly within lacustrine small eukaryotes. Environ. Microbiol. 2013, 15, 1745–1758. [Google Scholar] [CrossRef]
- Josephson, K.L.; Gerba, C.P.; Pepper, I.L. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 1993, 59, 3513–3515. [Google Scholar] [CrossRef] [PubMed]
- Dell’Anno, A. Extracellular DNA plays a key role in deep-sea ecosystem functioning. Science 2005, 309, 2179. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.K.; Campbell, V.; Connell, P.; Gellene, A.G.; Liu, Z.; Terrado, R.; Caron, D.A. Protistan diversity and activity inferred from RNA and DNA at a coastal ocean site in the eastern North Pacific. FEMS Microbiol. Ecol. 2016, 92, fiw050. [Google Scholar] [CrossRef]
- Charvet, S.; Vincent, W.F.; Lovejoy, C. Effects of light and prey availability on Arctic freshwater protist communities examined by high-throughput DNA and RNA sequencing. FEMS Microbiol. Ecol. 2014, 88, 550–564. [Google Scholar] [CrossRef]
- Sun, P.; Huang, L.; Xu, D.; Huang, B.; Chen, N.; Warren, A. Marked seasonality and high spatial variation in estuarine ciliates are driven by exchanges between the ‘abundant’ and ‘intermediate’ biospheres. Sci. Rep. 2017, 7, 1–12. [Google Scholar] [CrossRef]
- Dai, M.; Meng, F.; Tang, T.; Kao, S.-J.; Lin, J.; Chen, J.; Huang, J.-C.; Tian, J.; Gan, J.; Yang, S. Excess total organic carbon in the intermediate water of the South China Sea and its export to the North Pacific. Geochem. Geophys. Geosyst. 2009, 10. [Google Scholar] [CrossRef]
- Hu, J.; Kawamura, H.; Hong, H.; Qi, Y. A Review on the currents in the South China Sea: Seasonal circulation, South China Sea Warm Current and Kuroshio intrusion. J. Oceanogr. 2000, 56, 607–624. [Google Scholar] [CrossRef]
- Ning, X.; Chai, F.; Xue, H.; Cai, Y.; Liu, C.; Shi, J. Physical-biological oceanographic coupling influencing phytoplankton and primary production in the South China Sea. J. Geophys. Res. Space Phys. 2004, 109, 1–20. [Google Scholar] [CrossRef]
- He, L.; Yin, K.; Yuan, X.; Li, D.; Zhang, D.; Harrison, P.J. Spatial distribution of viruses, bacteria and chlorophyll in the northern South China Sea. Aquat. Microb. Ecol. 2009, 54, 153–162. [Google Scholar] [CrossRef]
- Hu, C.; Chen, X.; Yu, L.; Xu, D.; Jiao, N. Elevated contribution of low nucleic acid prokaryotes and viral lysis to the prokaryotic community along the nutrient gradient from an estuary to open ocean transect. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef]
- Jin, T.; Zhang, T.; Ye, L.; Lee, O.O.; Wong, Y.H.; Qian, P.Y. Diversity and quantity of ammonia-oxidizing Archaea and Bacteria in sediment of the Pearl River Estuary, China. Appl. Microbiol. Biotechnol. 2011, 90, 1137–1145. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; He, L.; Yin, K.; Pan, G.; Harrison, P.J. Bacterial distribution and nutrient limitation in relation to different water masses in the coastal and northwestern South China Sea in late summer. Cont. Shelf Res. 2011, 31, 1214–1223. [Google Scholar] [CrossRef]
- Li, J.; Jiang, X.; Jing, Z.; Li, G.; Chen, Z.; Zhou, L.; Zhao, C.; Liu, J.; Tan, Y. Spatial and seasonal distributions of bacterioplankton in the Pearl River Estuary: The combined effects of riverine inputs, temperature, and phytoplankton. Mar. Pollut. Bull. 2017, 125, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Zhang, C.; Zhou, X.; Wang, P. Salinity-dominated change in community structure and ecological function of Archaea from the lower Pearl River to coastal South China Sea. Appl. Microbiol. Biotechnol. 2014, 98, 7971–7982. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.; Huang, X.; Zhang, X. Picoplankton and virioplankton abundance and community structure in Pearl River Estuary and Daya Bay, South China. J. Environ. Sci. 2015, 32, 146–154. [Google Scholar] [CrossRef]
- Hu, A.; Jiao, N.; Zhang, R.; Hu, A. Community structure and function of planktonic Crenarchaeota: Changes with depth in the South China Sea. Microb. Ecol. 2011, 62, 549–563. [Google Scholar] [CrossRef]
- Xia, X.; Guo, W.; Liu, H. Dynamics of the bacterial and archaeal communities in the Northern South China Sea revealed by 454 pyrosequencing of the 16S rRNA gene. Deep. Sea Res. Part II Top. Stud. Oceanogr. 2015, 117, 97–107. [Google Scholar] [CrossRef]
- Hou, L.; Xie, X.; Wan, X.; Kao, S.; Jiao, N.; Zhang, Y. Niche differentiation of ammonia and nitrite oxidizers along a salinity gradient from the Pearl River estuary to the South China Sea. Biogeosciences 2018, 15, 5169–5187. [Google Scholar] [CrossRef]
- Lin, D.; Zhou, H.; Chen, Y.-Q.; Li, L.-Y.; Chen, J.-H.; Wu, S.-H.; Huang, Q.-J.; Qu, L.-H. Diversity and distribution of planktonic protists in the northern South China Sea. J. Plankton Res. 2010, 33, 445–456. [Google Scholar] [CrossRef][Green Version]
- Wu, W.; Huang, B.; Zhong, C. Photosynthetic picoeukaryote assemblages in the South China Sea from the Pearl River estuary to the SEATS station. Aquat. Microb. Ecol. 2014, 71, 271–284. [Google Scholar] [CrossRef]
- Wu, W.; Liu, H. Disentangling protist communities identified from DNA and RNA surveys in the Pearl River-South China Sea Continuum during the wet and dry seasons. Mol. Ecol. 2018, 27, 4627–4640. [Google Scholar] [CrossRef]
- Xu, D.; Jiao, N.; Ren, R.; Warren, A. Distribution and diversity of microbial eukaryotes in bathypelagic waters of the South China Sea. J. Eukaryot. Microbiol. 2017, 64, 370–382. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Chen, M.-Y.; Shao, P.; Zhou, H.; Chen, Y.-Q.; Qu, L. Genetic diversity of small eukaryotes from the coastal waters of Nansha Islands in China. FEMS Microbiol. Lett. 2004, 240, 163–170. [Google Scholar] [CrossRef]
- Wu, W.; Wang, L.; Liao, Y.; Huang, B. Microbial eukaryotic diversity and distribution in a river plume and cyclonic eddy-influenced ecosystem in the South China Sea. MicrobiologyOpen 2015, 4, 826–840. [Google Scholar] [CrossRef]
- Li, R.; Jiao, N.; Warren, A.; Xu, D. Changes in community structure of active protistan assemblages from the lower Pearl River to coastal Waters of the South China Sea. Eur. J. Protistol. 2018, 63, 72–82. [Google Scholar] [CrossRef]
- Jiang, Z.-Y.; Wang, Y.-S.; Sun, F.-L. Spatial structure of eukaryotic ultraplankton community in the northern South China Sea. Biologia 2014, 69, 557–565. [Google Scholar] [CrossRef]
- Welschmeyer, N.A. Fluorometric analysis of chlorophyll a in the presence of chlorophyll b and pheopigments. Limnol. Oceanogr. 1994, 39, 1985–1992. [Google Scholar] [CrossRef]
- Liang, Y.; Zhang, Y.; Zhang, Y.; Luo, T.; Rivkin, R.B.; Jiao, N. Distributions and relationships of virio- and picoplankton in the epi-, meso- and bathypelagic zones of the Western Pacific Ocean. FEMS Microbiol. Ecol. 2016, 93, fiw238. [Google Scholar] [CrossRef] [PubMed]
- Marie, D.; Partensky, F.; Vaulot, D.; Brussaard, C. Enumeration of phytoplankton, bacteria, and viruses in marine samples. Curr. Protoc. Cytom. 1999, 10, 11. [Google Scholar] [CrossRef]
- Jiao, N.; Yang, Y.Y.; Koshikawa, H.H.; Watanabe, M.M. Influence of hydrographic conditions on picoplankton distribution in the East China Sea. Aquat. Microb. Ecol. 2002, 30, 37–48. [Google Scholar] [CrossRef]
- Amaral-Zettler, L.A.; McCliment, E.A.; Ducklow, H.W.; Huse, S.M. A method for studying protistan diversity using mas-sively parallel sequencing of V9 hypervariable regions of small-subunit ribosomal RNA genes. PLoS ONE 2009, 4, e6372. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Magoč, T.; Magoč, M.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome sssemblies. Bioinform. 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; De Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote small sub-unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2012, 41, D597–D604. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Clarke, K.R.; Warwick, R.M. Change in Marine Communities: An Approach to Statistical Analysis and Interpretation, 2nd ed.; PRIMER-E: Plymouth, UK, 2001; p. 172. [Google Scholar]
- Clarke, K.R.; Gorley, R.N. PRIMER v6: User Manual/Tutorial; PRIMER-E: Plymouth, UK, 2006. [Google Scholar] [CrossRef]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D.A.; Ryan, P.D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 2001, 4, 9. [Google Scholar] [CrossRef]
- Legendre, P.; Legendre, L. Numerical Ecology; Elsevier: Amsterdam, The Netherlands, 1998. [Google Scholar]
- Stegen, J.C.; Lin, X.; Fredrickson, J.K.; Chen, X.; Kennedy, D.W.; Murray, C.J.; Rockhold, M.L.; Konopka, A. Quantifying community assembly processes and identifying features that impose them. ISME J. 2013, 7, 2069–2079. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Ning, D. Stochastic community assembly: Does it matter in microbial ecology? Microbiol. Mol. Biol. Rev. 2017, 81, e00002-17. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Huang, L.; Zhang, J.; Lin, S. Phytoplankton dynamics in and near the highly eutrophic Pearl River Estuary, South China Sea. Cont. Shelf Res. 2010, 30, 177–186. [Google Scholar] [CrossRef]
- Wu, W.; Wang, L.; Liao, Y.; Xu, S.; Huang, B. Spatial and seasonal distributions of photosynthetic picoeukaryotes along an estuary to basin transect in the northern South China Sea. J. Plankton Res. 2017, 39, 423–435. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Dai, M.; Jiao, N.; Herndl, G.J. Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Mol. Ecol. 2014, 23, 2260–2274. [Google Scholar] [CrossRef]
- Li, X.; Warren, A.; Jiao, N.; Xu, D. Distribution of protists in the deep South China Sea revealed by high-throughput sequencing. J. Ocean Univ. China 2020, 19, 161–170. [Google Scholar] [CrossRef]
- Countway, P.D.; Gast, R.J.; Dennett, M.R.; Savai, P.; Rose, J.M.; Caron, D.A. Distinct protistan assemblages characterize the euphotic zone and deep sea (2500 m) of the western North Atlantic (Sargasso Sea and Gulf Stream). Environ. Microbiol. 2007, 9, 1219–1232. [Google Scholar] [CrossRef]
- Schnetzer, A.; Moorthi, S.D.; Countway, P.D.; Gast, R.J.; Gilg, I.C.; Caron, D.A. Depth matters: Microbial eukaryote diversity and community structure in the eastern North Pacific revealed through environmental gene libraries. Deep. Sea Res. Part I Oceanogr. Res. Pap. 2011, 58, 16–26. [Google Scholar] [CrossRef]
- Countway, P.D.; Vigil, P.; Schnetzer, A.; Moorthi, S.D.; Caron, D.A. Seasonal analysis of protistan community structure and diversity at the USC Microbial Observatory (San Pedro Channel, North Pacific Ocean). Limnol. Oceanogr. 2010, 55, 2381–2396. [Google Scholar] [CrossRef]
- Choi, D.H.; An, S.M.; Chun, S.; Yang, E.C.; Selph, K.E.; Lee, C.M.; Noh, J.H. Dynamic changes in the composition of photosynthetic picoeukaryotes in the northwestern Pacific Ocean revealed by high-throughput tag sequencing of plastid 16S rRNA genes. FEMS Microbiol. Ecol. 2016, 92, fiv170. [Google Scholar] [CrossRef]
- Michaels, A.F.; Silver, M.W. Primary production, sinking fluxes and the microbial food web. Deep. Sea Res. Part A Oceanogr. Res. Pap. 1988, 35, 473–490. [Google Scholar] [CrossRef]
- Balzano, S.; Sarno, D.; Kooistra, W.H.C.F. Effects of salinity on the growth rate and morphology of ten Skeletonema strains. J. Plankton Res. 2010, 33, 937–945. [Google Scholar] [CrossRef]
- Balzano, S.; Abs, E.; Leterme, S. Protist diversity along a salinity gradient in a coastal lagoon. Aquat. Microb. Ecol. 2015, 74, 263–277. [Google Scholar] [CrossRef]
- Lee, J.E.; Chung, I.K.; Lee, S.-R. Dynamic genetic features of eukaryotic plankton diversity in the Nakdong River estuary of Korea. Chin. J. Oceanol. Limnol. 2016, 35, 844–857. [Google Scholar] [CrossRef]
- Sarno, D.; Kooistra, W.H.C.F.; Balzano, S.; Hargraves, P.E.; Zingone, A. Diversity in the genus Skeletonema (bacillariophyceae): III. Phylogenetic position and morphological variability of Skeletonema costatum and Skeletonema grevillei, with the description of Skeletonema ardens sp. nov. J. Phycol. 2007, 43, 156–170. [Google Scholar] [CrossRef]
- Łotocka, M. The first observed bloom of the diatom Dactyliosolen fragilissimus (Bergon) Hasle 1996 in the Gulf of Gdańsk [commun.]. Oceanologia 2006, 48, 447–452. [Google Scholar]
- Villareal, T.A.; Altabet, M.A.; Culver-Rymsza, K. Nitrogen transport by vertically migrating diatom mats in the North Pacific Ocean. Nat. Cell Biol. 1993, 363, 709–712. [Google Scholar] [CrossRef]
- Bass, D.; Cavalier-Smith, T. Phylum-specific environmental DNA analysis reveals remarkably high global biodiversity of Cercozoa (Protozoa). Int. J. Syst. Evol. Microbiol. 2004, 54, 2393–2404. [Google Scholar] [CrossRef]
- Berney, C.; Romac, S.; Mahé, F.; Santini, S.; Siano, R.; Bass, D. Vampires in the oceans: Predatory cercozoan amoebae in marine habitats. ISME J. 2013, 7, 2387–2399. [Google Scholar] [CrossRef] [PubMed]
- Fiore-Donno, A.M.; Richter-Heitmann, T.; Degrune, F.; Dumack, K.; Regan, K.M.; Marhan, S.; Boeddinghaus, R.S.; Rillig, M.C.; Friedrich, M.W.; Kandeler, E.; et al. Functional traits and spatio-temporal structure of a major group of soil protists (Rhizaria: Cercozoa) in a temperate grassland. Front. Microbiol. 2019, 10, 1332. [Google Scholar] [CrossRef] [PubMed]
- Reece, K.S.; Siddall, M.E.; Stokes, N.A.; Burreson, E.M. Molecular phylogeny of the haplosporidia based on two independent gene sequences. J. Parasitol. 2004, 90, 1111–1122. [Google Scholar] [CrossRef]
- Skovgaard, A.; Daugbjerg, N. Identity and systematic position of Paradinium poucheti and other Paradinium-like parasites of marine copepods based on morphology and nuclear-encoded SSU rDNA. Protist 2008, 159, 401–413. [Google Scholar] [CrossRef]
- Dumack, K.; Müller, M.E.; Bonkowski, M. Description of Lecythium terrestris sp. nov. (Chlamydophryidae, Cercozoa), a soil dwelling protist feeding on fungi and algae. Protist 2016, 167, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Thaler, M.; Lovejoy, C. Distribution and diversity of a protist predator Cryothecomonas (Cercozoa) in Arctic marine waters. J. Eukaryot. Microbiol. 2012, 59, 291–299. [Google Scholar] [CrossRef]
- Dolan, J.R.; Gallegos, C.L. Estuarine diversity of tintinnids (planktonic ciliates). J. Plankton Res. 2001, 23, 1009–1027. [Google Scholar] [CrossRef][Green Version]
- Massana, R.; Del Campo, J.; Sieracki, M.E.; Audic, S.; Logares, R. Exploring the uncultured microeukaryote majority in the oceans: Reevaluation of ribogroups within stramenopiles. ISME J. 2014, 8, 854–866. [Google Scholar] [CrossRef]
- Cheung, M.K.; Nong, W.; Kwan, H.S.; Wong, C.K. Composition of heterotrophic flagellates in coastal waters of different trophic status. Curr. Microbiol. 2013, 67, 351–355. [Google Scholar] [CrossRef]
- Not, F.; Campo, J.; Balagué, V.; De Vargas, C.; Massana, R. New insights into the diversity of marine picoeukaryotes. PLoS ONE 2009, 4, e7143. [Google Scholar] [CrossRef]
- Dupont, C.L.; McCrow, J.P.; Valas, R.; Moustafa, A.; Walworth, N.; Goodenough, U.; Roth, R.; Hogle, S.L.; Bai, J.; Johnson, Z.; et al. Genomes and gene expression across light and productivity gradients in eastern subtropical Pacific microbial communities. ISME J. 2015, 9, 1076–1092. [Google Scholar] [CrossRef]
- Shi, X.L.; Lepère, C.; Scanlan, D.J.; Vaulot, D. Plastid 16S rRNA Gene Diversity among eukaryotic picophytoplankton sorted by flow cytometry from the South Pacific Ocean. PLoS ONE 2011, 6, e18979. [Google Scholar] [CrossRef]
- Treusch, A.H.; Demir-Hilton, E.; Vergin, K.L.; Worden, A.Z.; Carlson, C.; Donatz, M.G.; Burton, R.M.; Giovannoni, S.J. Phytoplankton distribution patterns in the northwestern Sargasso Sea revealed by small subunit rRNA genes from plastids. ISME J. 2011, 6, 481–492. [Google Scholar] [CrossRef]
- Cabello, A.M.; Latasa, M.; Forn, I.; Morán, X.A.G.; Massana, R. Vertical distribution of major photosynthetic picoeukaryotic groups in stratified marine waters. Environ. Microbiol. 2016, 18, 1578–1590. [Google Scholar] [CrossRef] [PubMed]
- Unrein, F.; Gasol, J.M.; Not, F.; Forn, I.; Massana, R. Mixotrophic haptophytes are key bacterial grazers in oligotrophic coastal waters. ISME J. 2014, 8, 164–176. [Google Scholar] [CrossRef]
- Xu, D.; Sun, P.; Zhang, Y.; Li, R.; Huang, B.; Jiao, N.; Warren, A.; Wang, L. Pigmented microbial eukaryotes fuel the deep sea carbon pool in the tropical Western Pacific Ocean. Environ. Microbiol. 2018, 20, 3811–3824. [Google Scholar] [CrossRef] [PubMed]
- Richardson, T.L.; Jackson, G.A. Small Phytoplankton and carbon export from the surface ocean. Science 2007, 315, 838–840. [Google Scholar] [CrossRef]
- Iversen, M.; Ploug, H. Ballast minerals and the sinking carbon flux in the ocean: Carbon-specific respiration rates and sinking velocity of marine snow aggregates. Biogeosciences 2010, 7, 2613–2624. [Google Scholar] [CrossRef]
- Ploug, H.; Iversen, M.H.; Koski, M.; Buitenhuis, E.T. Production, oxygen respiration rates, and sinking velocity of copepod fecal pellets: Direct measurements of ballasting by opal and calcite. Limnol. Oceanogr. 2008, 53, 469–476. [Google Scholar] [CrossRef]
- Simon, M.; Grossart, H.; Schweitzer, B.; Ploug, H. Microbial ecology of organic aggregates in aquatic ecosystems. Aquat. Microb. Ecol. 2002, 28, 175–211. [Google Scholar] [CrossRef]
- Not, F.; Gausling, R.; Azam, F.; Heidelberg, J.F.; Worden, A.Z. Vertical distribution of picoeukaryotic diversity in the Sargasso Sea. Environ. Microbiol. 2007, 9, 1233–1252. [Google Scholar] [CrossRef]
- Zhang, Y.; Jiao, N.; Cottrell, M.T.; Kirchman, D.L. Contribution of major bacterial groups to bacterial biomass production along a salinity gradient in the South China Sea. Aquat. Microb. Ecol. 2006, 43, 233–241. [Google Scholar] [CrossRef]
- Xiong, J.; Ye, X.; Wang, K.; Chen, H.; Hu, C.; Zhu, J.; Zhang, D. Biogeography of the sediment bacterial community responds to a nitrogen pollution gradient in the East China Sea. Appl. Environ. Microbiol. 2014, 80, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Pandit, S.N.; Kolasa, J.; Cottenie, K. Contrasts between habitat generalists and specialists: An empirical extension to the basic metacommunity framework. Ecology 2009, 90, 2253–2262. [Google Scholar] [CrossRef] [PubMed]
- Grime, J.P. Benefits of plant diversity to ecosystems: Immediate, filter and founder effects. J. Ecol. 1998, 86, 902–910. [Google Scholar] [CrossRef]

| Pairs | ANOSIM | |
|---|---|---|
| r | p | |
| Surface vs. CS | 0.900 | 0.003 |
| Surface vs. DCM | 0.996 | 0.001 |
| Surface vs. Deep | 0.997 | 0.001 |
| CS vs. DCM | 0.761 | 0.006 |
| CS vs. Deep | 0.679 | 0.012 |
| DCM vs. Deep | 0.534 | 0.001 |
| Environmental Variables | OTU Richness | Shannon | ||
|---|---|---|---|---|
| r | p | r | p | |
| Latitude | −0.454 | 0.017 | −0.565 | 0.002 |
| Longitude | 0.454 | 0.017 | 0.565 | 0.002 |
| Depth | 0.483 | 0.011 | 0.554 | 0.003 |
| Temperature | −0.328 | 0.094 | −0.405 | 0.036 |
| Salinity | 0.479 | 0.011 | 0.482 | 0.011 |
| NO3- | 0.130 | 0.586 | 0.009 | 0.97 |
| NO2- | −0.100 | 0.722 | −0.175 | 0.534 |
| PO43- | 0.438 | 0.09 | 0.037 | 0.892 |
| SiO44- | 0.226 | 0.325 | 0.103 | 0.656 |
| Chl a | −0.153 | 0.507 | –0.215 | 0.349 |
| Prochlorococcus, abundance | −0.170 | 0.461 | –0.023 | 0.922 |
| Synechococcus, abundance | −0.482 | 0.027 | –0.292 | 0.200 |
| PPEs, abundance | –0.027 | 0.907 | –0.207 | 0.368 |
| Heterotrophic prokaryotes, abundance | −0.416 | 0.039 | −0.561 | 0.004 |
| Abundance of VLPs | –0.167 | 0.470 | −0.500 | 0.021 |
| Environmental Variables | Simple Mantel | Partial Mantel | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Abundant | Rare | CF | Total | Abundant | Rare | CF | Total | Abundant | Rare | ||||||||||
| r | p | r | p | r | p | r | p | r | p | r | p | r | p | r | p | r | p | |||
| Salinity | 0.387 | 0.001 | 0.426 | 0.001 | 0.144 | 0.005 | T | 0.319 | 0.001 | 0.348 | 0.001 | 0.059 | 0.130 | D | 0.335 | 0.001 | 0.368 | 0.001 | 0.077 | 0.073 |
| Depth | 0.276 | 0.001 | 0.326 | 0.001 | 0.254 | 0.001 | S | 0.187 | 0.008 | 0.236 | 0.001 | 0.224 | 0.001 | T | 0.081 | 0.077 | 0.110 | 0.026 | 0.108 | 0.017 |
| Temperature | 0.266 | 0.001 | 0.310 | 0.001 | 0.231 | 0.001 | S | 0.134 | 0.046 | 0.171 | 0.010 | 0.191 | 0.002 | D | 0.244 | 0.244 | 0.029 | 0.287 | −0.006 | 0.528 |
| Geo_distance | 0.090 | 0.097 | 0.047 | 0.200 | 0.039 | 0.237 | S | 0.065 | 0.153 | 0.015 | 0.334 | 0.028 | 0.282 | D | 0.102 | 0.047 | 0.059 | 0.163 | 0.048 | 0.176 |
| Latitude | 0.092 | 0.081 | 0.047 | 0.210 | 0.039 | 0.237 | S | 0.067 | 0.137 | 0.015 | 0.399 | 0.028 | 0.323 | D | 0.105 | 0.042 | 0.061 | 0.163 | 0.048 | 0.168 |
| Longitude | 0.095 | 0.060 | 0.053 | 0.181 | 0.038 | 0.222 | S | 0.072 | 0.117 | 0.023 | 0.308 | 0.028 | 0.287 | D | 0.106 | 0.051 | 0.064 | 0.121 | 0.046 | 0.184 |
| NO3- | 0.005 | 0.455 | 0.003 | 0.413 | −0.002 | 0.472 | S | −0.221 | 0.993 | −0.245 | 0.995 | −0.113 | 0.929 | D | −0.197 | 0.989 | −0.225 | 0.997 | −0.122 | 0.968 |
| NO2- | 0.159 | 0.124 | 0.085 | 0.248 | −0.001 | 0.465 | S | 0.044 | 0.356 | −0.046 | 0.578 | −0.059 | 0.718 | D | 0.153 | 0.156 | 0.063 | 0.315 | −0.019 | 0.552 |
| PO43- | 0.184 | 0.084 | 0.211 | 0.065 | 0.153 | 0.049 | S | 0.079 | 0.250 | 0.108 | 0.191 | 0.132 | 0.085 | D | −0.171 | 0.943 | −0.144 | 0.906 | 0.040 | 0.302 |
| SO44- | 0.050 | 0.304 | 0.043 | 0.304 | 0.008 | 0.426 | S | −0.131 | 0.909 | −0.157 | 0.964 | −0.061 | 0.798 | D | −0.217 | 0.992 | −0.258 | 1.000 | −0.130 | 0.981 |
| Chl a | 0.185 | 0.030 | 0.180 | 0.034 | 0.044 | 0.246 | S | 0.110 | 0.097 | 0.101 | 0.112 | 0.025 | 0.364 | D | 0.091 | 0.144 | 0.081 | 0.169 | −0.012 | 0.566 |
| Prochlorococcus, abundance | −0.052 | 0.736 | −0.059 | 0.770 | 0.021 | 0.368 | S | −0.111 | 0.940 | −0.121 | 0.983 | 0.002 | 0.478 | D | 0.008 | 0.415 | 0.001 | 0.432 | 0.052 | 0.242 |
| Synechococcus, abundance | 0.060 | 0.227 | 0.054 | 0.239 | −0.039 | 0.736 | S | −0.070 | 0.804 | −0.080 | 0.836 | −0.090 | 0.917 | D | 0.093 | 0.137 | 0.089 | 0.145 | −0.041 | 0.726 |
| PPEs, abundance | 0.156 | 0.047 | 0.140 | 0.057 | 0.003 | 0.444 | S | 0.063 | 0.185 | 0.043 | 0.259 | −0.035 | 0.708 | D | 0.146 | 0.043 | 0.126 | 0.067 | −0.026 | 0.639 |
| HP, abundance | 0.160 | 0.017 | 0.140 | 0.030 | 0.004 | 0.464 | S | 0.028 | 0.297 | −0.009 | 0.525 | −0.057 | 0.830 | D | 0.097 | 0.070 | 0.066 | 0.136 | −0.055 | 0.838 |
| VLPs, abundance | 0.059 | 0.204 | 0.070 | 0.166 | −0.007 | 0.520 | S | 0.036 | 0.287 | 0.047 | 0.229 | −0.020 | 0.600 | D | −0.036 | 0.676 | −0.032 | 0.648 | −0.080 | 0.887 |
| Environmental Variables | Simple Mantel | Partial Mantel | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Abundant | Rare | Control for | Total | Abundant | Rare | |||||||
| r | p | r | p | r | p | r | p | r | p | r | p | ||
| Geo_distance | 0.461 | 0.005 | 0.503 | 0.004 | 0.209 | 0.079 | |||||||
| Latitude | 0.458 | 0.008 | 0.499 | 0.002 | 0.201 | 0.093 | Geo_distance | −0.017 | 0.552 | −0.035 | 0.610 | −0.092 | 0.72 |
| Longitude | 0.423 | 0.012 | 0.461 | 0.008 | 0.182 | 0.108 | Geo_distance | −0.210 | 0.909 | −0.241 | 0.959 | −0.146 | 0.821 |
| Salinity | −0.024 | 0.518 | 0.093 | 0.360 | 0.163 | 0.075 | Geo_distance | −0.224 | 0.814 | −0.103 | 0.634 | 0.098 | 0.158 |
| Temperature | 0.282 | 0.151 | 0.404 | 0.060 | 0.184 | 0.052 | Geo_distance | 0.154 | 0.291 | 0.291 | 0.139 | 0.124 | 0.116 |
| NO3- | 0.253 | 0.182 | 0.371 | 0.081 | 0.057 | 0.305 | Geo_distance | 0.103 | 0.319 | 0.234 | 0.175 | 0.000 | 0.505 |
| NO2- | 0.212 | 0.301 | 0.406 | 0.137 | 0.127 | 0.236 | Geo_distance | 0.166 | 0.323 | 0.360 | 0.187 | 0.104 | 0.300 |
| PO43- | 0.467 | 0.066 | 0.593 | 0.027 | −0.033 | 0.651 | Geo_distance | 0.429 | 0.105 | 0.575 | 0.032 | −0.096 | 0.836 |
| SiO44- | 0.483 | 0.036 | 0.496 | 0.017 | −0.021 | 0.592 | Geo_distance | 0.441 | 0.052 | 0.458 | 0.035 | −0.069 | 0.743 |
| Chl a | 0.195 | 0.230 | 0.321 | 0.115 | 0.137 | 0.183 | Geo_distance | −0.002 | 0.473 | 0.155 | 0.255 | 0.060 | 0.349 |
| Prochlorococcus, abundance | 0.122 | 0.203 | 0.069 | 0.323 | 0.140 | 0.190 | Geo_distance | 0.095 | 0.234 | 0.031 | 0.388 | 0.126 | 0.191 |
| Synechococcus, abundance | −0.052 | 0.548 | 0.097 | 0.314 | 0.040 | 0.338 | Geo_distance | −0.141 | 0.688 | 0.022 | 0.429 | 0.008 | 0.458 |
| PPEs, abundance | 0.31 | 0.129 | 0.449 | 0.046 | −0.074 | 0.782 | Geo_distance | 0.209 | 0.233 | 0.368 | 0.089 | −0.143 | 0.931 |
| HP, abundance | 0.107 | 0.294 | 0.192 | 0.220 | 0.016 | 0.439 | Geo_distance | 0.078 | 0.329 | 0.176 | 0.225 | −0.001 | 0.506 |
| VLPs, abundance | −0.167 | 0.778 | −0.097 | 0.621 | −0.017 | 0.515 | Geo_distance | −0.166 | 0.735 | −0.088 | 0.604 | −0.004 | 0.517 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Hu, C.; Wang, J.; Sun, J.; Wang, Y.; Jiao, N.; Xu, D. Biogeographical Distribution and Community Assembly of Active Protistan Assemblages along an Estuary to a Basin Transect of the Northern South China Sea. Microorganisms 2021, 9, 351. https://doi.org/10.3390/microorganisms9020351
Li R, Hu C, Wang J, Sun J, Wang Y, Jiao N, Xu D. Biogeographical Distribution and Community Assembly of Active Protistan Assemblages along an Estuary to a Basin Transect of the Northern South China Sea. Microorganisms. 2021; 9(2):351. https://doi.org/10.3390/microorganisms9020351
Chicago/Turabian StyleLi, Ran, Chen Hu, Jianning Wang, Jun Sun, Ying Wang, Nianzhi Jiao, and Dapeng Xu. 2021. "Biogeographical Distribution and Community Assembly of Active Protistan Assemblages along an Estuary to a Basin Transect of the Northern South China Sea" Microorganisms 9, no. 2: 351. https://doi.org/10.3390/microorganisms9020351
APA StyleLi, R., Hu, C., Wang, J., Sun, J., Wang, Y., Jiao, N., & Xu, D. (2021). Biogeographical Distribution and Community Assembly of Active Protistan Assemblages along an Estuary to a Basin Transect of the Northern South China Sea. Microorganisms, 9(2), 351. https://doi.org/10.3390/microorganisms9020351








