Dynamics of the Apple Fruit Microbiome after Harvest and Implications for Fruit Quality
Abstract
1. Introduction
2. Materials and Methods
2.1. Fruit Growing and Storage Conditions
2.2. Microbiological Sampling and Culturing Conditions
2.3. Metabarcoding
2.4. Quantitative PCR
2.5. Data Analyses
3. Results
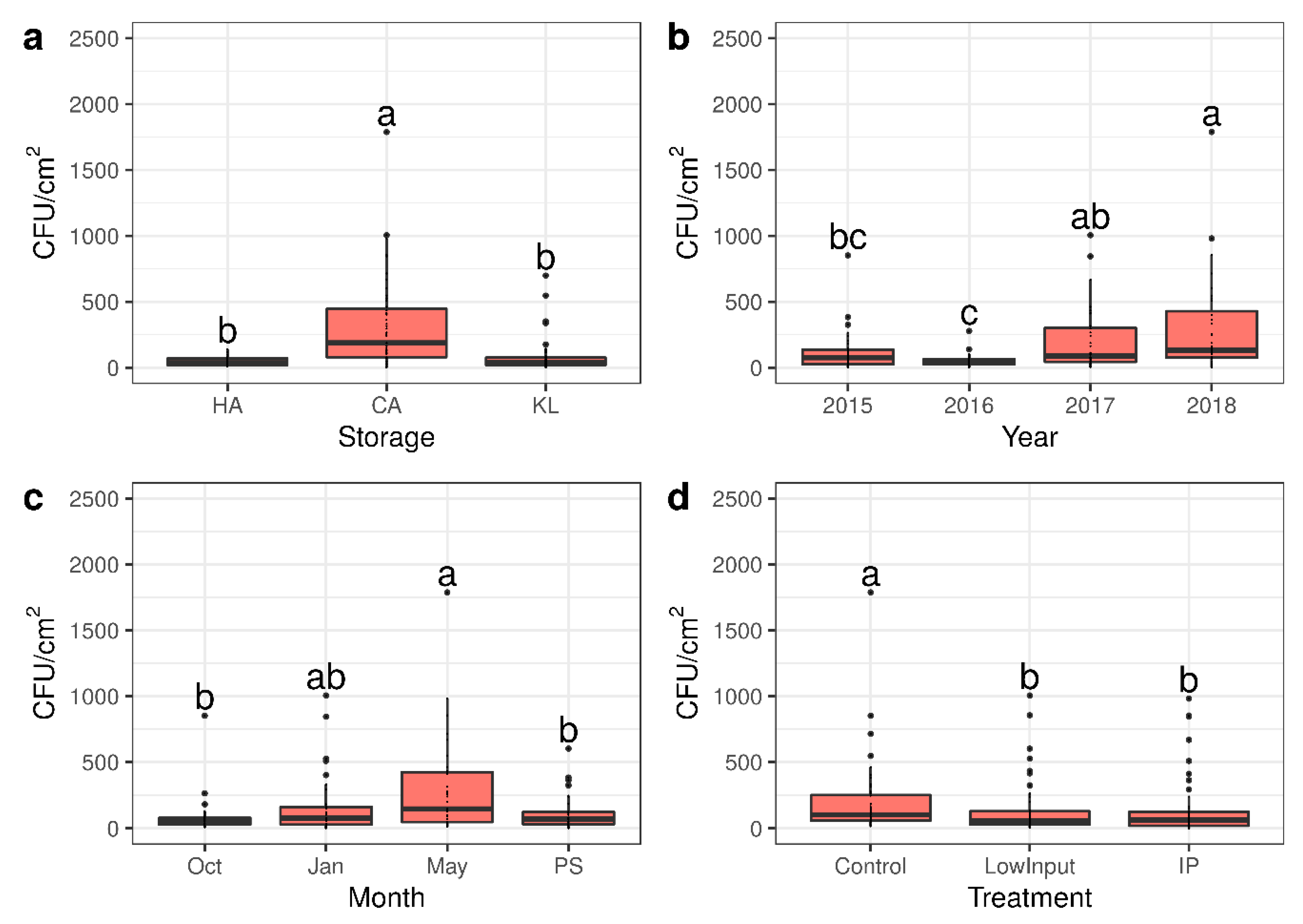
3.1. Characterisation of Mesophilic Aeorbic Microbiome on Apple Skins
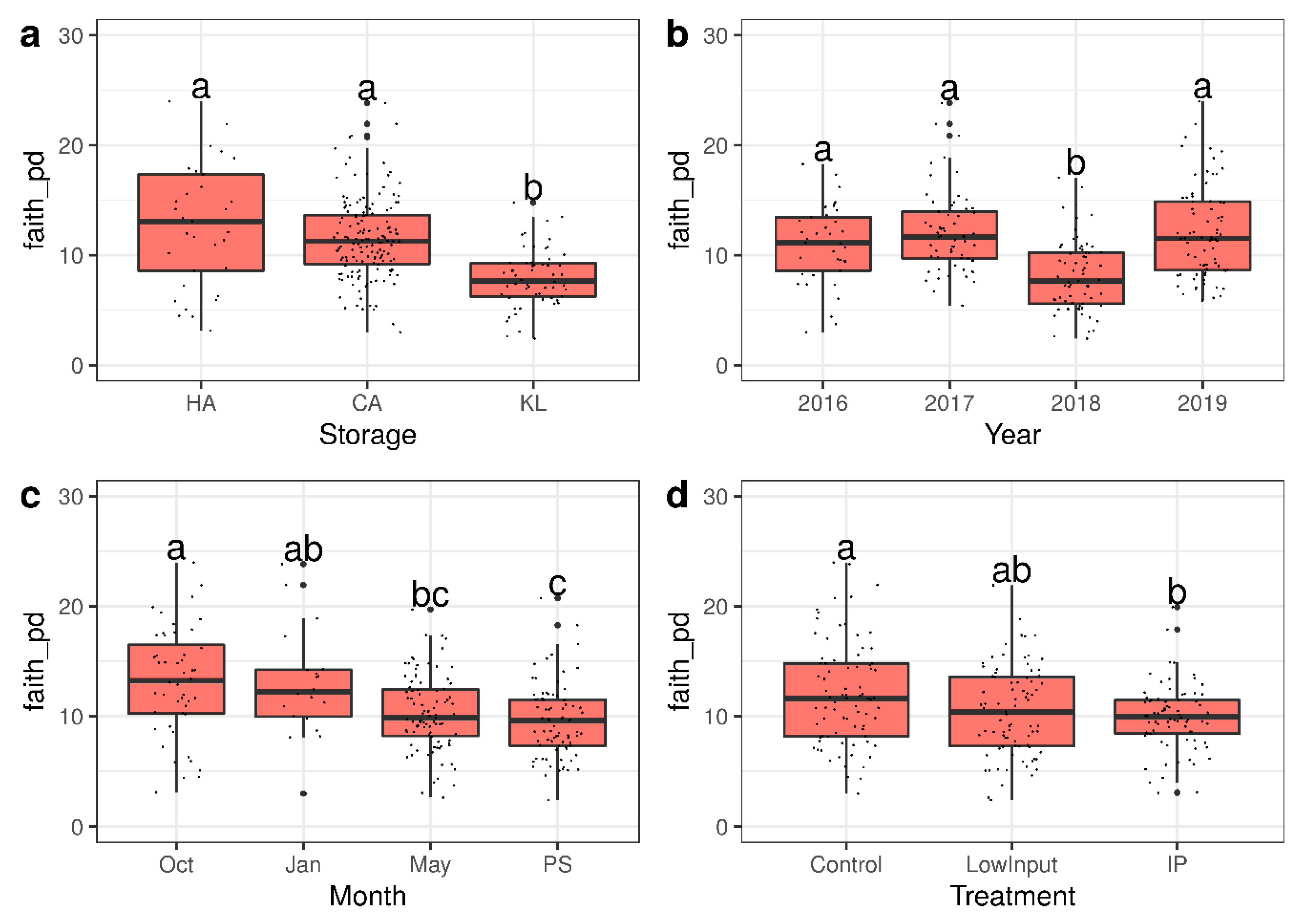
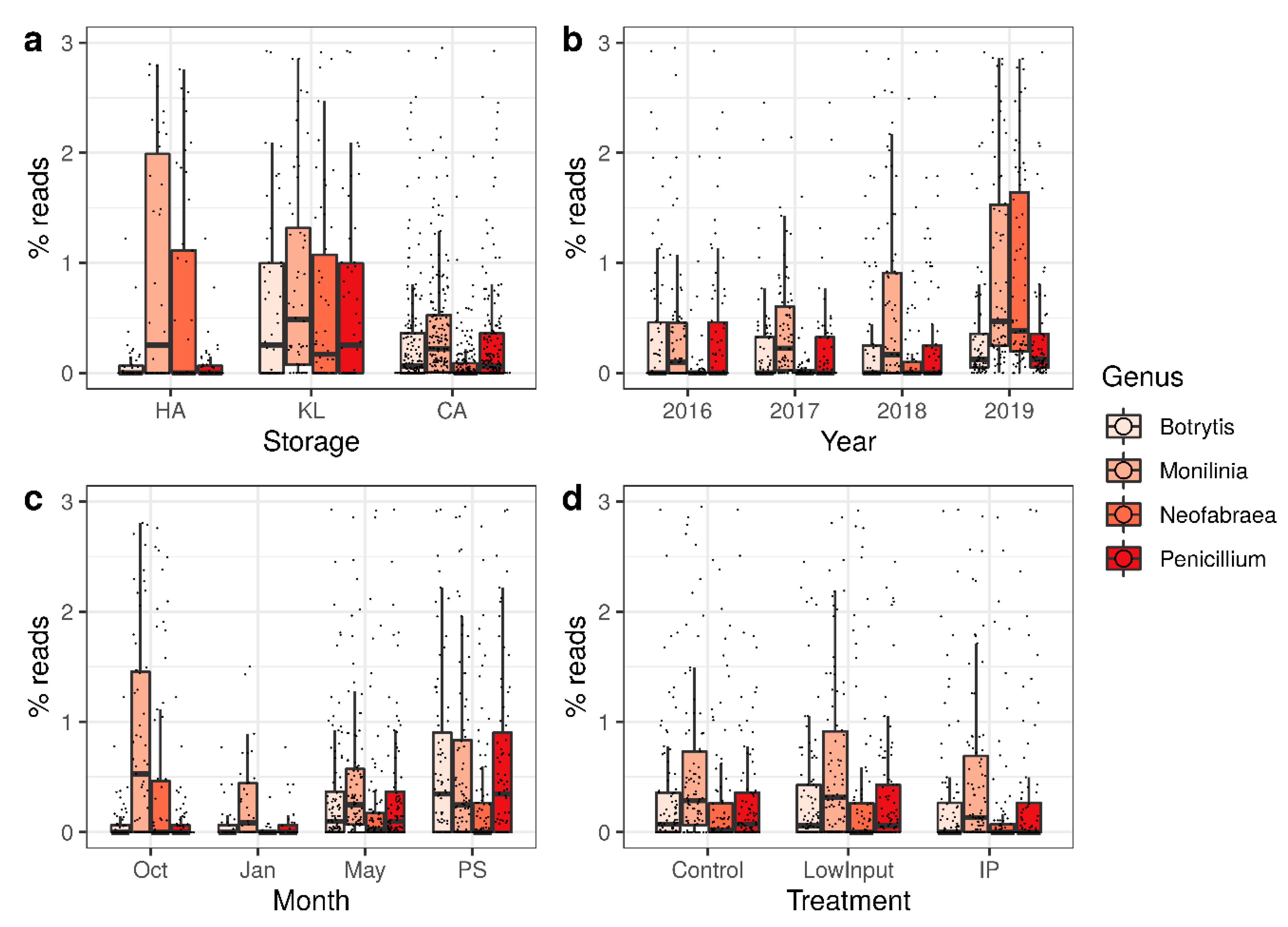
3.2. Characterization of the Total Microbiome on Apple Skins
3.3. qPCR of Neofabraea Alba DNA
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vorholt, J.A. Microbial life in the phyllosphere. Nat. Rev. Micro 2012, 10, 828–840. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Rott, M.; Schlaeppi, K.; Ver Loren van Themaat, E.; Ahmadinejad, N.; Assenza, F.; Rauf, P.; Huettel, B.; Reinhardt, R.; Schmelzer, E.; et al. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 2012, 488, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Rio, T.G.D.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [PubMed]
- Ritpitakphong, U.; Falquet, L.; Vimoltust, A.; Berger, A.; Métraux, J.-P.; L’Haridon, F. The microbiome of the leaf surface of Arabidopsis protects against a fungal pathogen. New Phytol. 2016, 210, 1033–1043. [Google Scholar] [CrossRef]
- Diskin, S.; Feygenberg, O.; Maurer, D.; Droby, S.; Prusky, D.; Alkan, N. Microbiome alterations are correlated with occurrence of postharvest stem-end rot in mango fruit. Phytobiom. J. 2017, 1, 117–127. [Google Scholar] [CrossRef]
- Kaushal, M.; Swennen, R.; Mahuku, G. Unlocking the microbiome communities of Banana (Musa spp.) under disease stressed (Fusarium wilt) and non-stressed conditions. Microorganisms 2020, 8, 443. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Droby, S.; Wisniewski, M.; Teixidó, N.; Spadaro, D.; Jijakli, M.H. The science, development, and commercialization of postharvest biocontrol products. Postharvest Biol. Technol. 2016, 122, 22–29. [Google Scholar] [CrossRef]
- Massart, S.; Martinez-Medina, M.; Jijakli, M.H. Biological control in the microbiome era: Challenges and opportunities. Biol. Contr. 2015, 89, 98–108. [Google Scholar] [CrossRef]
- FAO. Agricultural Crop Production Statistics; Food and Agriculture Organization: Rome, Italy, 2019. [Google Scholar]
- Yashiro, E.; Spear, R.N.; McManus, P.S. Culture-dependent and culture-independent assessment of bacteria in the apple phyllosphere. J. App. Microbiol. 2011, 110, 1284–1296. [Google Scholar] [CrossRef] [PubMed]
- Shade, A.; McManus, P.S.; Handelsman, J. Unexpected diversity during community succession in the apple flower microbiome. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Abdelfattah, A.; Norelli, J.; Burchard, E.; Schena, L.; Droby, S.; Wisniewski, M. Apple endophytic microbiota of different rootstock/scion combinations suggests a genotype-specific influence. Microbiome 2018, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, A.; Wisniewski, M.; Droby, S.; Schena, L. Spatial and compositional variation in the fungal communities of organic and conventionally grown apple fruit at the consumer point-of-purchase. Hortic. Res. 2016, 3, 16047. [Google Scholar] [CrossRef] [PubMed]
- Wassermann, B.; Müller, H.; Berg, G. An apple a day: Which bacteria do we eat with organic and conventional apples? Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Wassermann, B.; Kusstatscher, P.; Berg, G. Microbiome response to hot water treatment and potential synergy with biological control on stored apples. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef]
- Abdelfattah, A.; Whitehead, S.R.; Macarisin, D.; Liu, J.; Burchard, E.; Freilich, S.; Dardick, C.; Droby, S.; Wisniewski, M. Effect of washing, waxing and low-temperature storage on the postharvest microbiome of apple. Microorganisms 2020, 8, 944. [Google Scholar] [CrossRef]
- Weber, R.W.S. Lagerfäulen an Äpfeln: Aktuelles aus Europa. Mitt. Obstbauvers. Alten Landes 2009, 64, 227–231. [Google Scholar]
- Lolas, M.; Díaz, G.; Mendez, R.; Cáceres, M.; Neubauer, L. Evaluation of the efficacy of fungicide fludioxonil in the postharvest control of bull’s eye rot (Neofabraea alba) in Chile. In Proceedings of the III International Symposium on Postharvest Pathology: Using Science to Increase Food Availability 1144, Bari, Italy, 7–11 June 2015; pp. 461–464. [Google Scholar]
- Maxin, P.; Weber, R.W.S.; Pedersen, H.L.; Williams, M. Control of a wide range of storage rots in naturally infected apples by hot-water dipping and rinsing. Postharv. Biol. Technol. 2012, 70, 25–31. [Google Scholar] [CrossRef]
- Vanwalleghem, T.; Dekeyser, D.; Nuyttens, D.; Tsige, A.; Verboven, P.; Van Hemelrijck, W.; Bylemans, D. Vaporization of biological control organisms in cold storage rooms to control postharvest diseases. In Proceedings of the III International Symposium on Postharvest Pathology: Using Science to Increase Food Availability 1144, Bari, Italy, 7–11 June 2015; pp. 121–128. [Google Scholar]
- Henriquez, J.L.; Sugar, D.; Spotts, R.A. Effects of environmental factors and cultural practices on Bull’s Eye Rot of pear. Plant. Dis. 2008, 92, 421–424. [Google Scholar] [CrossRef]
- Henriquez, J.L.; Sugar, D.; Spotts, R.A. Induction of cankers on pear tree branches by Neofabraea alba and N. perennans, and fungicide effects on conidial production on cankers. Plant. Dis. 2006, 90, 481–486. [Google Scholar] [CrossRef]
- Cameldi, I.; Neri, F.; Ventrucci, D.; Ceredi, G.; Muzzi, E.; Mari, M. Influence of harvest date on Bull’s Eye Rot of ‘Cripps Pink’ apple and control chemical strategies. Plant. Dis. 2016, 100, 2287–2293. [Google Scholar] [CrossRef] [PubMed]
- Roubal, C.; Nicot, P.C. Apple scab: Numerical optimization of a new thermal time scale and application for modelling ascospore release in southern France. Plant. Pathol. 2016, 65, 79–91. [Google Scholar] [CrossRef]
- Gölles, M.; Naef, A.; Kuske, S. Möglichkeiten zur Vermeidung von Rückständen im integrierten Apfelanbau. Schweiz. Zeitschr. Obs. Weinb. 2014, 8, 9–13. [Google Scholar]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108, 4516. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.D.; Lee, S.B.; Taylor, J.W. Amplification and direct sequencing of fungal ribosomal RNA Genes for phylogenetics. In PCR—Protocols and Applications—A Laboratory Manual; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: Ann Arbor, MI, USA, 1990; pp. 315–322. [Google Scholar]
- Köhl, J.; Wenneker, M.; Groenenboom-de Haas, B.H.; Anbergen, R.; Goossen-van de Geijn, H.M.; Lombaers-van der Plas, C.H.; Pinto, F.A.M.F.; Kastelein, P. Dynamics of post-harvest pathogens Neofabraea spp. and Cadophora spp. in plant residues in Dutch apple and pear orchards. Plant. Pathol. 2018, 67, 1264–1277. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. QIIME 2: Reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Prepr. 2018, 6, e27295v27292. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Meth. 2016, 13, 581. [Google Scholar] [CrossRef]
- Nilsson, R.H.; Glöckner, F.O.; Saar, I.; Tedersoo, L.; Kõljalg, U.; Abarenkov, K.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Schigel, D.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucl. Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Gerken, J.; Peplies, J.; Yarza, P.; Yilmaz, P.; Schweer, T.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucl. Acids Res. 2012, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Paulson, J.N. Biomformat: An Interface Package for the BIOM File Format. Available online: http://dx.doi.org/10.1371%2Fjournal.pone.0061217 (accessed on 1 January 2020).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Research 2016, 5, 1492. [Google Scholar] [CrossRef] [PubMed]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive metagenomic visualization in a Web browser. BMC Bioinform. 2011, 12, 385. [Google Scholar] [CrossRef] [PubMed]
- Bencheqroun, S.K.; Bajji, M.; Massart, S.; Bentata, F.; Labhilili, M.; Achbani, H.; El Jaafari, S.; Jijakli, M.H. Biocontrol of blue mold on apple fruits by Aureobasidium pullulans (strain Ach 1-1): In vitro and in situ evidence for the possible involvement of competition for nutrients. Commun. Agric. Appl. Biol. Sci. 2006, 71, 1151–1157. [Google Scholar] [PubMed]
- Di Francesco, A.; Di Foggia, M.; Baraldi, E. Aureobasidium pullulans volatile organic compounds as alternative postharvest method to control brown rot of stone fruits. Food Microbiol. 2020, 87, 103395. [Google Scholar] [CrossRef] [PubMed]
- Freimoser, F.M.; Rueda-Mejia, M.P.; Tilocca, B.; Migheli, Q. Biocontrol yeasts: Mechanisms and applications. World J. Microbiol. Biotechnol. 2019, 35, 154. [Google Scholar] [CrossRef]
- Cui, Z.; Huntley, R.B.; Zeng, Q.; Steven, B. Temporal and spatial dynamics in the apple flower microbiome in the presence of the phytopathogen Erwinia amylovora. ISME J. 2020. [Google Scholar] [CrossRef]
- Liu, J.; Ridgway, H.J.; Jones, E.E. Apple endophyte community is shaped by tissue type, cultivar and site and has members with biocontrol potential against Neonectria ditissima. J. Appl. Microbiol. 2020, 128, 1735–1753. [Google Scholar] [CrossRef]
- Yu, T.; Chen, Y. Effects of elevated carbon dioxide on environmental microbes and its mechanisms: A review. Sci. Total Environ. 2019, 655, 865–879. [Google Scholar] [CrossRef]
- Brizzolara, S.; Manganaris, G.A.; Fotopoulos, V.; Watkins, C.B.; Tonutti, P. Primary metabolism in fresh fruits during storage. Front. Plant. Sci. 2020, 11. [Google Scholar] [CrossRef]
- Wright, A.H.; Delong, J.M.; Arul, J.; Prange, R.K. The trend toward lower oxygen levels during apple (Malus × domestica Borkh) storage. J. Hortic. Sci. Biotechnol. 2015, 90, 1–13. [Google Scholar] [CrossRef]
- Nguyen Van Long, N.; Vasseur, V.; Couvert, O.; Coroller, L.; Burlot, M.; Rigalma, K.; Mounier, J. Modeling the effect of modified atmospheres on conidial germination of fungi from dairy foods. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.R.B.; Forauer, E.C.; D’Amico, D.J. Effect of modified atmosphere packaging on the growth of spoilage microorganisms and Listeria monocytogenes on fresh cheese. J. Dairy Sci. 2018, 101, 7768–7779. [Google Scholar] [CrossRef] [PubMed]
- Angeli, D.; Sare, A.R.; Jijakli, M.H.; Pertot, I.; Massart, S. Insights gained from metagenomic shotgun sequencing of apple fruit epiphytic microbiota. Postharvest Biol. Technol. 2019, 153, 96–106. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Collins, T.S.; Masarweh, C.; Allen, G.; Heymann, H.; Ebeler, S.E.; Mills, D.A. Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics. mBio 2016, 7, e00631-16. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139. [Google Scholar] [CrossRef]
- Gostinčar, C.; Ohm, R.A.; Kogej, T.; Sonjak, S.; Turk, M.; Zajc, J.; Zalar, P.; Grube, M.; Sun, H.; Han, J.; et al. Genome sequencing of four Aureobasidium pullulans varieties: Biotechnological potential, stress tolerance, and description of new species. BMC Genom. 2014, 15, 549. [Google Scholar] [CrossRef]
- Köhl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant. Sci. 2019, 10. [Google Scholar] [CrossRef]
- Woo, S.L.; Pepe, O. Microbial consortia: Promising probiotics as plant biostimulants for sustainable agriculture. Front. Plant. Sci. 2018, 9. [Google Scholar] [CrossRef]
- Macarisin, D.; Sheth, I.; Hur, M.; Wooten, A.; Kwon, H.J.; Gao, Z.; De Jesus, A.; Jurick, W.; Chen, Y. Survival of outbreak, food, and environmental strains of Listeria monocytogenes on whole apples as affected by cultivar and wax coating. Sci. Rep. 2019, 9, 12170. [Google Scholar] [CrossRef]
- Simonin, M.; Dasilva, C.; Terzi, V.; Ngonkeu, E.L.M.; Diouf, D.; Kane, A.; Béna, G.; Moulin, L. Influence of plant genotype and soil on the wheat rhizosphere microbiome: Evidences for a core microbiome across eight African and European soils. FEMS Microbiol. Ecol. 2020, 96. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.R.; Lundberg, D.S.; del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bösch, Y.; Britt, E.; Perren, S.; Naef, A.; Frey, J.E.; Bühlmann, A. Dynamics of the Apple Fruit Microbiome after Harvest and Implications for Fruit Quality. Microorganisms 2021, 9, 272. https://doi.org/10.3390/microorganisms9020272
Bösch Y, Britt E, Perren S, Naef A, Frey JE, Bühlmann A. Dynamics of the Apple Fruit Microbiome after Harvest and Implications for Fruit Quality. Microorganisms. 2021; 9(2):272. https://doi.org/10.3390/microorganisms9020272
Chicago/Turabian StyleBösch, Yvonne, Elisabeth Britt, Sarah Perren, Andreas Naef, Jürg E. Frey, and Andreas Bühlmann. 2021. "Dynamics of the Apple Fruit Microbiome after Harvest and Implications for Fruit Quality" Microorganisms 9, no. 2: 272. https://doi.org/10.3390/microorganisms9020272
APA StyleBösch, Y., Britt, E., Perren, S., Naef, A., Frey, J. E., & Bühlmann, A. (2021). Dynamics of the Apple Fruit Microbiome after Harvest and Implications for Fruit Quality. Microorganisms, 9(2), 272. https://doi.org/10.3390/microorganisms9020272





