Identification of Novel phoP-phoQ Regulated Genes that Contribute to Polymyxin B Tolerance in Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals, Bacterial Strains, and Plasmids
2.2. RNA Extraction, Reverse Transcription, and Quantitative Real-Time PCR
2.3. Construction of lacZ Transcriptional Fusions and β-Galactosidase Activity Measurement
2.4. ChIP-Seq and Data Analysis
2.5. Expression and Purification of the 6×His-Tagged PhoP Protein
2.6. EMSA
2.7. Minimum Inhibitory Concentration (MIC) Measurement
2.8. Bacterial Survival Assay
2.9. SDS Susceptibility Assay
2.10. Ethidium Bromide Influx Assay
2.11. Dansyl-Polymyxin B Binding Assay
2.12. Outer Membrane Permeability Assay
2.13. Inner Membrane Integrity Assay
2.14. Statistical Analysis
3. Results
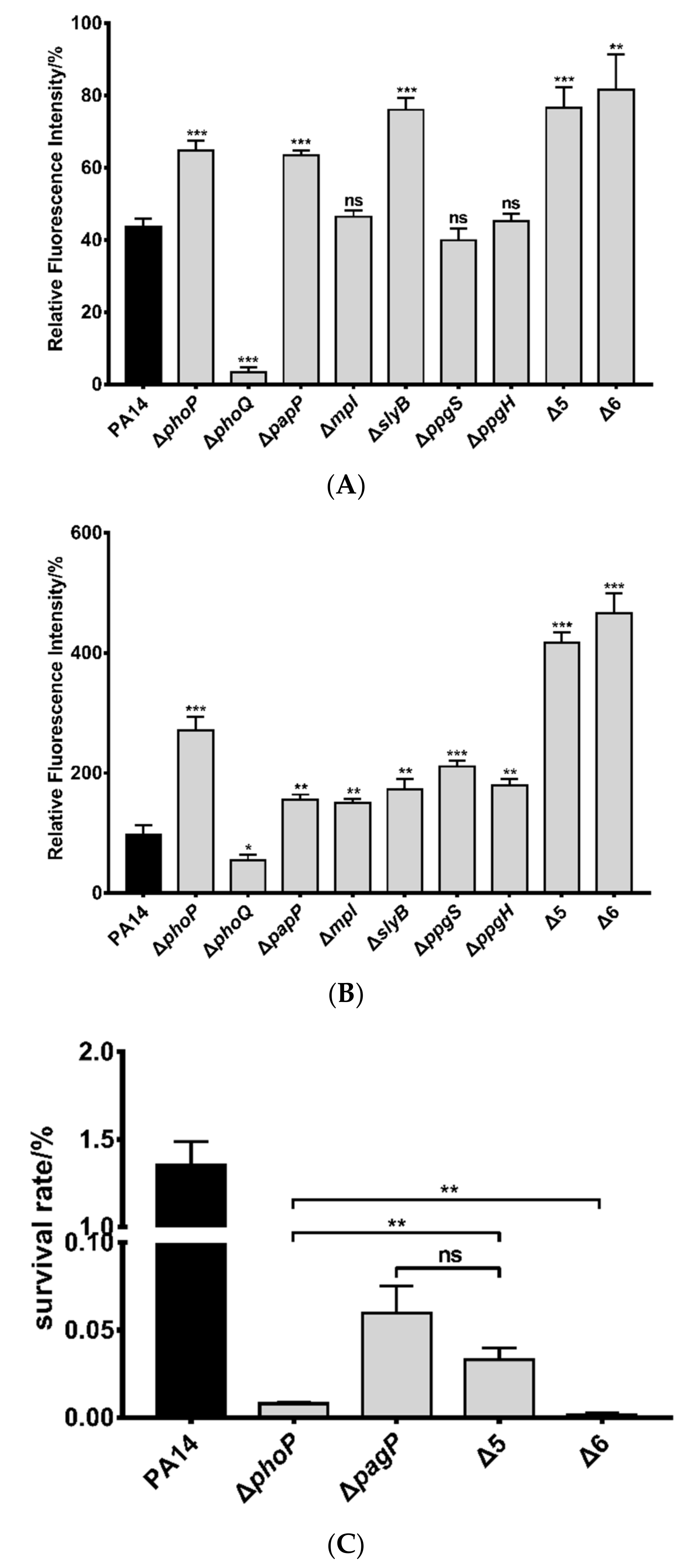
3.1. Identification of Genes Directly Regulated by PhoP in P. aeruginosa
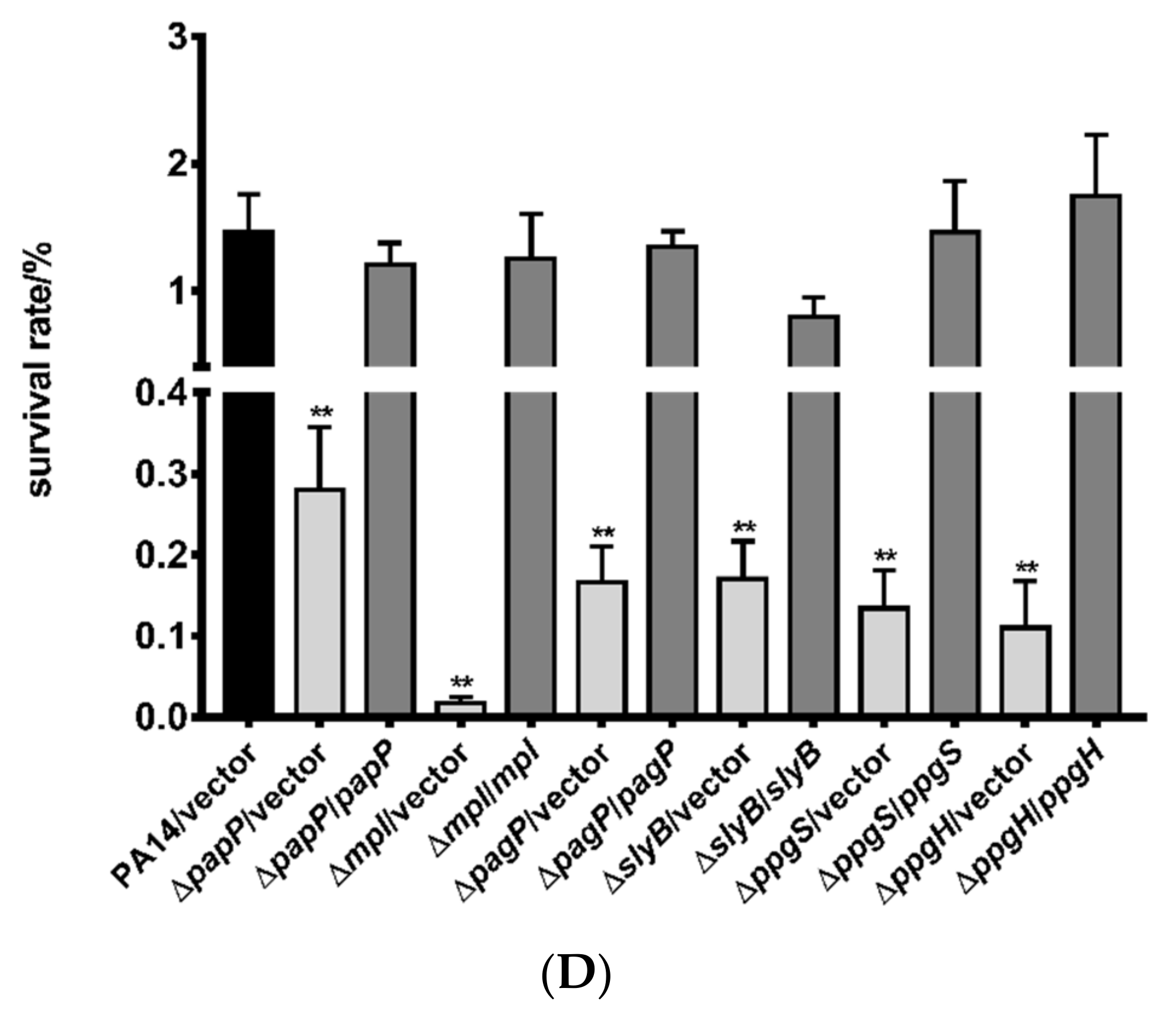
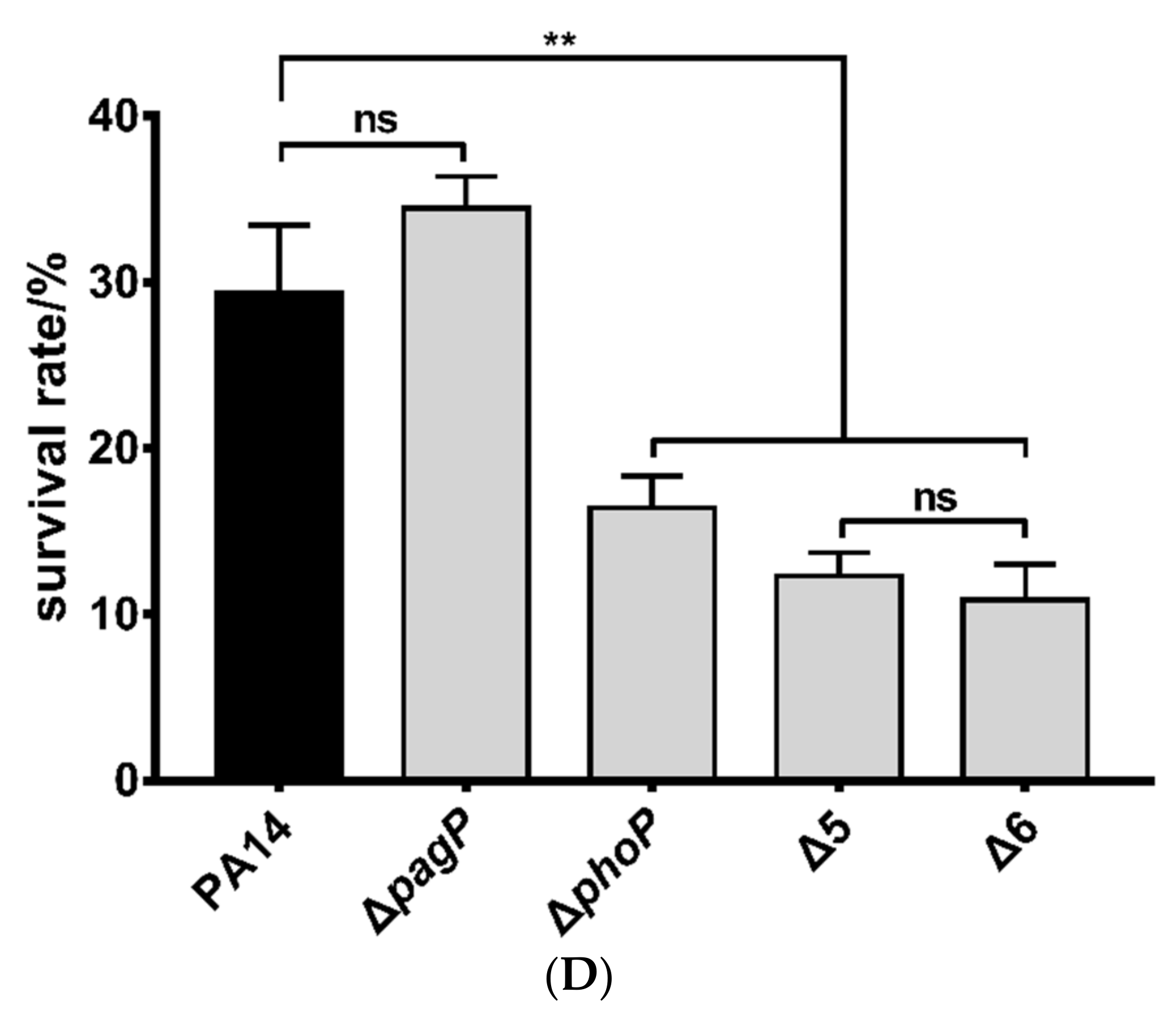
3.2. Roles of PhoP-PhoQ Regulated Genes in the Bacterial Tolerance to Polymyxin B
3.3. Mechanisms of Polymyxin B Tolerance Mediated by the Identified Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zanin, M.; Baviskar, P.; Webster, R.; Webby, R. The interaction between respiratory pathogens and mucus. Cell Host Microbe 2016, 19, 159–168. [Google Scholar] [CrossRef]
- Anantharajah, A.; Mingeot-Leclercq, M.P.; Bambeke, V.F. Targeting the type three secretion system in Pseudomonas aeruginosa. Trends Pharmacol. Sci. 2016, 37, 734–749. [Google Scholar] [CrossRef] [PubMed]
- Lister, P.D.; Wolter, D.J.; Hanson, N.D. Antibacterial-resistant Pseudomonas aeruginosa: Clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 2009, 22, 582–610. [Google Scholar] [CrossRef]
- Zhao, S.; Adamiak, J.W.; Bonifay, V.; Mehla, J.; Zgurskaya, H.I.; Tan, D.S. Defining new chemical space for drug penetration into Gram-negative bacteria. Nat. Chem. Biol. 2020, 12, 1293–1302. [Google Scholar] [CrossRef] [PubMed]
- Hulen, C.; Racine, P.J.; Chevalier, S.; Feuilloley, M.; Lomri, N.E. Identification of the PA1113 gene product as an ABC transporter involved in the uptake of carbenicillin in Pseudomonas aeruginosa PAO1. Antibiotics 2020, 9, 596. [Google Scholar] [CrossRef]
- Azimi, L.; Fallah, F.; Karimi, A.; Shirdoust, M.; Azimi, T.; Sedighi, I.; Rahbar, M.; Armin, S. Survey of various carbapenem-resistant mechanisms of Acinetobacter baumannii and Pseudomonas aeruginosa isolated from clinical samples in Iran. Iran. J. Basic Med. Sci. 2020, 23, 1396–1400. [Google Scholar] [CrossRef] [PubMed]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef]
- Puja, H.; Bolard, A.; Noguès, A.; Plésiat, P.; Jeannot, K. The efflux pump MexXY/OprM contributes to the tolerance and acquired resistance of Pseudomonas aeruginosa to colistin. Antimicrob. Agents Chemother. 2020, 64, e02033-19. [Google Scholar] [CrossRef]
- Hao, G.; Chen, A.I.; Liu, M.; Zhou, H.; Egan, M.; Yang, X.; Kan, B.; Wang, H.; Goulian, M.; Zhu, J. Colistin-resistance-mediated bacterial surface modification sensitizes phage infection. Antimicrob. Agents Chemother. 2019, 63, e01609-19. [Google Scholar] [CrossRef]
- Genteluci, G.L.; de Souza, P.A.; Gomes, D.B.C.; Sousa, V.S.; de Souza, M.J.; Abib, J.R.L.; de Castro, E.A.R.; Rangel, K.; Bôas, M.H.S.V. Polymyxin B heteroresistance and adaptive resistance in multidrug- and extremely drug-resistant Acinetobacter baumannii. Curr. Microbiol. 2020, 77, 2300–2306. [Google Scholar] [CrossRef]
- Soon, R.L.; Velkov, T.; Chiu, F.; Thompson, P.E.; Kancharla, R.; Roberts, K.; Larson, I.; Nation, R.L.; Li, J. Design, synthesis, and evaluation of a new fluorescent probe for measuring polymyxin-lipopolysaccharide binding interactions. Anal. Biochem. 2011, 409, 273–283. [Google Scholar] [CrossRef] [PubMed]
- Domingues, M.M.; Inácio, R.G.; Raimundo, J.M.; Martins, M.; Castanho, M.A.R.B.; Santos, N.C. Biophysical characterization of polymyxin B interaction with LPS aggregates and membrane model systems. Biopolymers 2012, 98, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Shao, X.; Xiao, X.; Dai, Y.; Wang, Y.; Xie, J.; Jiang, W.; Sun, Y.; Cong, Z.; Qiao, Z.; et al. Host defense peptide mimicking peptide polymer exerting fast, broad spectrum, and potent activities toward clinically isolated multidrug resistant bacteria. ACS Infect. Dis. 2020, 6, 479–488. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Deris, Z.Z.; Huang, J.X.; Azad, M.A.K.; Butler, M.; Sivanesan, S.; Kaminskas, L.M.; Dong, Y.; Boyd, B.; Baker, M.A.; et al. Surface changes and polymyxin interactions with a resistant strain of Klebsiella pneumoniae. Innate Immun. 2014, 20, 350–363. [Google Scholar] [CrossRef] [PubMed]
- Velkov, T.; Soon, R.L.; Chong, P.L.; Huang, J.X.; Cooper, M.A.; Azad, M.A.K.; Baker, M.A.; Thompson, P.E.; Roberts, K.; Nation, R.L.; et al. Molecular basis for the increased polymyxin susceptibility of Klebsiella pneumoniae strains with under-acylated lipid A. Innate Immun. 2013, 19, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Krzyzanski, W.; Rao, G.G. Multi-scale model of drug induced adaptive resistance of Gram-negative bacteria to polymyxin B. PLoS ONE 2017, 12, e0171834. [Google Scholar] [CrossRef]
- Trimble, M.J.; Mlynárčik, P.; Kolář, M.; Hancock, R.E.W. Polymyxin: Alternative mechanisms of action and resistance. Cold Spring Harb. Perspect. Med. 2016, 6, a025288. [Google Scholar] [CrossRef]
- Macfarlane, E.L.A.; Kwasnicka, A.; Hancock, R.E.W. Role of Pseudomonas aeruginosa PhoP-phoQ in resistance to antimicrobial cationic peptides and aminoglycosides. Microbiology 2000, 146, 2543–2554. [Google Scholar] [CrossRef]
- Macfarlane, E.L.A.; Kwasnicka, A.; Ochs, M.M.; Hancock, R.E.W. PhoP ± PhoQ homologues in Pseudomonas aeruginosa regulate expression of the outer membrane protein OprH and polymyxin B resistance. Mol. Microbiol. 1999, 34, 305–316. [Google Scholar] [CrossRef]
- Lesley, J.A.; Waldburger, C.D. Comparison of the Pseudomonas aeruginosa and Escherichia coli PhoQ sensor domains: Evidence for distinct mechanisms of signal detection. J. Biol. Chem. 2001, 276, 30827–30833. [Google Scholar] [CrossRef] [PubMed]
- Brinkman, F.S.; Macfarlane, E.L.; Warrener, P.; Hancock, R.E.W. Evolutionary relationships among virulence-associated histidine kinases. Infect. Immun. 2001, 69, 5207–5211. [Google Scholar] [CrossRef]
- Bell, A.; Bains, M.; Hancock, R.E.W. Pseudomonas aeruginosa outer membrane protein OprH: Expression from the cloned gene and function in EDTA and gentamicin resistance. J. Bacteriol. 1991, 173, 6657–6664. [Google Scholar] [CrossRef] [PubMed]
- Conrad, R.S.; Galanos, C. Fatty acid alterations and polymyxin B binding by lipopolysaccharides from Pseudomonas aeruginosa adapted to polymyxin B resistance. Antimicrob. Agents Chemother. 1989, 33, 1724–1728. [Google Scholar] [CrossRef] [PubMed]
- Guo, L.; Lim, K.B.; Gunn, J.S.; Bainbridge, B.; Darveau, R.P.; Hackett, M.; Miller, S.I. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP–phoQ. Science 1997, 276, 250–253. [Google Scholar] [CrossRef] [PubMed]
- Landman, D.; Bratu, S.; Alam, M.; Quale, J. Citywide emergence of Pseudomonas aeruginosa strains with reduced susceptibility to polymyxin B. J. Antimicrob. Chemother. 2005, 55, 954–957. [Google Scholar] [CrossRef]
- Lee, M.; Sousa, M.C. Structural basis for substrate specificity in ArnB. A key enzyme in the polymyxin resistance pathway of Gram-negative bacteria. Biochemistry 2014, 53, 796–805. [Google Scholar] [CrossRef]
- Miller, A.K.; Brannon, M.K.; Stevens, L.; Johansen, H.K.; Selgrade, S.E.; Miller, S.I.; Høiby, N.; Moskowitz, S.M. PhoQ mutations promote lipid A modification and polymyxin resistance of Pseudomonas aeruginosa found in colistin-treated cystic fibrosis patients. Antimicrob. Agents Chemother. 2011, 55, 5761–5769. [Google Scholar] [CrossRef] [PubMed]
- Schurek, K.N.; Sampaio, J.L.M.; Kiffer, C.R.V.; Sinto, S.; Mendes, C.M.F.; Hancock, R.E.W. Involvement of pmrAB and phoPQ in polymyxin B adaptation and inducible resistance in non-cystic fibrosis clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 4345–4351. [Google Scholar] [CrossRef]
- Lee, J.Y.; Chung, E.S.; Na, I.N.; Kim, H.; Shin, D.; Ko, K.S. Development of colistin resistance in pmrA-, phoP-, parR- and cprR-inactivated mutants of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2014, 69, 2966–2971. [Google Scholar] [CrossRef]
- Kreamer, N.N.K.; Wilks, J.C.; Marlow, J.J.; Coleman, M.L.; Newman, D.K. BqsR/BqsS constitute a two-component system that senses extracellular Fe(II) in Pseudomonas aeruginosa. J. Bacteriol. 2012, 194, 1195–1204. [Google Scholar] [CrossRef]
- Thaipisuttikul, I.; Hittle, L.E.; Chandra, R.; Zangari, D.; Dixon, C.L.; Garrett, T.A.; Rasko, D.A.; Dasgupta, N.; Moskowitz, S.M.; Malmström, L.; et al. A divergent Pseudomonas aeruginosa palmitoyltransferase essential for cystic fibrosis-specific lipid A. Mol. Microbiol. 2014, 91, 158–174. [Google Scholar] [CrossRef] [PubMed]
- McPhee, J.B.; Bains, M.; Winsor, G.; Lewenza, S.; Kwasnicka, A.; Brazas, M.D.; Brinkman, F.S.L.; Hancock, R.E.W. Contribution of the PhoP-PhoQ and PmrA-PmrB two-component regulatory systems to Mg2+-induced gene regulation in Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 3995–4006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Dhillon, P.; Yan, H.; Farmer, S.; Hancock, R.E.W. Interactions of bacterial cationic peptide antibiotics with outer and cytoplasmic membranes of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2000, 44, 3317–3321. [Google Scholar] [CrossRef] [PubMed]
- Gooderham, W.J.; Gellatly, S.L.; Sanschagrin, F.; McPhee, J.B.; Bains, M.; Cosseau, C.; Levesque, R.C.; Hancock, R.E.W. The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 2009, 155, 699–711. [Google Scholar] [CrossRef]
- Wu, W.; Jin, S. PtrB of Pseudomonas aeruginosa suppresses the type III secretion system under the stress of DNA damage. J. Bacteriol. 2005, 187, 6058–6068. [Google Scholar] [CrossRef]
- Xia, Y.; Wang, D.; Pan, X.; Xia, B.; Weng, Y.; Long, Y.; Ren, H.; Zhou, J.; Jin, Y.; Bai, F.; et al. TpiA is a key metabolic enzyme that affects virulence and resistance to aminoglycoside antibiotics through CrcZ in Pseudomonas aeruginosa. mBio 2020, 11, e02079-19. [Google Scholar] [CrossRef]
- Deng, X.; Li, M.; Pan, X.; Zheng, R.; Liu, C.; Chen, F.; Liu, X.; Cheng, Z.; Jin, S.; Wu, W. Fis regulates type III secretion system by influencing the transcription of exsA in Pseudomonas aeruginosa strain PA14. Front. Microbiol. 2017, 8, 669. [Google Scholar] [CrossRef]
- Kawasaki, K.; China, K.; Nishijima, M. Release of the lipopolysaccharide deacylase PagL from latency compensates for a lack of lipopolysaccharide aminoarabinose modification-dependent resistance to the antimicrobial peptide polymyxin B in Salmonella enterica. J. Bacteriol. 2007, 189, 4911–4919. [Google Scholar] [CrossRef]
- Klebensberger, J.; Rui, O.; Fritz, E.; Schink, B.; Philipp, B. Cell aggregation of Pseudomonas aeruginosa strain PAO1 as an energy-dependent stress response during growth with sodium dodecyl sulfate. Arch. Microbiol. 2006, 185, 417–427. [Google Scholar] [CrossRef]
- Akhoundsadegh, N.; Belanger, C.R.; Hancock, R.E.W. Outer membrane interaction kinetics of new polymyxin B analogs in gram-negative Bacilli. Antimicrob. Agents Chemother. 2019, 63, e00935-19. [Google Scholar] [CrossRef]
- Ma, B.; Fang, C.; Lu, L.; Wang, M.; Xue, X.; Zhou, Y.; Li, M.; Hu, Y.; Luo, X.; Hou, Z. The antimicrobial peptide thanatin disrupts the bacterial outer membrane and inactivates the NDM-1 metallo-β-lactamase. Nat. Commun. 2019, 10, 3517. [Google Scholar] [CrossRef] [PubMed]
- Loh, B.; Grant, C.; Hancock, R.E.W. Use of the fluorescent probe 1-N-phenylnaphthylamine to study the interactions of aminoglycoside antibiotics with the outer membrane of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1984, 26, 546–551. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Liu, Y.; Huang, X.; Ding, S.; Wang, Y.; Shen, J.; Zhu, K. A broad spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat. Microbiol. 2020, 5, 1040–1050. [Google Scholar] [CrossRef] [PubMed]
- Yarlagadda, V.; Akkapeddi, P.; Manjunath, G.B.; Haldar, J. Membrane active vancomycin analogues: A strategy to combat bacterial resistance. J. Med. Chem. 2014, 57, 4558–4568. [Google Scholar] [CrossRef] [PubMed]
- Gilleland, H.E.; Stinnett, J.D.; Eagon, R.G. Ultrastructural and chemical alteration of the cell envelope of Pseudomonas aeruginosa, associated with resistance to ethylenediaminetetraacetate resulting from growth in a Mg2+-deficient medium. J. Bacteriol. 1974, 117, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Torrens, G.; Barceló, I.M.; Gallego, M.P.; Salom, M.E.; Gracia, S.T.; Bestard, M.M.; Nicolau, M.D.M.G.; Venegas, Y.J.C.; Rumbos, E.N.R.; Cabot, G.; et al. Publisher Correction: Profiling the susceptibility of Pseudomonas aeruginosa strains from acute and chronic infections to cell-wall-targeting immune proteins. Sci. Rep. 2020, 10, 4356. [Google Scholar] [CrossRef] [PubMed]
- Lejona, S.; Aguirre, A.; Cabeza, M.L.; Véscovi, E.G.; Soncini, F.C. Molecular characterization of the Mg2+-responsive PhoP-PhoQ regulon in Salmonella enterica. J. Bacteriol. 2003, 185, 6287–6294. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, S.; Ogasawara, H.; Kato, A.; Yamamoto, K.; Eguchi, Y.; Oshima, T.; Mori, H.; Ishihama, A.; Utsumi, R. Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 2003, 185, 3696–3702. [Google Scholar] [CrossRef] [PubMed]
- Plesa, M.; Hernalsteens, J.P.; Vandenbussche, G.; Ruysschaert, J.M.; Cornelis, P. The SlyB outer membrane lipoprotein of Burkholderia multivorans contributes to membrane integrity. Res. Microbiol. 2006, 157, 582–592. [Google Scholar] [CrossRef]
- Arendt, W.; Hebecker, S.; Jäger, S.; Nimtz, M.; Moser, J. Resistance phenotypes mediated by aminoacyl-phosphatidylglycerol synthases. J. Bacteriol. 2012, 194, 1401–1416. [Google Scholar] [CrossRef]
- Barrow, K.; Kwon, D.H. Alterations in two-component regulatory systems of phoPQ and pmrAB are associated with polymyxin B resistance in clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 2009, 53, 5150–5154. [Google Scholar] [CrossRef] [PubMed]
- Fernández, L.; Gooderham, W.J.; Bains, M.; McPhee, J.B.; Wiegand, I.; Hancock, R.E.W. Adaptive resistance to the "last hope" antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR–ParS. Antimicrob. Agents Chemother. 2010, 54, 3372–3382. [Google Scholar] [CrossRef]
- Liberati, N.T.; Urbach, J.M.; Miyata, S.; Lee, D.G.; Drenkard, E.; Wu, G.; Villanueva, J.; Wei, T.; Ausubel, F.M. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc. Natl. Acad. Sci. USA 2006, 103, 2833–2838. [Google Scholar] [CrossRef]
- Lee, C.C.; Sun, Y.; Qian, S.; Huang, H.W. Transmembrane pores formed by human antimicrobial peptide LL-37. Biophys. J. 2011, 100, 1688–1696. [Google Scholar] [CrossRef]
- Darveau, R.P. Lipid A diversity and the innate host response to bacterial infection. Curr. Opin. Microbiol. 1998, 1, 36–42. [Google Scholar] [CrossRef]
- Schindler, P.R.; Teuber, M. Action of polymyxin B on bacterial membranes: Morphological changes in the cytoplasm and in the outer membrane of Salmonella typhimurium and Escherichia coli. Antimicrob. Agents Chemother. 1975, 8, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Yu, N.Y.; Wagner, J.R.; Laird, M.R.; Melli, G.; Rey, S.; Lo, R.; Dao, P.; Sahinalp, S.C.; Ester, M.; Foster, L.J.; et al. PSORTb 3.0: Improved protein subcellular localization prediction with refined localization subcategories and predictive capabilities for all prokaryotes. Bioinformatics 2010, 26, 1608–1615. [Google Scholar] [CrossRef]
- Winsor, G.L.; Griffiths, E.J.; Lo, R.; Dhillon, B.K.; Shay, J.A.; Brinkman, F.S.L. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic. Acids Res. 2016, 44, D646–D653. [Google Scholar] [CrossRef]
- Qadi, M.; Rabassa, S.I.; Borrás, M.M.; Sánchez, A.D.; Juan, C.; Goldberg, J.B.; Hancock, R.E.W.; Albertí, S. Sensing Mg2+ contributes to the resistance of Pseudomonas aeruginosa to complement-mediated opsonophagocytosis. Environ. Microbiol. 2017, 19, 4278–4286. [Google Scholar] [CrossRef]
- Ludwig, A.; Tengel, C.; Bauer, S.; Bubert, A.; Benz, R.; Mollenkopf, H.J.; Goebel, W. SlyA, a regulatory protein from Salmonella typhimurium, induces a haemolytic and pore-forming protein in Escherichia coli. Mol. Gen. Genet. 1995, 249, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Firoved, A.M.; Deretic, V. Microarray analysis of global gene expression in mucoid Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Firoved, A.M.; Boucher, J.C.; Deretic, V. Global genomic analysis of AlgU (sigma(E))-dependent promoters (sigmulon) in Pseudomonas aeruginosa and implications for inflammatory processes in cystic fibrosis. J. Bacteriol. 2002, 184, 1057–1064. [Google Scholar] [CrossRef] [PubMed]
- Klein, S.; Lorenzo, C.; Hoffmann, S.; Walther, G.M.; Storbeck, S.; Piekarski, T.; Tindall, B.J.; Wray, V.; Nimtz, M.; Moser, J. Adaptation of Pseudomonas aeruginosa to various conditions includes tRNA-dependent formation of alanyl-phosphatidylglycerol. Mol. Microbiol. 2009, 71, 551–565. [Google Scholar] [CrossRef] [PubMed]
- Fields, R.N.; Roy, H. Deciphering the tRNA-dependent lipid aminoacylation systems in bacteria: Novel components and structural advances. RNA Biol. 2018, 15, 480–491. [Google Scholar] [CrossRef] [PubMed]
- Arendt, W.; Groenewold, M.K.; Hebecker, S.; Dickschat, J.S.; Moser, J. Identification and characterization of a periplasmic aminoacyl-phosphatidylglycerol hydrolase responsible for Pseudomonas aeruginosa lipid homeostasis. J. Biol. Chem. 2013, 288, 24717–24730. [Google Scholar] [CrossRef] [PubMed]

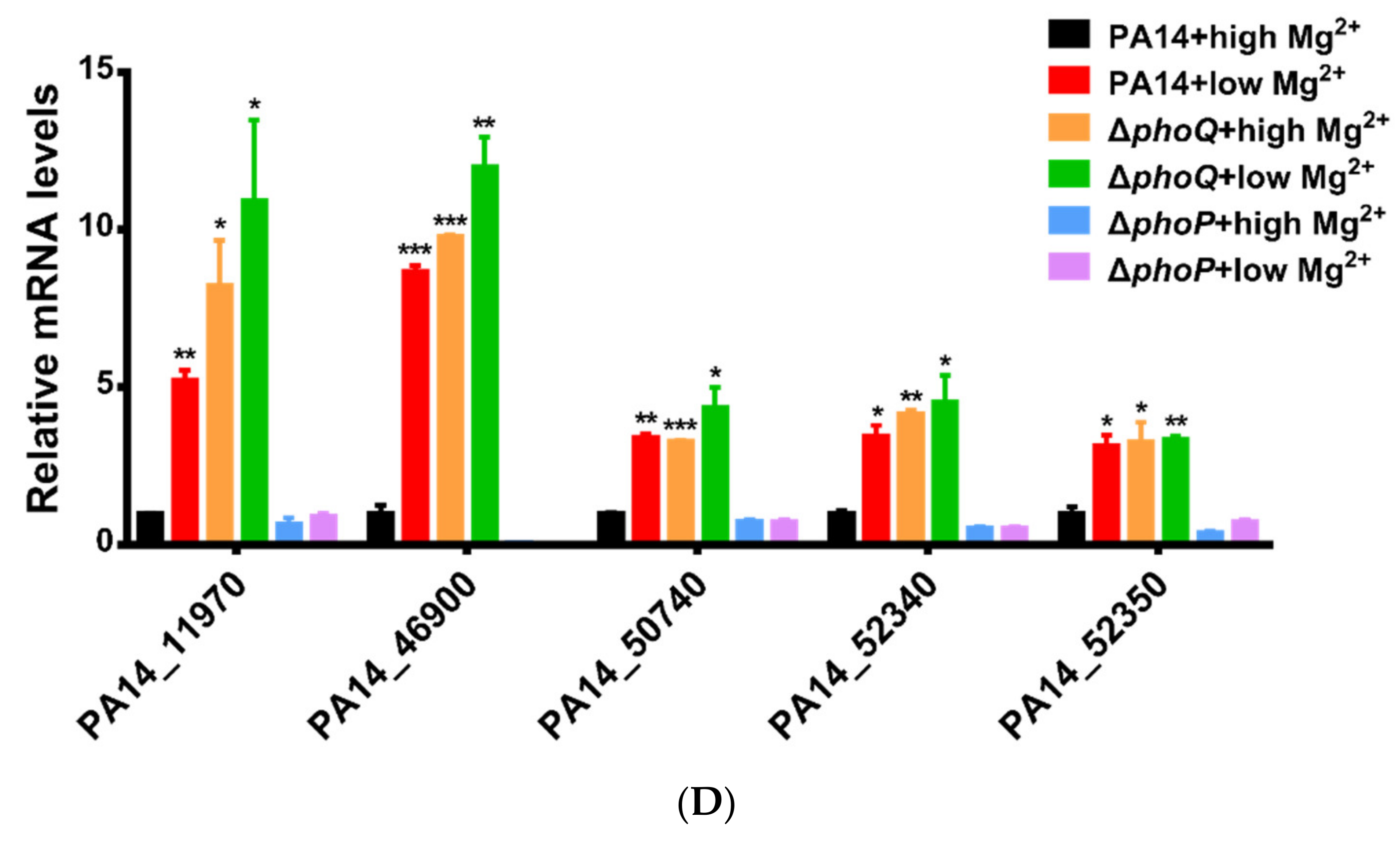
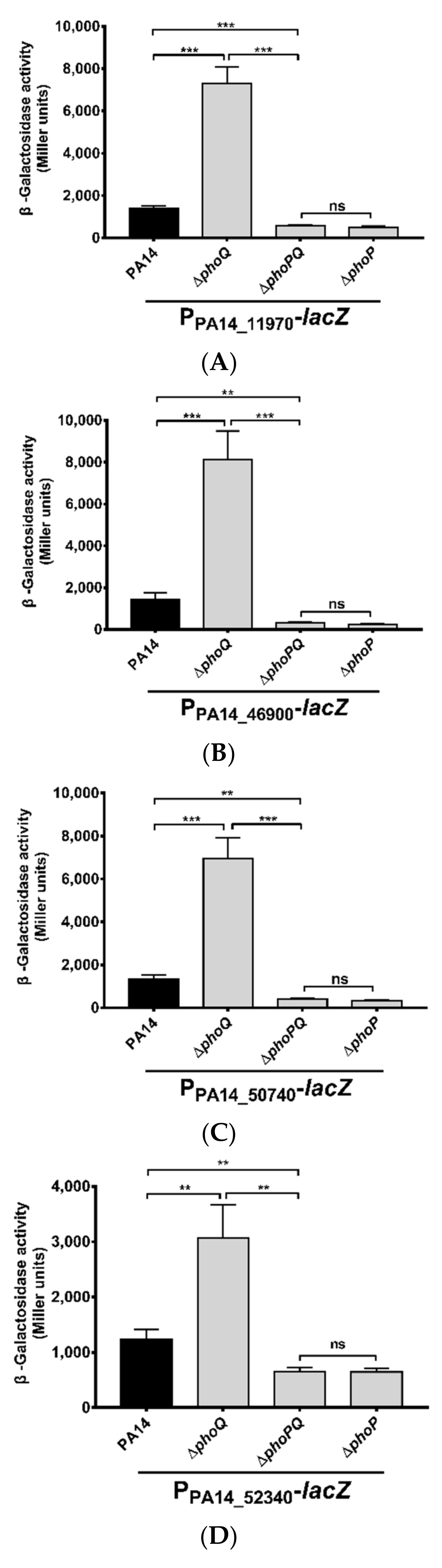
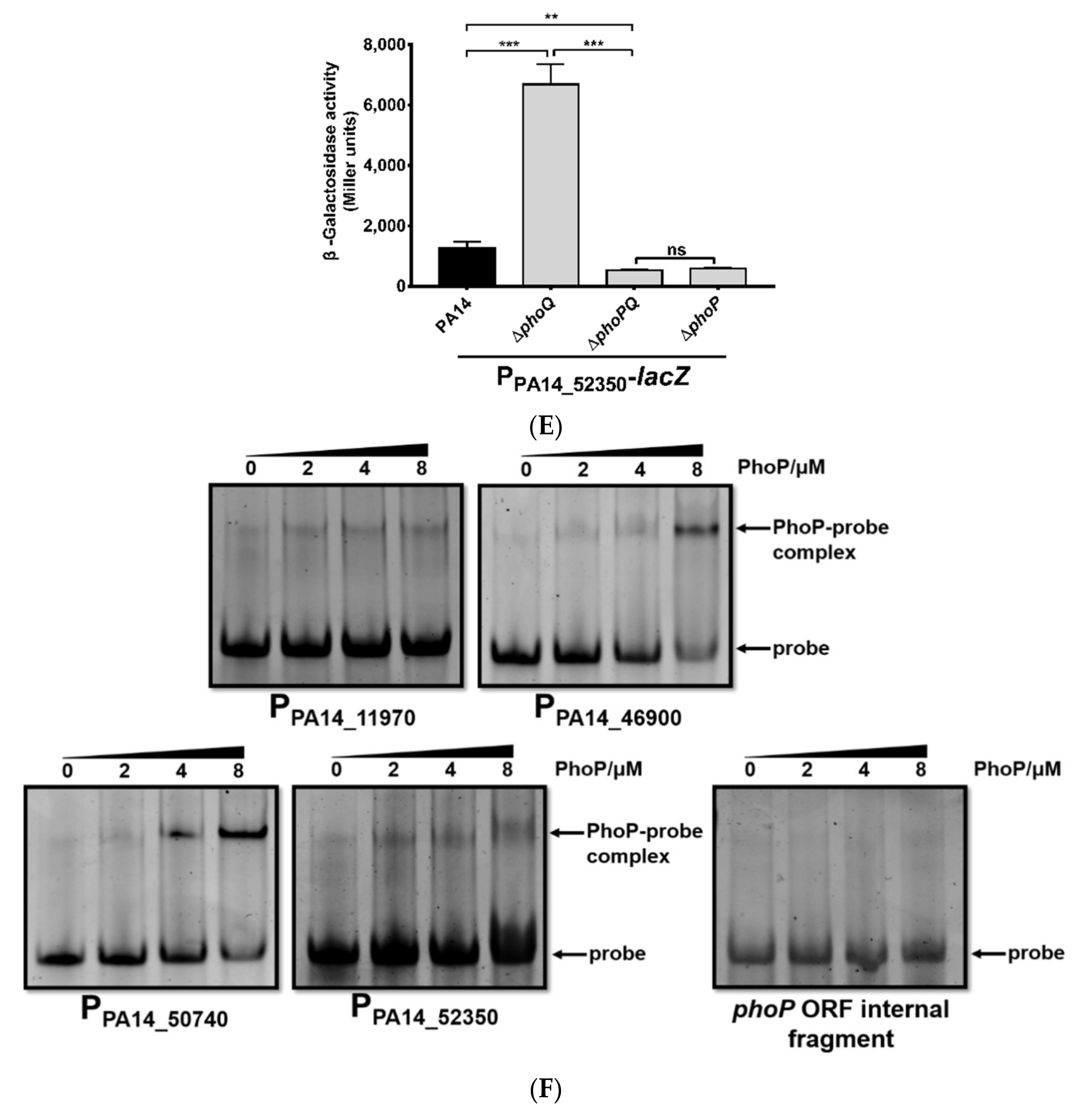



| Genes | Summits in PA14 Chromosome | Fold Enrichment |
|---|---|---|
| oprH | 4372078 | 17.23 |
| PA14_46900, PA14_46910 | 4177576 | 15.03 |
| arnB | 1578361 | 6.85 |
| PA14_50740, PA14_50750 | 4508762 | 6.24 |
| PA14_21860, PA14_21870 | 1900312 | 5.29 |
| PA14_52340, PA14_52350 | 4644776 | 4.87 |
| PA14_11970, PA14_11980 | 1035748 | 4.17 |
| pilY1 | 5372861 | 2.85 |
| PA14 Locus Tag | PAO1 Locus Tag | Functional Description | Product Name | Preliminary Phenotypic Analysis |
|---|---|---|---|---|
| PA14_11960 | PA4011 | PAP2 superfamily protein/DedA family protein | papP * (phosphatidic acid phosphatase in P.a) | |
| PA14_11970 | PA4010 | 3-methyladenine DNA glycosylase | mpl * | |
| PA14_46900 | PA1343 | palmitoyltransferase | pagP | |
| PA14_50740 | PA1053 | outer membrane lipoprotein | slyB |
|
| PA14_52340 | PA0921 | hypothetical protein | / | Directly regulated by PhoP-PhoQ [32] |
| PA14_52350 | PA0920 | alanyl-phosphatidylglycerol synthase | ppgS * (phosphatidylglycerol synthases) | Involved in polymyxin E MIC and might altered bacterial membrane potential (a conjecture) [50] |
| PA14_52370 | PA0919 | alanyl-phosphatidylglycerol hydrolase | ppgH * (phosphatidylglycerol hydrolase) |
| Strain | MIC (μg/mL) |
|---|---|
| PA14 | 0.3125 |
| ΔphoP | 0.1563 |
| ΔpapP::Tn | 0.3125 |
| Δmpl::Tn | 0.3125 |
| ΔPA14_11980::Tn | 0.3125 |
| ΔpagP::Tn | 0.3125 |
| ΔslyB::Tn | 0.3125 |
| ΔPA14_52340::Tn | 0.3125 |
| ΔppgS::Tn | 0.3125 |
| ΔppgH::Tn | 0.3125 |
| ΔpapP | 0.3125 |
| Δmpl | 0.3125 |
| ΔpagP | 0.3125 |
| ΔslyB | 0.3125 |
| ΔppgS | 0.3125 |
| ΔppgH | 0.3125 |
| Δ5 a | 0.3125 |
| Δ6 b | 0.1563 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, B.; Liu, C.; Pan, X.; Fu, W.; Fan, Z.; Jin, Y.; Bai, F.; Cheng, Z.; Wu, W. Identification of Novel phoP-phoQ Regulated Genes that Contribute to Polymyxin B Tolerance in Pseudomonas aeruginosa. Microorganisms 2021, 9, 344. https://doi.org/10.3390/microorganisms9020344
Yang B, Liu C, Pan X, Fu W, Fan Z, Jin Y, Bai F, Cheng Z, Wu W. Identification of Novel phoP-phoQ Regulated Genes that Contribute to Polymyxin B Tolerance in Pseudomonas aeruginosa. Microorganisms. 2021; 9(2):344. https://doi.org/10.3390/microorganisms9020344
Chicago/Turabian StyleYang, Baopeng, Chang Liu, Xiaolei Pan, Weixin Fu, Zheng Fan, Yongxin Jin, Fang Bai, Zhihui Cheng, and Weihui Wu. 2021. "Identification of Novel phoP-phoQ Regulated Genes that Contribute to Polymyxin B Tolerance in Pseudomonas aeruginosa" Microorganisms 9, no. 2: 344. https://doi.org/10.3390/microorganisms9020344
APA StyleYang, B., Liu, C., Pan, X., Fu, W., Fan, Z., Jin, Y., Bai, F., Cheng, Z., & Wu, W. (2021). Identification of Novel phoP-phoQ Regulated Genes that Contribute to Polymyxin B Tolerance in Pseudomonas aeruginosa. Microorganisms, 9(2), 344. https://doi.org/10.3390/microorganisms9020344







