Xerotolerance: A New Property in Exiguobacterium Genus
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Strains and Culture Conditions
2.3. Xerotolerance Test for the Isolation of Xerotolerant Bacterial Strains
2.4. Identification of Xerotolerant Isolated Strains
2.5. Sequencing, Assembly, and Bioinformatic Analyses of Genome
2.6. Genetic Manipulations
2.7. Microscopy
2.8. Metal Resistance and Formation of SeNPs
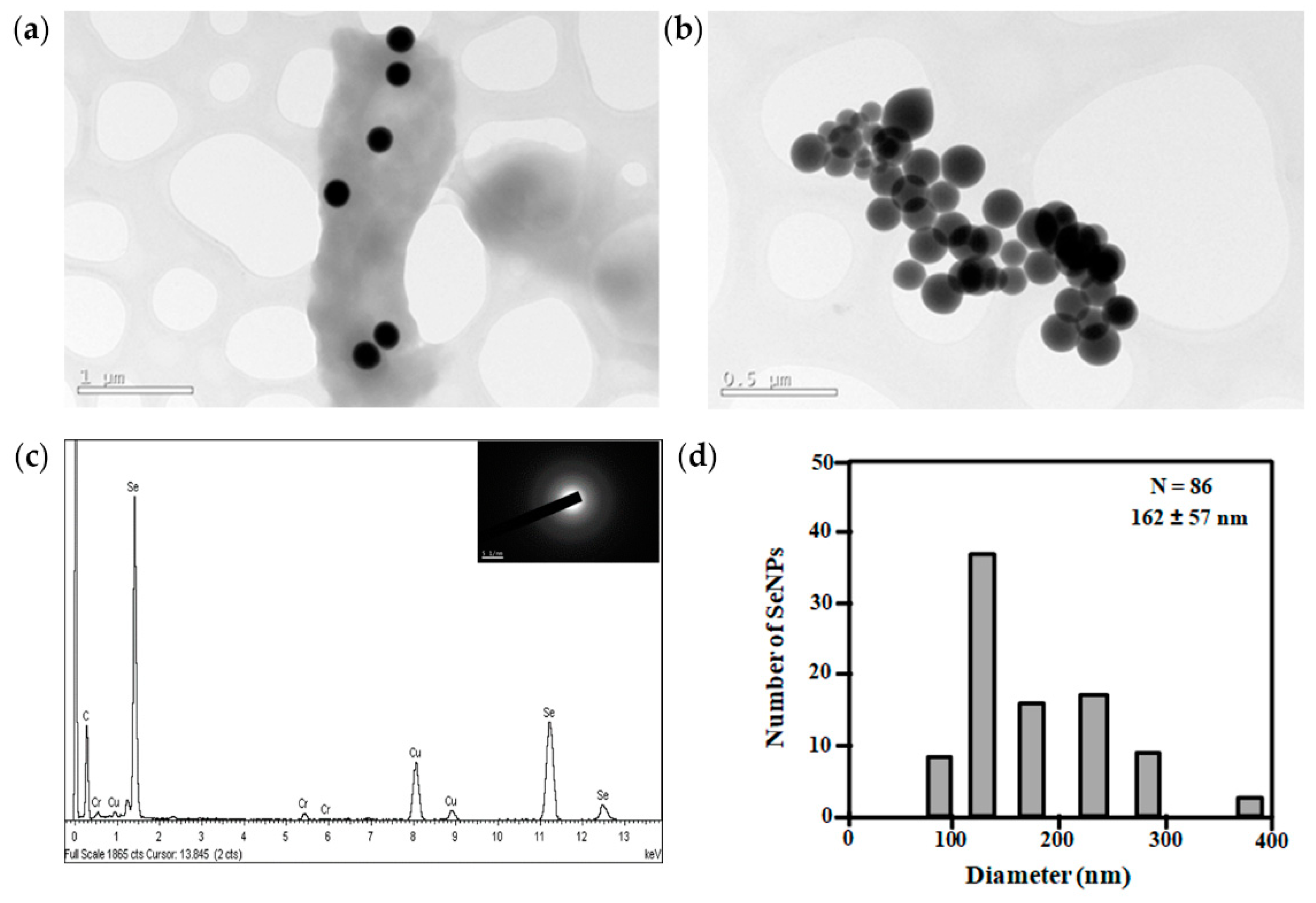
2.9. Characterization of Selenium Nanoparticles
3. Results
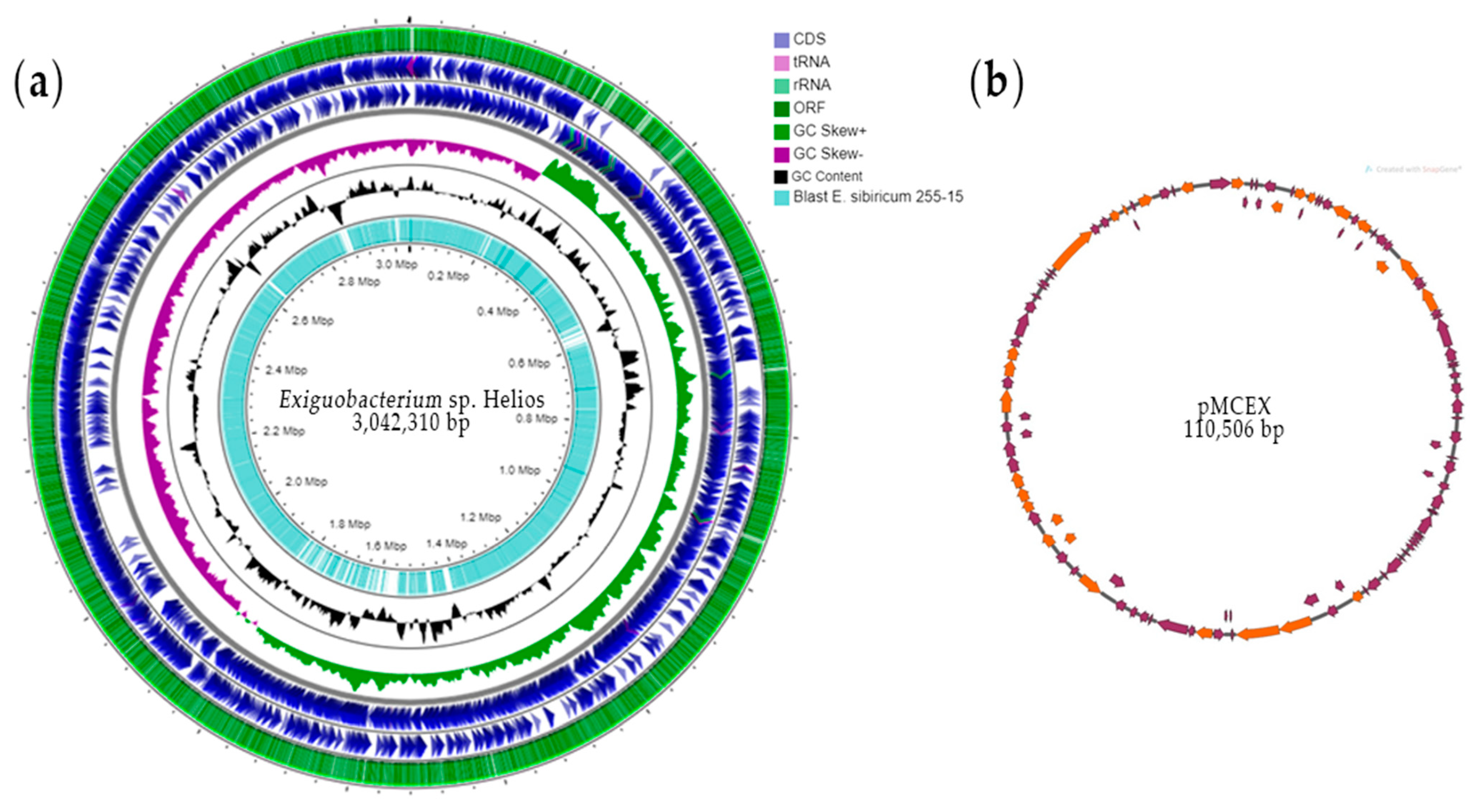
3.1. Analysis of Exiguobacterium sp. Helios Genome
3.2. Genome Comparison of Exiguobacterium sp. Helios and E. sibiricum 255-15
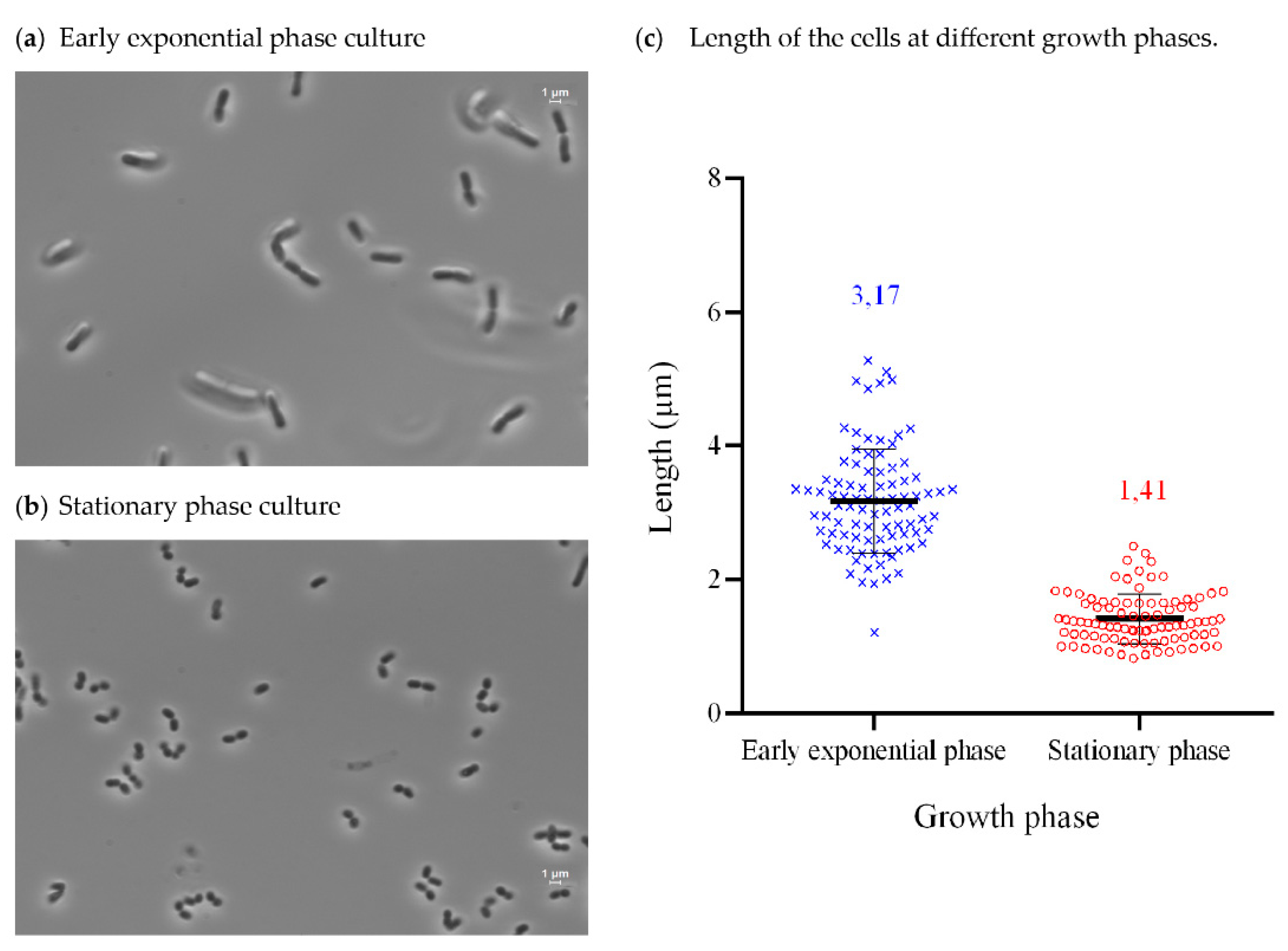
3.3. Isolation, Identification, Growth Conditions, and Phenotypical Characterization of Exiguobacterium sp. Helios
3.4. Metal Resistance of Exiguobacterium sp. Helios
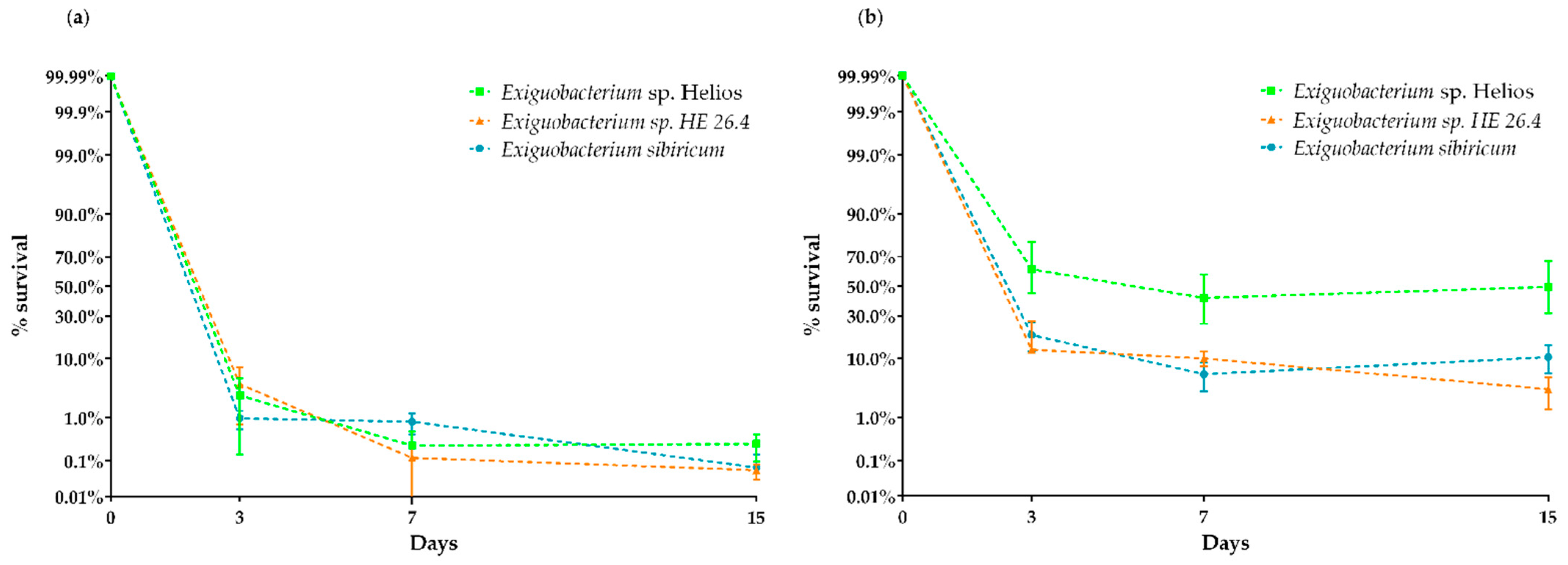
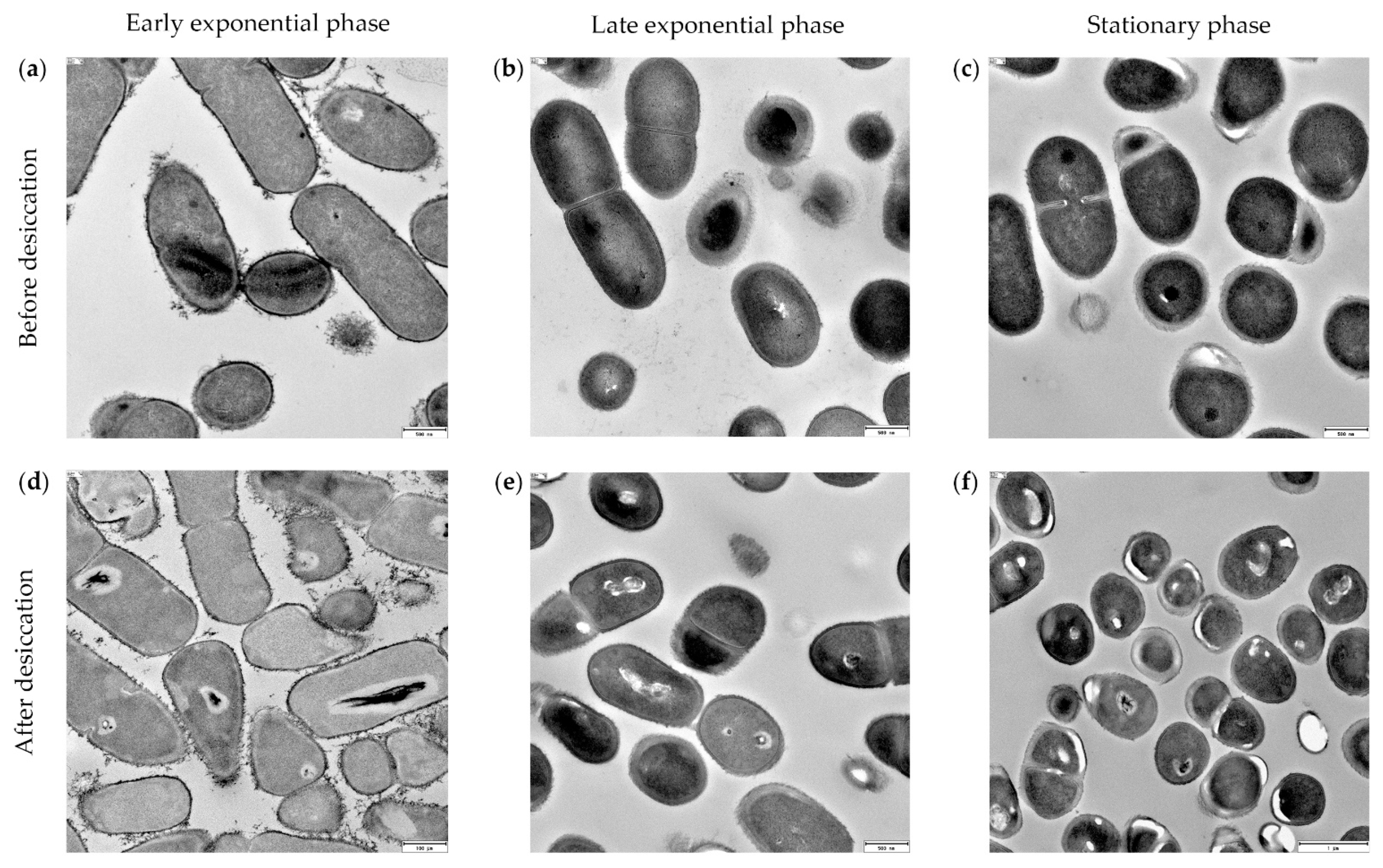
3.5. Xerotolerance of Exiguobacterium sp. Helios
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Collins, M.D.; Lund, B.M.; Farrow, J.A.E.; Schleifer, K.H. Chemotaxonomic Study of an Alkalophilic Bacterium, Exiguobacterium aurantiacum gen. nov., sp. nov. J. Gen. Microbiol. 1983, 129, 2037–2042. [Google Scholar] [CrossRef]
- Vishnivetskaya, T.A.; Kathariou, S.; Tiedje, J.M. The Exiguobacterium Genus: Biodiversity and Biogeography. Extremophiles 2009, 13, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Kasana, R.C.; Pandey, C.B. Exiguobacterium: An Overview of a Versatile Genus with Potential in Industry and Agriculture. Crit. Rev. Biotechnol. 2018, 38, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Goris, J.; Vishnivetskaya, T.; Gilichinsky, D.; Thomashow, M.F.; Tiedje, J.M. Characterization of Exiguobacterium Isolates from the Siberian Permafrost. Description of Exiguobacterium sibiricum sp. nov. Extremophiles 2006, 10, 285–294. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Shivaji, S. Exiguobacterium Indicum sp. nov., a Psychrophilic Bacterium from the Hamta Glacier of the Himalayan Mountain Ranges of India. Int. J. Syst. Evol. Microbiol. 2006, 56, 2765–2770. [Google Scholar] [CrossRef]
- Crapart, S.; Fardeau, M.L.; Cayol, J.L.; Thomas, P.; Sery, C.; Ollivier, B.; Combet-Blanc, Y. Exiguobacterium profundum sp. nov., a Moderately Thermophilic, Lactic Acid-Producing Bacterium Isolated from a Deep-Sea Hydrothermal Vent. Int. J. Syst. Evol. Microbiol. 2007, 57, 287–292. [Google Scholar] [CrossRef]
- Kasana, R.C.; Yadav, S.K. Isolation of a Psychrotrophic Exiguobacterium sp. SKPB5 (MTCC 7803) and Characterization of Its Alkaline Protease. Curr. Microbiol. 2007, 54, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Alam, M.Z.; Malik, A. Chromate Resistance, Transport and Bioreduction by Exiguobacterium sp. ZM-2 Isolated from Agricultural Soil Irrigated with Tannery Effluent. J. Basic Microbiol. 2008, 48, 416–420. [Google Scholar] [CrossRef]
- Chaturvedi, P.; Prabahar, V.; Manorama, R.; Pindi, P.K.; Bhadra, B.; Begum, Z.; Shivaji, S. Exiguobacterium soli sp. nov., a Psychrophilic Bacterium from the McMurdo Dry Valleys, Antarctica. Int. J. Syst. Evol. Microbiol. 2008, 58, 2447–2453. [Google Scholar] [CrossRef]
- Carneiro, A.R.; Ramos, R.T.J.; Dall’Agnol, H.; Pinto, A.C.; Soares, S.d.C.; Santos, A.R.; Guimarães, L.C.; Almeida, S.S.; Baraúna, R.A.; das Graças, D.A.; et al. Genome Sequence of Exiguobacterium antarcticum B7, Isolated from a Biofilm in Ginger Lake, King George Island, Antarctica. J. Bacteriol. 2012, 194, 6689–6690. [Google Scholar] [CrossRef]
- Singh, N.K.; Raichand, R.; Kaur, I.; Kaur, C.; Pareek, S.; Mayilraj, S. Exiguobacterium himgiriensis sp. nov. A Novel Member of the Genus Exiguobacterium, Isolated from the Indian Himalayas. Antonie Van Leeuwenhoek 2013, 103, 789–796. [Google Scholar] [CrossRef]
- Dastager, S.G.; Mawlankar, R.; Sonalkar, V.V.; Thorat, M.N.; Mual, P.; Verma, A.; Krishnamurthi, S.; Tang, S.K.; Li, W.J. Exiguobacterium enclense sp. nov., Isolated from Sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 1611–1616. [Google Scholar] [CrossRef]
- Etemadifar, Z.; Gholami, M.; Derikvand, P. UV-Resistant Bacteria with Multiple-Stress Tolerance Isolated from Desert Areas in Iran. Geomicrobiol. J. 2016, 33, 1–7. [Google Scholar] [CrossRef]
- Hanaoka, Y.; Takebe, F.; Nodasaka, Y.; Hara, I.; Matsuyama, H.; Yumoto, I. Growth-Dependent Catalase Localization in Exiguobacterium oxidotolerans T-2-2T Reflected by Catalase Activity of Cells. PLoS ONE 2013, 8, e76862. [Google Scholar] [CrossRef]
- Remonsellez, F.; Castro-Severyn, J.; Pardo-Esté, C.; Aguilar, P.; Fortt, J.; Salinas, C.; Barahona, S.; León, J.; Fuentes, B.; Areche, C.; et al. Characterization and Salt Response in Recurrent Halotolerant Exiguobacterium sp. SH31 Isolated from Sediments of Salar de Huasco, Chilean Altiplano. Front. Microbiol. 2018, 9, 2228. [Google Scholar] [CrossRef] [PubMed]
- Castro-Severyn, J.; Pardo-Esté, C.; Mendez, K.N.; Morales, N.; Marquez, S.L.; Molina, F.; Remonsellez, F.; Castro-Nallar, E.; Saavedra, C.P. Genomic Variation and Arsenic Tolerance Emerged as Niche Specific Adaptations by Different Exiguobacterium Strains Isolated From the Extreme Salar de Huasco Environment in Chilean—Altiplano. Front. Microbiol. 2020, 11, 1632. [Google Scholar] [CrossRef] [PubMed]
- Selvakumar, G.; Joshi, P.; Nazim, S.; Mishra, P.K.; Kundu, S.; Gupta, H.S. Exiguobacterium acetylicum Strain 1P (MTCC 8707) a Novel Bacterial Antagonist from the North Western Indian Himalayas. World J. Microbiol. Biotechnol. 2009, 25, 131–137. [Google Scholar] [CrossRef]
- Selvakumar, G.; Kundu, S.; Joshi, P.; Nazim, S.; Gupta, A.D.; Gupta, H.S. Growth Promotion of Wheat Seedlings by Exiguobacterium acetylicum 1P (MTCC 8707) a Cold Tolerant Bacterial Strain from the Uttarakhand Himalayas. Indian J. Microbiol. 2010, 50, 50–56. [Google Scholar] [CrossRef] [PubMed]
- Gangoiti, J.; Pijning, T.; Dijkhuizen, L. The Exiguobacterium sibiricum 255-15 GtfC Enzyme Represents a Novel Glycoside Hydrolase 70 Subfamily of 4,6-α-Glucanotransferase Enzymes. Appl. Environ. Microbiol. 2016, 82, 756–766. [Google Scholar] [CrossRef] [PubMed]
- Qiao, Y.; Peng, Q.; Yan, J.; Wang, H.; Ding, H.; Shi, B. Gene Cloning and Enzymatic Characterization of Alkali-Tolerant Type I Pullulanase from Exiguobacterium acetylicum. Lett. Appl. Microbiol. 2015, 60, 52–59. [Google Scholar] [CrossRef]
- Crespim, E.; Zanphorlin, L.M.; de Souza, F.H.M.; Diogo, J.A.; Gazolla, A.C.; Machado, C.B.; Figueiredo, F.; Sousa, A.S.; Nóbrega, F.; Pellizari, V.H.; et al. A Novel Cold-Adapted and Glucose-Tolerant GH1 β-Glucosidase from Exiguobacterium antarcticum B7. Int. J. Biol. Macromol. 2016, 82, 375–380. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Kwon, S.; Park, S.H.; Kim, B.Y.; Yoo, W.; Ryu, B.H.; Kim, H.W.; Shin, S.C.; Kim, S.; Park, H.; et al. Crystal Structure and Functional Characterization of an Esterase (EaEST) from Exiguobacterium antarcticum. PLoS ONE 2017, 12, e0169540. [Google Scholar] [CrossRef]
- Castro-Severyn, J.; Remonsellez, F.; Valenzuela, S.L.; Salinas, C.; Fortt, J.; Aguilar, P.; Pardo-Esté, C.; Dorador, C.; Quatrini, R.; Molina, F.; et al. Comparative Genomics Analysis of a New Exiguobacterium Strain from Salar de Huasco Reveals a Repertoire of Stress-Related Genes and Arsenic Resistance. Front. Microbiol. 2017, 8, 456. [Google Scholar] [CrossRef] [PubMed]
- Aburto, C.; Castillo, C.; Cornejo, F.; Arenas-Salinas, M.; Vásquez, C.; Guerrero, C.; Arenas, F.; Illanes, A.; Vera, C. Β-Galactosidase from Exiguobacterium acetylicum: Cloning, Expression, Purification and Characterization. Bioresour. Technol. 2019, 277, 211–215. [Google Scholar] [CrossRef]
- Yin, B.; Gu, H.; Mo, X.; Xu, Y.; Yan, B.; Li, Q.; Ou, Q.; Wu, B.; Guo, C.; Jiang, C. Identification and Molecular Characterization of a Psychrophilic GH1 β-Glucosidase from the Subtropical Soil Microorganism Exiguobacterium sp. GXG2. AMB Expr. 2019, 9, 159. [Google Scholar] [CrossRef]
- Wang, Y.; Le, L.T.H.L.; Yoo, W.; Lee, C.W.; Kim, K.K.; Lee, J.H.; Kim, T.D. Characterization, Immobilization, and Mutagenesis of a Novel Cold-Active Acetylesterase (EaAcE) from Exiguobacterium antarcticum B7. Int. J. Biol. Macromol. 2019, 136, 1042–1051. [Google Scholar] [CrossRef]
- Arora, P.K.; Bae, H. Biodegradation of 4-Chloroindole by Exiguobacterium sp. PMA. J. Hazard. Mater. 2015, 284, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Yang, J.; Wu, W.M.; Zhao, J.; Song, Y.; Gao, L.; Yang, R.; Jiang, L. Biodegradation and Mineralization of Polystyrene by Plastic-Eating Mealworms: Part 1. Chemical and Physical Characterization and Isotopic Tests. Environ. Sci. Technol. 2015, 49, 12080–12086. [Google Scholar] [CrossRef]
- Chauhan, D.; Agrawal, G.; Deshmukh, S.; Roy, S.S.; Priyadarshini, R. Biofilm Formation by Exiguobacterium sp. DR11 and DR14 Alter Polystyrene Surface Properties and Initiate Biodegradation. RSC Adv. 2018, 8, 37590–37599. [Google Scholar] [CrossRef]
- García, A.H. Anhydrobiosis in Bacteria: From Physiology to Applications. J. Biosci. 2011, 36, 939–950. [Google Scholar] [CrossRef]
- Gerber, E.; Bernard, R.; Castang, S.; Chabot, N.; Coze, F.; Dreux-Zigha, A.; Hauser, E.; Hivin, P.; Joseph, P.; Lazarelli, C.; et al. Deinococcus as New Chassis for Industrial Biotechnology: Biology, Physiology and Tools. J. Appl. Microbiol. 2015, 119, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Mattimore, V.; Battista, J.R. Radioresistance of Deinococcus Radiodurans: Functions Necessary to Survive Ionizing Radiation Are Also Necessary to Survive Prolonged Desiccation. J. Bacteriol. 1996, 178, 633–637. [Google Scholar] [CrossRef]
- Dorado-Morales, P.; Vilanova, C.; Peretó, J.; Codoñer, F.M.; Ramón, D.; Porcar, M. A Highly Diverse, Desert-like Microbial Biocenosis on Solar Panels in a Mediterranean City. Sci. Rep. 2016, 6, 29235. [Google Scholar] [CrossRef] [PubMed]
- Porcar, M.; Louie, K.B.; Kosina, S.M.; van Goethem, M.W.; Bowen, B.P.; Tanner, K.; Northen, T.R. Microbial Ecology on Solar Panels in Berkeley, CA, United States. Front. Microbiol. 2018, 9, 3043. [Google Scholar] [CrossRef] [PubMed]
- Tanner, K.; Martí, J.M.; Belliure, J.; Fernández-Méndez, M.; Molina-Menor, E.; Peretó, J.; Porcar, M. Polar Solar Panels: Arctic and Antarctic Microbiomes Display Similar Taxonomic Profiles. Environ. Microbiol. Rep. 2018, 10, 75–79. [Google Scholar] [CrossRef]
- Sambrook, J.; Russell, D.W. Molecular Cloning: A Laboratory Manual, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2001; Volume 1, ISBN 978-087969577-4. [Google Scholar]
- Cohen, G.N.; Rickenberg, R.H. Reversible Specific Concentration of Amino Acids in Escherichia coli. Ann. Inst. Pasteur Paris. 1956, 91, 693–720. [Google Scholar]
- Uhía, I.; Galán, B.; Morales, V.; García, J.L. Initial Step in the Catabolism of Cholesterol by Mycobacterium smegmatis MC2155. Environ. Microbiol. 2011, 13, 943–959. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-R, L.M.; Konstantinidis, K.T. The Enveomics Collection: A Toolbox for Specialized Analyses of Microbial Genomes and Metagenomes. PeerJ. Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Russo, P.; Iturria, I.; Mohedano, M.L.; Caggianiello, G.; Rainieri, S.; Fiocco, D.; Angel Pardo, M.; López, P.; Spano, G. Zebrafish Gut Colonization by MCherry-Labelled Lactic Acid Bacteria. Appl. Microbiol. Biotechnol. 2015, 99, 3479–3490. [Google Scholar] [CrossRef]
- Collins, T.J. ImageJ for Microscopy. Biotechniques 2007, 43, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Mergeay, M.; Nies, D.; Schlegel, H.G.; Gerits, J.; Charles, P.; van Gijsegem, A.F. Alcaligenes eutrophus CH34 Is a Facultative Chemolithotroph with Plasmid-Bound Resistance to Heavy Metals. J. Bacteriol. 1985, 162, 328–334. [Google Scholar] [CrossRef] [PubMed]
- Li, D.B.; Cheng, Y.Y.; Wu, C.; Li, W.W.; Li, N.; Yang, Z.C.; Tong, Z.H.; Yu, H.Q. Selenite Reduction by Shewanella oneidensis MR-1 Is Mediated by Fumarate Reductase in Periplasm. Sci. Rep. 2014, 4, 3735. [Google Scholar] [CrossRef] [PubMed]
- Abecasis, A.B.; Serrano, M.; Alves, R.; Quintais, L.; Pereira-Leal, J.B.; Henriques, A.O. A Genomic Signature and the Identification of New Sporulation Genes. J. Bacteriol. 2013, 195, 2101–2115. [Google Scholar] [CrossRef]
- Kappes, R.M.; Kempf, B.; Bremer, E. Three Transport Systems for the Osmoprotectant Glycine Betaine Operate in Bacillus subtilis: Characterization of OpuD. J. Bacteriol. 1996, 178, 5071–5079. [Google Scholar] [CrossRef]
- Hou, S.; Larsen, R.W.; Boudko, D.; Riley, C.W.; Karatan, E.; Zimmer, M.; Ordal George, W.; Alam, M. Myoglobin-like Aerotaxis Transducers in Archaea and Bacteria. Nature 2000, 403, 540–544. [Google Scholar] [CrossRef]
- Poole, R.K.; Hughes, M.N. MicroReview New Functions for the Ancient Globin Family: Bacterial Responses to Nitric Oxide and Nitrosative Stress. Mol. Microbiol. 2000, 36, 775–783. [Google Scholar] [CrossRef]
- Appukuttan, D.; Seo, H.S.; Jeong, S.; Im, S.; Joe, M.; Song, D.; Choi, J.; Lim, S. Expression and Mutational Analysis of DinB-like Protein DR0053 in Deinococcus radiodurans. PLoS ONE 2015, 10, e0118275. [Google Scholar] [CrossRef][Green Version]
- Thurotte, A.; Brüser, T.; Mascher, T.; Schneider, D. Membrane Chaperoning by Members of the PspA/IM30 Protein Family. Commun. Integr. Biol. 2017, 10, e1264546. [Google Scholar] [CrossRef]
- Keto-Timonen, R.; Hietala, N.; Palonen, E.; Hakakorpi, A.; Lindström, M.; Korkeala, H. Cold Shock Proteins: A Minireview with Special Emphasis on Csp-Family of Enteropathogenic yersinia. Front. Microbiol. 2016, 7, 1151. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriag, G. Insights on the Evolution of Trehalose Biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Chen, N.; Shi, M.; Xian, M.; Song, Y.; Liu, J. The Metabolism and Biotechnological Application of Betaine in Microorganism. Appl. Microbiol. Biotechnol. 2016, 100, 3865–3876. [Google Scholar] [CrossRef]
- Billi, D.; Wright, D.J.; Helm, R.F.; Prickett, T.; Potts, M.; Crowe, J.H. Engineering Desiccation Tolerance in Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 1680–1684. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Salazar, J.; Moreno, S.; Espín, G. LEA Proteins Are Involved in Cyst Desiccation Resistance and Other Abiotic Stresses in Azotobacter vinelandii. Cell Stress Chaperones 2017, 22, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, C.; Wang, Z.; Lin, M.; Wang, J.; Wu, M. Pleiotropic Roles of Late Embryogenesis Abundant Proteins of Deinococcus radiodurans against Oxidation and Desiccation. Comput. Struct. Biotechnol. J. 2021, 19, 3407–3415. [Google Scholar] [CrossRef] [PubMed]
- Èliassi, A.; Modarress, H.; Ali Mansoori, G. Measurement of Activity of Water in Aqueous Poly(Ethylene Glycol) Solutions (Effect of Excess Volume on the Flory-Huggins χ-Parameter). J. Chem. Eng. Data 1999, 44, 52–55. [Google Scholar] [CrossRef]
- Zheng, S.; Su, J.; Wang, L.; Yao, R.; Wang, D.; Deng, Y.; Wang, R.; Wang, G.; Rensing, C. Selenite Reduction by the Obligate Aerobic Bacterium Comamonas testosteroni S44 Isolated from a Metal-Contaminated Soil. BMC Microbiol. 2014, 14, 204. [Google Scholar] [CrossRef]
- Staicu, L.C.; Ackerson, C.J.; Cornelis, P.; Ye, L.; Berendsen, R.L.; Hunter, W.J.; Noblitt, S.D.; Henry, C.S.; Cappa, J.J.; Montenieri, R.L.; et al. Pseudomonas moraviensis subsp. stanleyae, a Bacterial Endophyte of Hyperaccumulator Stanleya pinnata, Is Capable of Efficient Selenite Reduction to Elemental Selenium under Aerobic Conditions. J. Appl. Microbiol. 2015, 119, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Speeding up Bioproduction of Selenium Nanoparticles by Using Vibrio natriegens as Microbial Factory. Sci. Rep. 2017, 7, 16046. [Google Scholar] [CrossRef]
- Lebre, P.H.; de Maayer, P.; Cowan, D.A. Xerotolerant Bacteria: Surviving through a Dry Spell. Nat. Rev. Microbiol. 2017, 15, 285–296. [Google Scholar] [CrossRef]
- Seckbach, J.; Rampelotto, P.H. Polyextremophiles. In Microbial Evolution under Extreme Conditions; Bakermans, C., Ed.; De Gruyter Editorial: Berlin, Germany, 2015; pp. 153–170. ISBN 978-3-11-033506-4. [Google Scholar]
- Billi, D.; Potts, M. Life and Death of Dried Prokaryotes. Res. Microbiol. 2002, 153, 7–12. [Google Scholar] [CrossRef]
- Ponder, M.A.; Thomashow, M.F.; Tiedje, J.M. Metabolic Activity of Siberian Permafrost Isolates, Psychrobacter arcticus and Exiguobacterium sibiricum, at Low Water Activities. Extremophiles 2008, 12, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Ivanova, N.; He, Z.; Huebner, M.; Zhou, J.; Tiedje, J.M. Architecture of Thermal Adaptation in an Exiguobacterium sibiricum Strain Isolated from 3 Million Year Old Permafrost: A Genome and Transcriptome Approach. BMC Genom. 2008, 9, 547. [Google Scholar] [CrossRef] [PubMed]
- Caiola, M.G.; Billi, D.; Friedmann, E.I. Effect of Desiccation on Envelopes of the Cyanobacterium Chroococcidiopsis sp. (Chroococcales). Eur. J. Phycol. 1996, 31, 97–105. [Google Scholar] [CrossRef][Green Version]

| Gene | Locus | Function Defined in B. subtilis subsp. subtilis Str. 168 | %ID aa | Accession # |
|---|---|---|---|---|
| clcC/mecB | HNY42_RS01815 | Negative regulator of genetic competence clcC/mecB | 80.62% | P37571.1 |
| comEC | HNY42_RS05440 | DNA internalization-related competence protein ComEC/Rec2 | 28.05% | P39695.2 |
| comK | HNY42_RS05215 HNY42_RS13920 | Competence transcription factor | 24.84% 27.70% | P40396.1 |
| addABb | HNY42_RS05235 | ATP-dependent helicase/nuclease AddAB, subunit B | 36.23% | P23477.2 |
| addABa | HNY42_RS05240 | ATP-dependent helicase/nuclease AddAB, subunit A | 40.87% | P23478.2 |
| comEA | HNY42_RS05435 | Late competence protein ComEA, DNA receptor | 37.44% | P39694.1 |
| comGA | HNY42_RS05970 | Late competence protein ComGA, access of DNA to ComEA | 30.68% | P25953.2 |
| comGB | HNY42_RS05975 | Late competence protein ComGB, access of DNA to ComEA | ND | |
| comGC | HNY42_RS05980 | Late competence protein ComGC, access of DNA to ComEA | ND | |
| mecA1 | HNY42_RS12035 | Adapter protein MecA 1 | 42.67% | P37958.1 |
| mecA2 | HNY42_RS10860 | Adapter protein MecA 2 | 29.29% | P50734.1 |
| coiA | HNY42_RS12020 | Competence protein CoiA | ND | |
| yhgH | HNY42_RS14090 | Competence protein F homolog. Protein YhgH required for utilization of DNA as sole source of carbon and energy | 32.61% | P39147.1 |
| comF | HNY42_RS14095 | ComF operon protein A, DNA transporter ATPase. | 41.42% | P39145.1 |
| cinA | HNY42_RS06685 | competence/damage-inducible protein A | 46.68% | P46323.3 |
| Gene | Locus | Accession # | Function Defined in B. subtilis subsp. subtilis Str. 168 | %ID aa | Accession # |
|---|---|---|---|---|---|
| sigB | HNY42_RS00075 | QNR19439.1 | RNA polymerase σB factor. General stress protein | 63.18% | P06574.3 |
| spoIIAA | HNY42_RS00085 | QNR19441.1 | Regulation of the sigma factor (σF) activity. | 48.62% | P17903.1 |
| ykvl | HNY42_RS00585 | QNR19540.1 | 7-carboxy-7-deazaguanine (CDG) synthase. tRNA modification. | 49.79% | O31677.1 |
| yyaC | HNY42_RS01240 | QNR22337.1 | Spore specific protease. | 48.90% | P37521.1 |
| spoVT/abrB | HNY42_RS01560 | QNR19702.1 | Transition state regulatory protein AbrB | 81.11% | P08874.1 |
| spoVG | HNY42_RS01605 | QNR19711.1 | Regulator required for spore cortex synthesis. | 65.59% | P28015.1 |
| mcsA | HNY42_RS01805 | QNR19736.1 | Activator of protein kinase McsB. | 26.70% | P37569.1 |
| mcsB | HNY42_RS01810 | QNR19737.1 | Protein arginine kinase. | 49.13% | P37570.1 |
| sigH | HNY42_RS01870 | QNR19748.1 | RNA polymerase σH. Transcription of early stationary phase genes (sporulation, competence). | 71.50% | P17869.1 |
| dapG | HNY42_RS03930 | QNR20129.1 | Aspartokinase | 40.30% | P08495.2 |
| yqhQ | HNY42_RS06055 | QNR20509.1 | Survival to stress conditions. σB and σF regulons. | 55.16% | P54515.2 |
| spo0A | HNY42_RS06130 | QNR20524.1 | Coordinates DNA replication and initiation of sporulation by binding to sites close to the oriC. | 47.47% | P06534.1 |
| spoVS | HNY42_RS06705 | QNR20638.1 | Regulator required for dehydration of the spore core and assembly of the coat. | 87.21% | P45693.1 |
| spoIIIAA | HNY42_RS12180 | QNR21664.1 | Uncharacterized AAA domain-containing protein YrvN | 48.12% | O34528.1 |
| ytvI | HNY42_RS13260 | QNR21869.1 | Unknown | 38.58% | O34991.1 |
| cwlD | HNY42_RS14150 | QNR22035.1 | N-acetylmuramoyl-L-alanine amidase, spore cortex peptidoglycan synthesis. | 24.19% | O32041.1 |
| Substrates | Exiguobacterium sp. Helios | E. sibiricum 255-15 |
|---|---|---|
| Arabinose | + | − |
| Fructose | + | + |
| Galactose | + | + |
| Glucose | + | + |
| Lactose | + | +/− |
| Maltose | + | + |
| Ribose | + | + |
| Sucrose | + | + |
| Xylose | + | − |
| 3,4-Dihydroxybenzoic acid | + | + |
| 3-Hydroxybenzoic acid | − | − |
| 4-Hydroxybenzoic acid | + | + |
| Benzoic acid | + | + |
| Gentisic acid | − | − |
| Phenylacetic acid | + | + |
| Catechol | − | − |
| Citric acid | + | + |
| Pyruvic acid | + | + |
| Succinic acid | + | + |
| Antibiotics | MIC (µg/mL) |
|---|---|
| Kanamycin | 6 |
| Gentamycin | 10 |
| Ampicillin | 50 |
| Chloramphenicol | 8 |
| Spectinomycin | 75 |
| Metal and Metalloid | MIC (mM) |
|---|---|
| NiCl2 | 1.25 |
| ZnCl2 | 2.50 |
| K2TeO3 | 0.62 |
| K2SeO3 | 20.00 |
| NaAsO2 | 1.25 |
| Na3AsO4 | 25.00 |
| CdCl2 | 0.62 |
| AgNO3 | 0.62 |
| Pb(NO3)2 | 5.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
López, M.C.; Galán, B.; Carmona, M.; Navarro Llorens, J.M.; Peretó, J.; Porcar, M.; Getino, L.; Olivera, E.R.; Luengo, J.M.; Castro, L.; et al. Xerotolerance: A New Property in Exiguobacterium Genus. Microorganisms 2021, 9, 2455. https://doi.org/10.3390/microorganisms9122455
López MC, Galán B, Carmona M, Navarro Llorens JM, Peretó J, Porcar M, Getino L, Olivera ER, Luengo JM, Castro L, et al. Xerotolerance: A New Property in Exiguobacterium Genus. Microorganisms. 2021; 9(12):2455. https://doi.org/10.3390/microorganisms9122455
Chicago/Turabian StyleLópez, María Castillo, Beatriz Galán, Manuel Carmona, Juana María Navarro Llorens, Juli Peretó, Manuel Porcar, Luis Getino, Elías R. Olivera, José M. Luengo, Laura Castro, and et al. 2021. "Xerotolerance: A New Property in Exiguobacterium Genus" Microorganisms 9, no. 12: 2455. https://doi.org/10.3390/microorganisms9122455
APA StyleLópez, M. C., Galán, B., Carmona, M., Navarro Llorens, J. M., Peretó, J., Porcar, M., Getino, L., Olivera, E. R., Luengo, J. M., Castro, L., & García, J. L. (2021). Xerotolerance: A New Property in Exiguobacterium Genus. Microorganisms, 9(12), 2455. https://doi.org/10.3390/microorganisms9122455









