Insights into the Bacterial and Nitric Oxide-Induced Salt Tolerance in Sugarcane and Their Growth-Promoting Abilities
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Soil Samples and Isolation of Rhizobacteria
2.2. Intrinsic Salt (NaCl) Tolerance Assay
2.3. Morphological and Biochemical Characterization
2.4. Screening of Plant-Growth-Promoting (PGP) Attributes and Exopolysaccharide Production
2.4.1. Indole-3-Acetic Acid (IAA) Production
2.4.2. ACC Deaminase Activity
2.4.3. Phosphate (P) Solubilization
2.4.4. Siderophore Production
2.4.5. Hydrogen Cyanide (HCN) Production
2.4.6. Ammonia Production
2.4.7. Exopolysaccharide (EPS) Production Assay
2.5. Analysis of Biofilm Formation
2.6. Molecular Identification Based on 16S rRNA Gene Sequencing Analysis
2.7. Greenhouse Study for Plant Growth Promotion
2.8. Measurement of Photosynthetic Characteristics
2.9. Relative Water Content (RWC) Measurement
2.10. Electrolyte Leakage (EL) Assay
2.11. Salt Tolerance Index (STI)
2.12. Analysis of Na+/K+ Ion Content
2.13. Chlorophyll Content (SPAD Meter Values)
2.14. Proline Content
2.15. Total Soluble Sugar (TSS) Content
2.16. ROS Scavenging Antioxidant Enzymes Activity Assay
2.16.1. CAT Activity
2.16.2. SOD Activity
2.16.3. PPO Activity
2.16.4. POD Activity
2.16.5. MDA Content
2.17. Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR) of Stress-Related Gene Expression
2.18. Statistical Analysis
3. Results
3.1. Isolation and Characterization of the Salt-Tolerant Rhizobacterial Strain
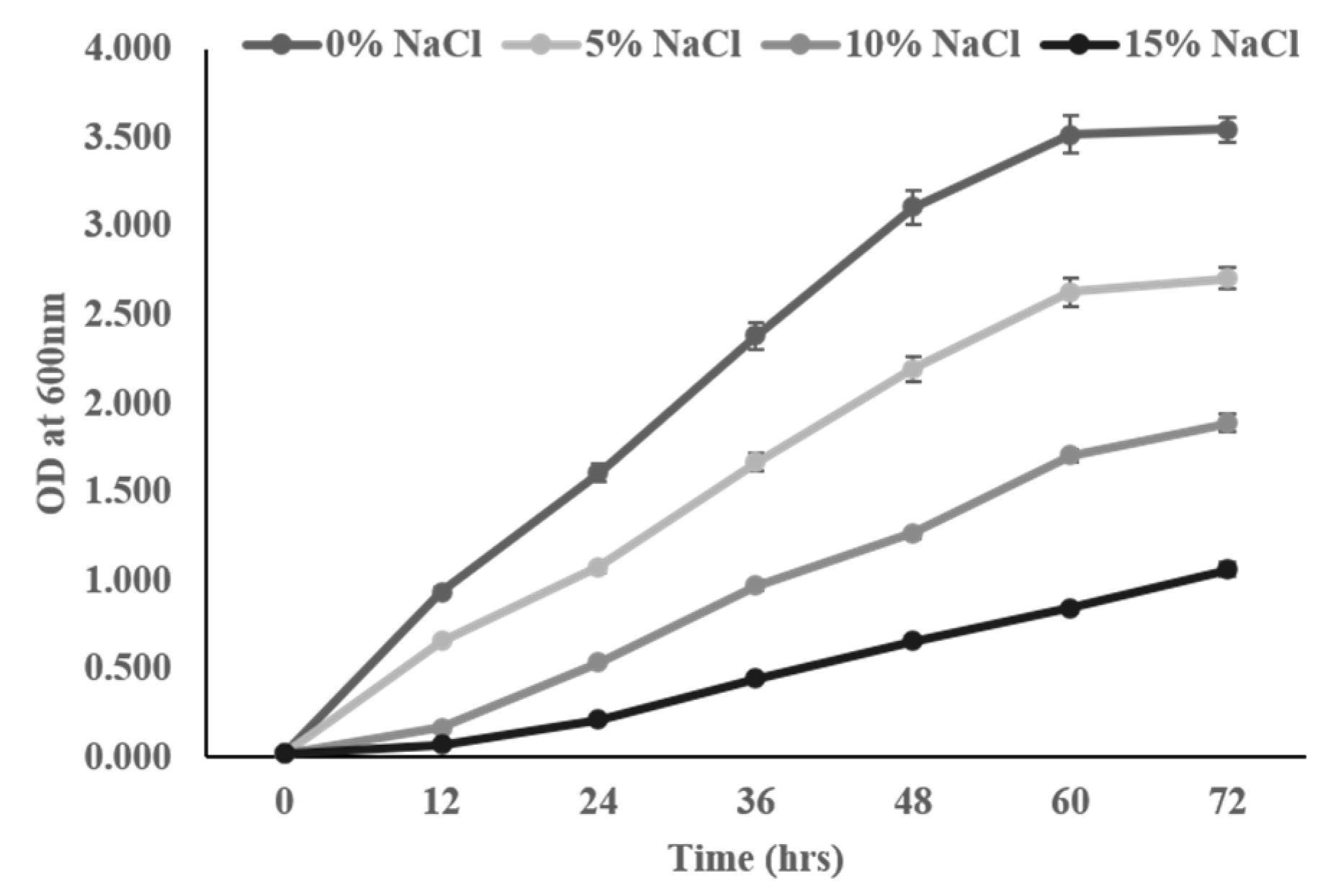
3.2. Intrinsic Salt Stress Tolerance Assay
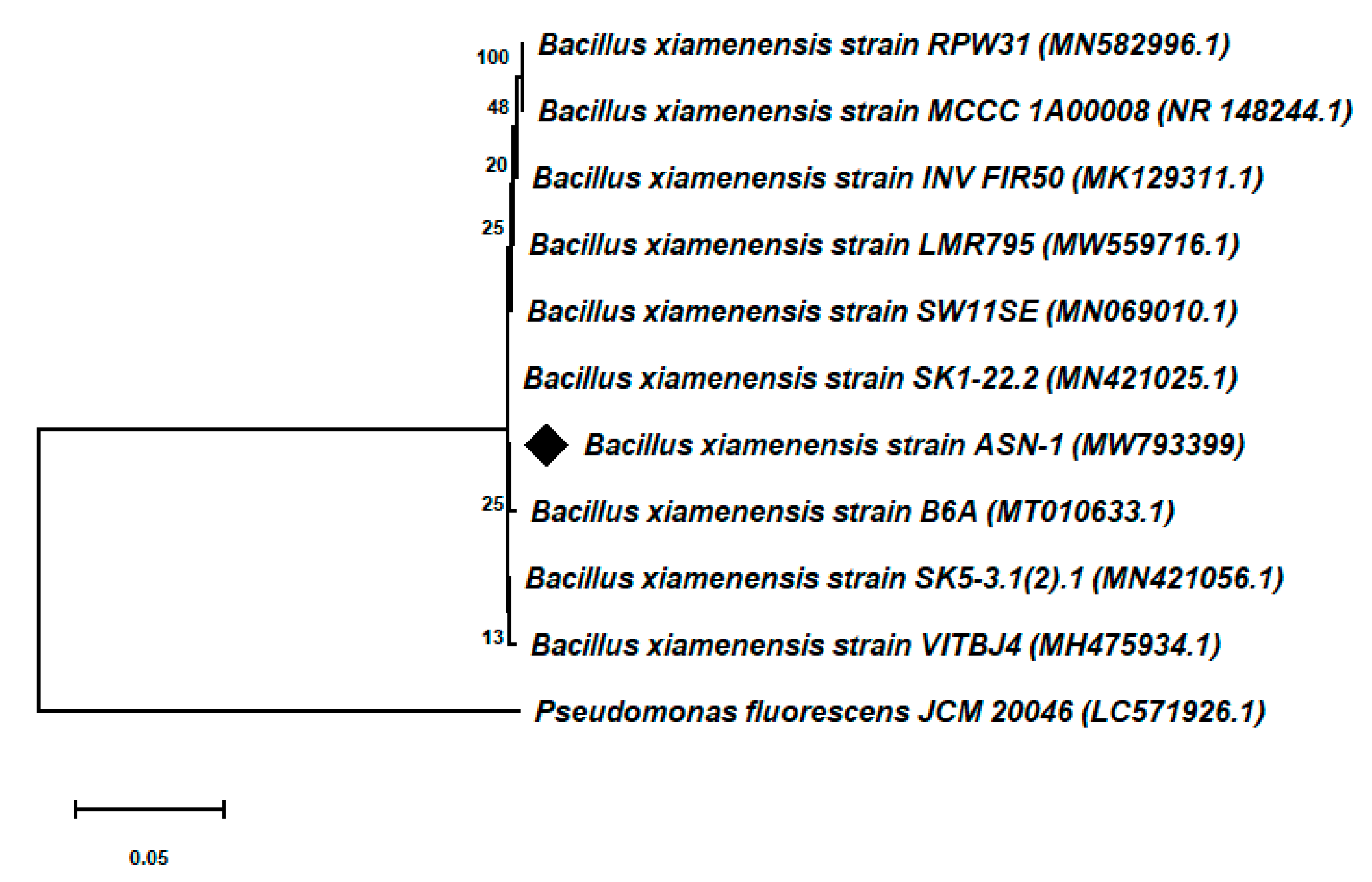
3.3. Molecular Identification and Phylogenetic Analysis
3.4. Screening for Plant-Growth-Promoting Attributes
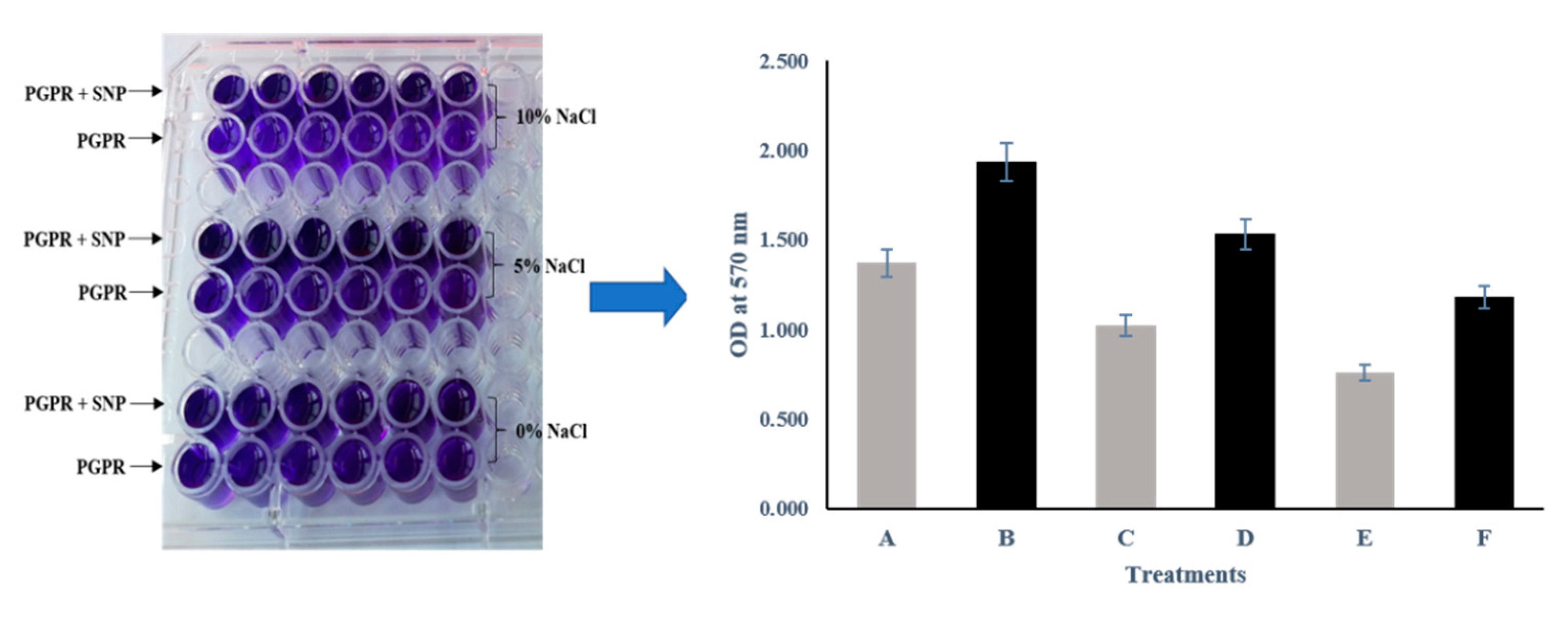
3.5. Biofilm Formation
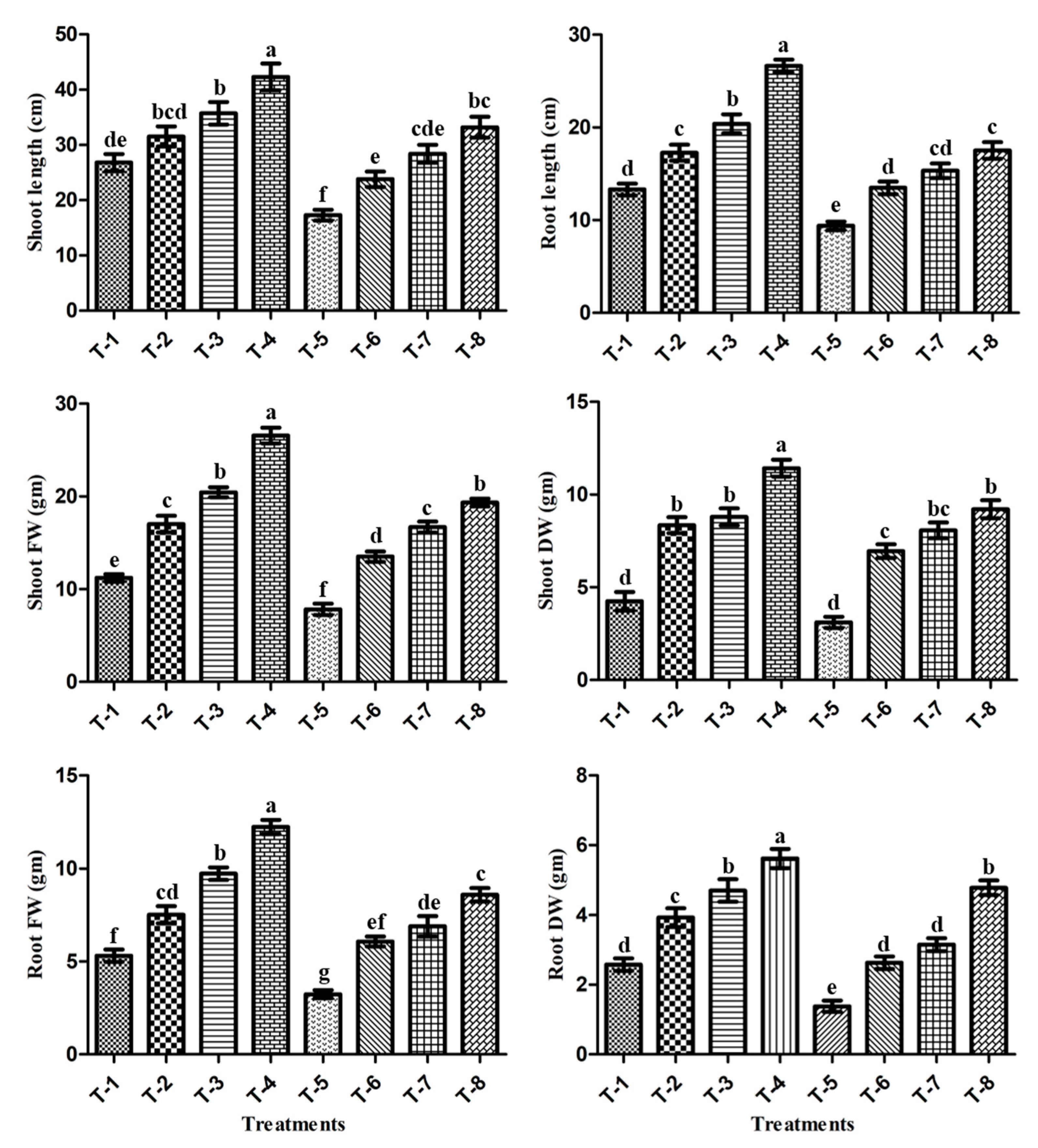
3.6. Effect of PGPR Isolates ASN-1 and SNP (NO Donor) on the Mitigation of Salt Stress and the Growth Promotion of Sugarcane under Greenhouse Conditions
3.7. RWC and Electrolytic Leakage (EL)
3.8. STI Index
3.9. Analysis of Ionic Content
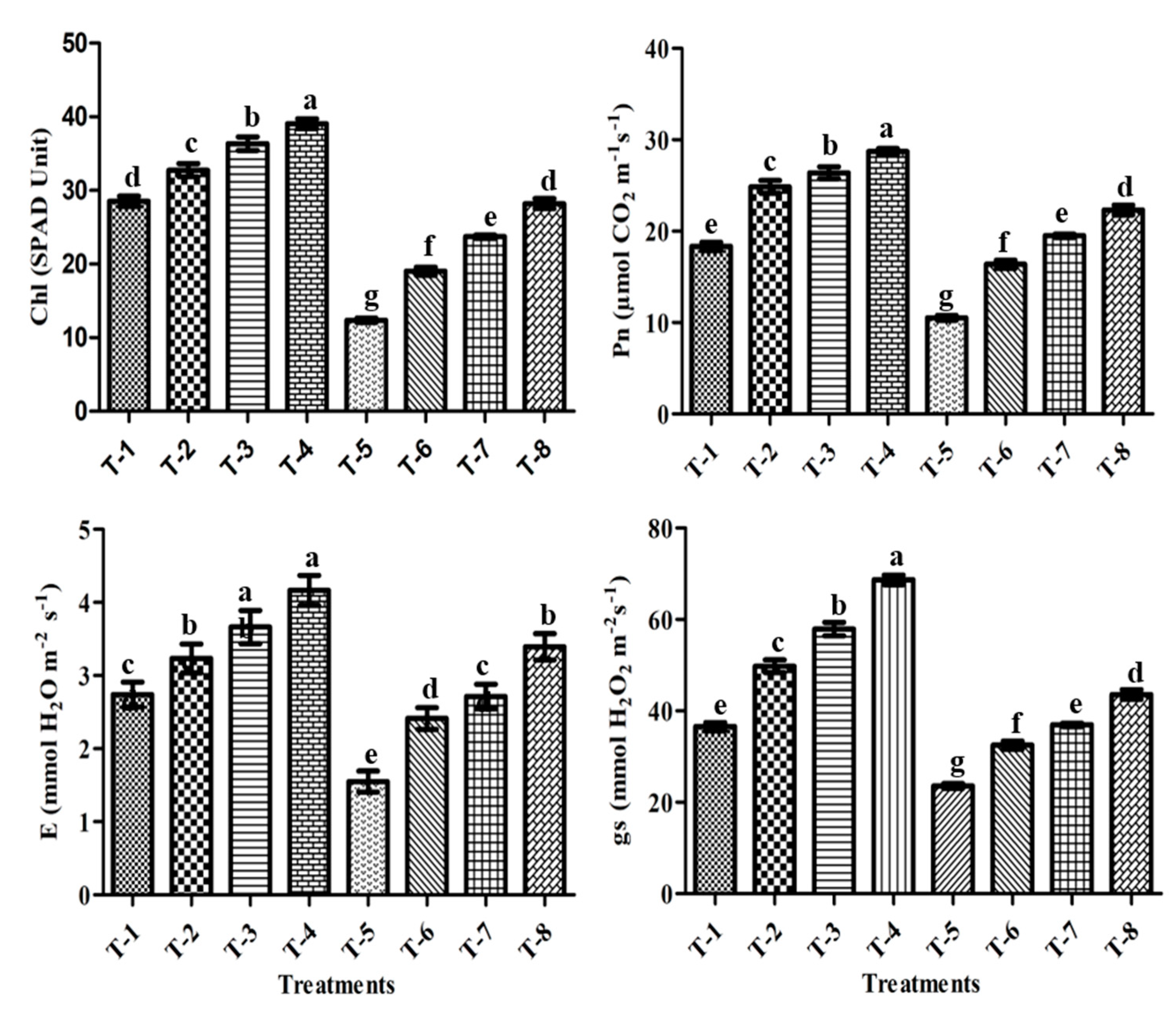
3.10. Chlorophyll Content (SPAD Value)
3.11. Gas-Exchange Parameters
3.12. Proline and TSS Contents
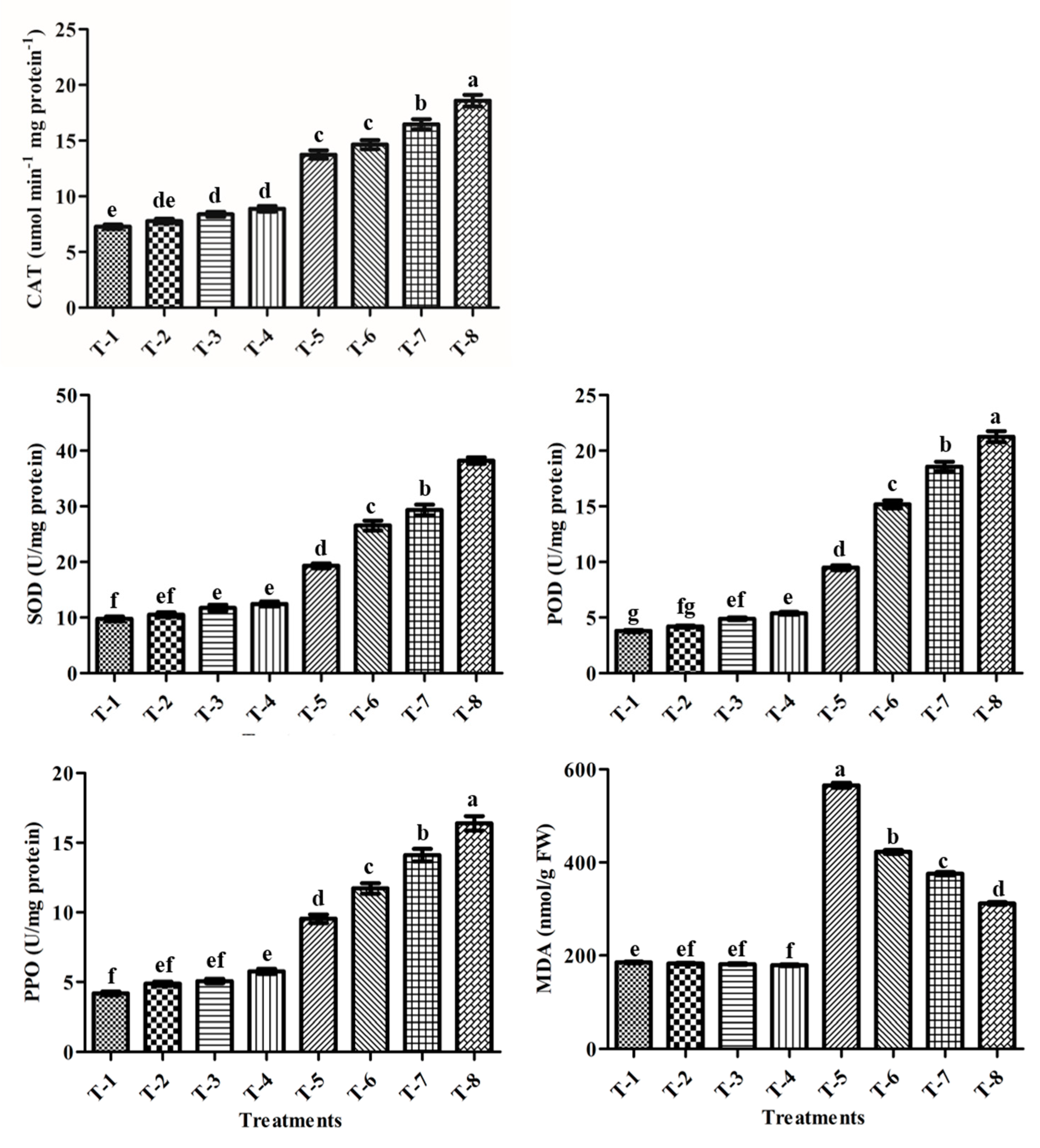
3.13. Antioxidants Enzyme Activities
3.14. MDA Contents
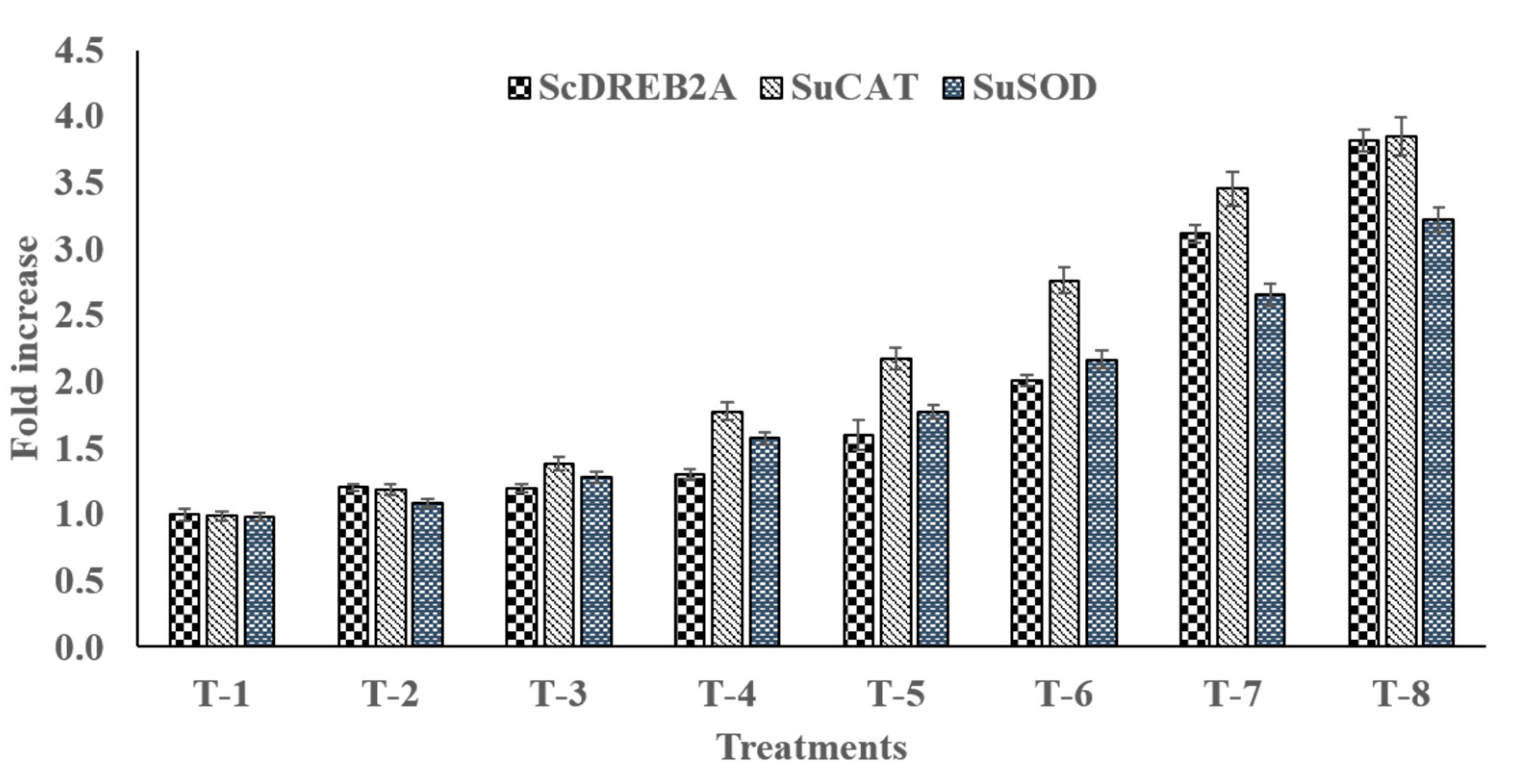
3.15. qRT-PCR Gene Expression Analysis
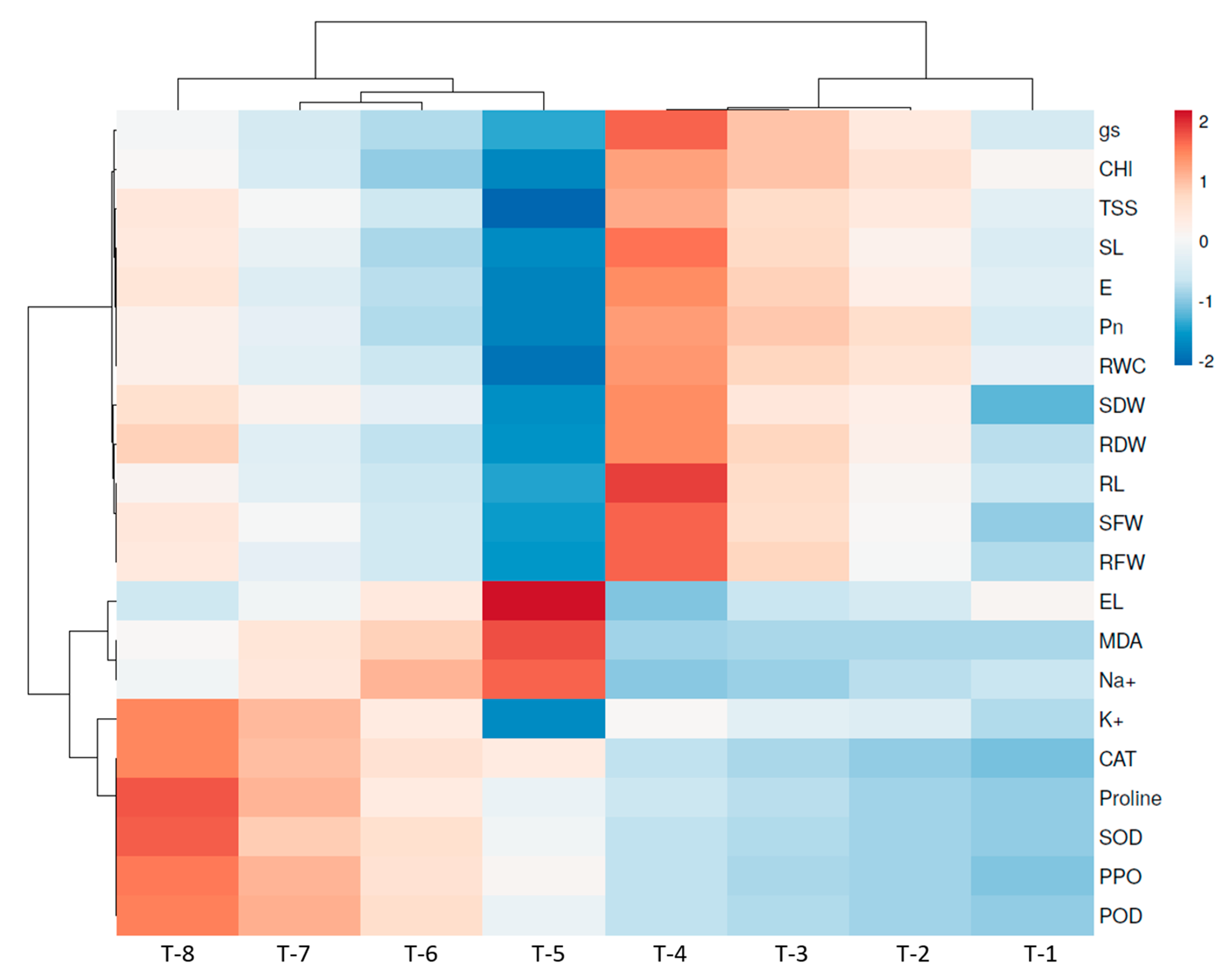
3.16. Heatmap of Pearson’s Correlation Analysis
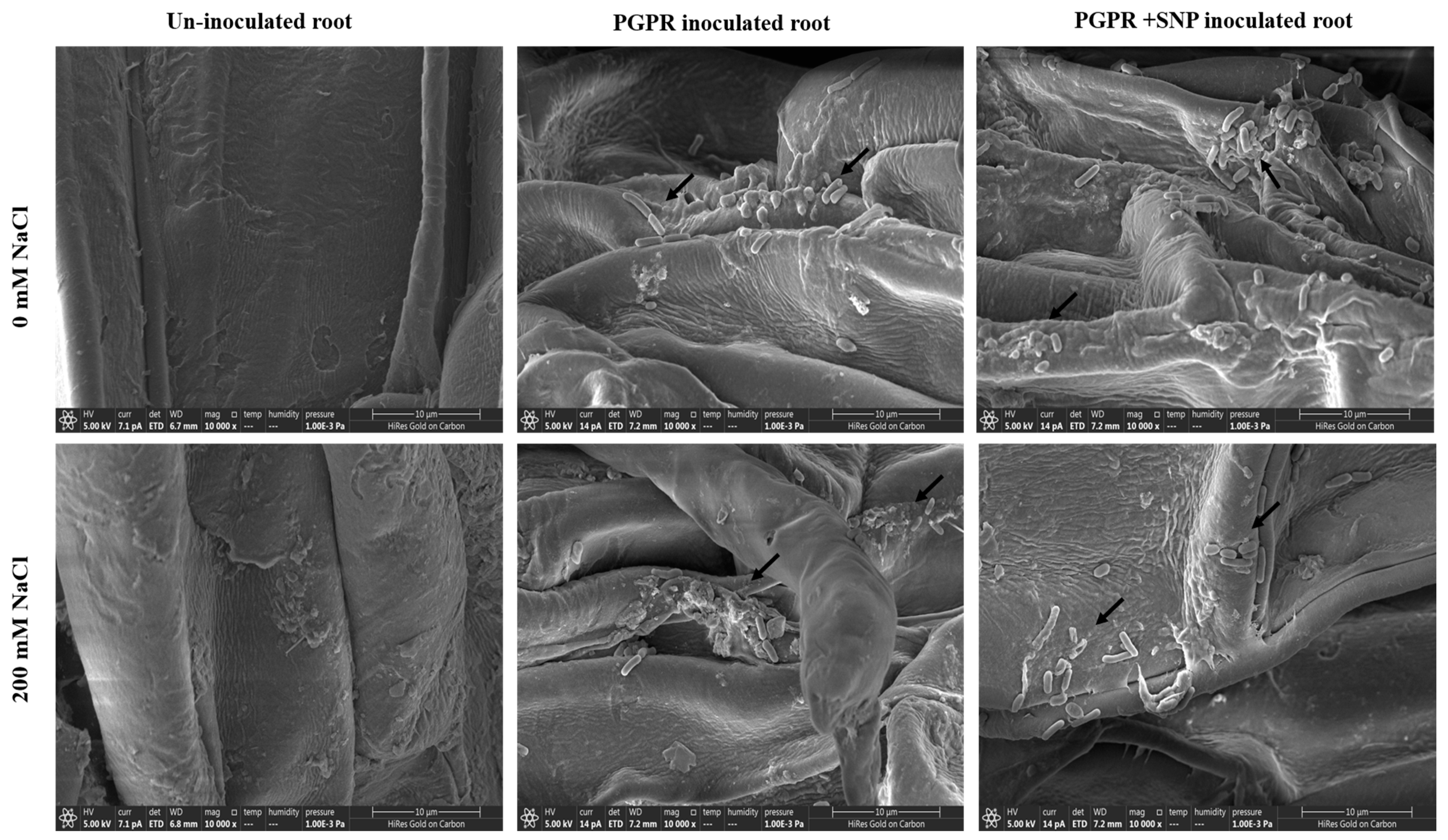
3.17. Root Colonization of PGPR Isolates ASN-1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dos Reis, S.P.; Lima, A.M.; de Souza, C.R. Recent molecular advances on downstream plant responses to abiotic stress. Int. J. Mol. Sci. 2012, 13, 8628–8647. [Google Scholar] [CrossRef]
- Vaishnav, A.; Singh, J.; Singh, P.; Rajput, R.S.; Singh, H.B.; Sarma, B.K. Sphingobacterium sp. BHU-AV3 Induces Salt Tolerance in Tomato by Enhancing Antioxidant Activities and Energy Metabolism. Front. Microbiol. 2020, 11, 443. [Google Scholar] [CrossRef]
- FAO. The State of Food Insecurity in the World 2015. Meeting the 2015 International Hunger Targets: Taking Stock of Uneven Progress; FAO: Rome, Italy, 2015. [Google Scholar]
- Parihar, P.; Singh, S.; Singh, R.; Singh, V.P.; Prasad, S.M. Effect of salinity stress on plants and its tolerance strategies: A review. Environ. Sci. Pollut. Res. Int. 2015, 22, 4056–4075. [Google Scholar] [CrossRef]
- Kumar, A.; Singh, S.; Gaurav, A.K.; Srivastava, S.; Verma, J.P. Plant Growth-Promoting Bacteria: Biological Tools for the Mitigation of Salinity Stress in Plants. Front. Microbiol. 2020, 11, 1216. [Google Scholar] [CrossRef] [PubMed]
- Hassani, A.; Azapagic, A.; Shokri, N. Predicting long-term dynamics of soil salinity and sodicity on a global scale. Proc. Natl. Acad. Sci. USA 2020, 117, 33017–33027. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Adl, S.M. Can interaction between silicon and non–rhizobial bacteria benefit in improving nodulation and nitrogen fixation in salinity–stressed legumes? A review. Rhizosphere 2020, 100229. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [Green Version]
- Arora, N.K.; Fatima, T.; Mishra, J.; Mishra, I.; Verma, S.; Verma, R.; Verma, M.; Bhattacharya, A.; Verma, P.; Mishra, P.; et al. Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 2020, 26, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 2018, 217, 523–539. [Google Scholar] [CrossRef] [Green Version]
- Akyol, T.Y.; Yilmaz, O.; Uzilday, B.; Uzilday, R.Ö.; Türkan, İ. Plant response to salinity: An analysis of ROS formation, signaling, and antioxidant defense. Turk. J. Bot. 2020, 44, 1–13. [Google Scholar]
- Egamberdieva, D.; Wirth, S.; Bellingrath-Kimura, S.D.; Mishra, J.; Arora, N.K. Salt-Tolerant Plant Growth Promoting Rhizobacteria for Enhancing Crop Productivity of Saline Soils. Front. Microbiol. 2019, 10, 2791. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nabi, R.B.S.; Tayade, R.; Hussain, A.; Kulkarni, K.P.; Imran, Q.M.; Mun, B.G.; Yun, B.W. Nitric oxide regulates plant responses to drought, salinity, and heavy metal stress. Environ. Exp. Bot. 2019, 161, 120–133. [Google Scholar] [CrossRef]
- Fita, A.; Rodríguez-Burruezo, A.; Boscaiu, M.; Prohens, J.; Vicente, O. Breeding and domesticating crops adapted to drought and salinity: A new paradigm for increasing food production. Front. Plant Sci. 2015, 6, 978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, T.M.L.; Tran, T.N.; Nguyen, T.K.T.; Williams, B.; Wurm, P.; Bellairs, S.; Mundree, S. Improvement of salinity stress tolerance in rice: Challenges and opportunities. Agron 2016, 6, 54. [Google Scholar] [CrossRef]
- Dadshani, S.; Sharma, R.C.; Baum, M.; Ogbonnaya, F.C.; Léon, J.; Ballvora, A. Multi-dimensional evaluation of response to salt stress in wheat. PLoS ONE 2019, 14, e0222659. [Google Scholar] [CrossRef]
- Billah, M.; Khan, M.; Bano, A.; Nisa, S.; Hussain, A.; Dawar, K.M.; Munir, A.; Khan, N. Rock Phosphate-Enriched Compost in Combination with Rhizobacteria; A Cost-Effective Source for Better Soil Health and Wheat (Triticum aestivum) Productivity. Agronomy 2020, 10, 1390. [Google Scholar] [CrossRef]
- Nawaz, A.; Shahbaz, M.; Imran, A.; Marghoob, M.U.; Imtiaz, M.; Mubeen, F. Potential of salt tolerant PGPR in growth and yield augmentation of wheat (Triticum aestivum L.) under saline conditions. Front. Microbiol. 2020, 11, 2019. [Google Scholar] [CrossRef]
- Numan, M.; Bashir, S.; Khan, Y.; Mumtaz, R.; Shinwari, Z.K.; Khan, A.L.; Khan, A.; Ahmed, A.H. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: A review. Microbiol. Res. 2018, 209, 21–32. [Google Scholar] [CrossRef]
- Bhat, M.A.; Kumar, V.; Bhat, M.A.; Wani, I.A.; Dar, F.L.; Farooq, I.; Jan, A.T. Mechanistic insights of the interaction of plant growth-promoting rhizobacteria (PGPR) with plant roots toward enhancing plant productivity by alleviating salinity stress. Front. Microbiol. 2020, 11, 1952. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, A.; Shukla, A.K.; Sharma, A.; Kumar, R.; Choudhary, D.K. Endophytic bacteria in plant salt stress tolerance: Current and future prospects. J. Plant Growth Regul. 2019, 38, 650–668. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A.; Curá, J.A. Role of Beneficial Microorganisms and Salicylic Acid in Improving Rainfed Agriculture and Future Food Safety. Microorganisms 2020, 8, 1018. [Google Scholar] [CrossRef] [PubMed]
- Jamali, H.; Sharma, A.; Srivastava, A.K. Biocontrol potential of Bacillus subtilis RH5 against sheath blight of rice caused by Rhizoctonia solani. J. Basic Microbiol. 2020, 60, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Mokrani, S.; Nabti, E.H.; Cruz, C. Current advances in plant growth promoting bacteria alleviating salt stress for sustainable agriculture. Appl. Sci. 2020, 10, 7025. [Google Scholar] [CrossRef]
- Khan, N. Application of Plant Growth Promoting Microorganism and Plant Growth Regulators in Agricultural Production and Research. Agronomy 2021, 11, 524. [Google Scholar] [CrossRef]
- Gupta, B.; Huang, B. Mechanism of salinity tolerance in plants: Physiological, biochemical, and molecular characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhu, W.; Zhang, H.; Ding, H.; Zhang, H.J. Exogenous nitric oxide protects against salt-induced oxidative stress in the leaves from two genotypes of tomato (Lycopersicom esculentum Mill.). Acta Physiol. Plant. 2011, 33, 1199–1209. [Google Scholar] [CrossRef]
- Ghadakchiasl, A.; Mozafari, A.A.; Ghaderi, N. Mitigation by sodium nitroprusside of the effects of salinity on the morpho-physiological and biochemical characteristics of Rubus idaeus under in vitro conditions. Physiol. Mol. Biol. Plants 2017, 23, 73–83. [Google Scholar] [CrossRef] [Green Version]
- Prakash, V.; Vishwakarma, K.; Singh, V.P.; Rai, P.; Ramawat, N.; Tripathi, D.K.; Sharma, S. NO and ROS implications in the organization of root system architecture. Physiol. Plant 2020, 168, 473–489. [Google Scholar] [CrossRef]
- Parankusam, S.; Adimulam, S.S.; Bhatnagar-Mathur, P.; Sharma, K.K. Nitric oxide (NO) in plant heat stress tolerance: Current knowledge and perspectives. Front. Plant Sci. 2017, 8, 1582. [Google Scholar] [CrossRef]
- Siddiqui, M.H.; Al-Whaibi, M.H.; Basalah, M.O. Role of nitric oxide in tolerance of plants to abiotic stress. Protoplasma 2011, 248, 447–455. [Google Scholar] [CrossRef]
- Ahmad, P.; Ahanger, M.A.; Alyemeni, M.N.; Wijaya, L.; Alam, P. Exogenous application of nitric oxide modulates osmolyte metabolism, antioxidants, enzymes of ascorbate-glutathione cycle and promotes growth under cadmium stress in tomato. Protoplasma 2018, 255, 79–93. [Google Scholar] [CrossRef]
- Rather, B.A.; Mir, I.R.; Masood, A.; Anjum, N.A.; Khan, N.A. Nitric oxide pre-treatment advances seed germination and alleviates copper-induced photosynthetic inhibition in Indian mustard. Plants 2020, 9, 776. [Google Scholar] [CrossRef]
- Tang, M.; Li, Z.; Luo, L.; Cheng, B.; Zhang, Y.; Zeng, W.; Peng, Y. Nitric Oxide Signal, Nitrogen Metabolism, and Water Balance Affected by γ-Aminobutyric Acid (GABA) in Relation to Enhanced Tolerance to Water Stress in Creeping Bentgrass. Int. J. Mol. Sci. 2020, 21, 7460. [Google Scholar] [CrossRef]
- FAO. Top Sugarcane Production: Food and Agriculture Organization of the United Nations; FAO: Rome, Italy, 2016. [Google Scholar]
- Li, Y.R.; Yang, L.T. Sugarcane agriculture and sugar industry in China. Sugar Tech. 2015, 17, 1–8. [Google Scholar] [CrossRef]
- Gomathi, R.; Thandapani, P. Influence of salinity stress on growth parameters and yield of sugarcane. IOSR J. Pharm. Biol. Sci. 2014, 93, 28–32. [Google Scholar] [CrossRef]
- Simões, W.L.; Calgaro, M.; Coelho, D.S.; Santos, D.B.D.; Souza, M.A.D. Growth of sugar cane varieties under salinity. Rev. Ceres 2016, 632, 265–271. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Zhu, K.; Momotaz, A.; Gao, X. Sugarcane Plant Growth and Physiological Responses to Soil Salinity during Tillering and Stalk Elongation. Agriculture 2020, 1012, 608. [Google Scholar] [CrossRef]
- Khan, N.; Zandi, P.; Ali, S.; Mehmood, A.; Shahid, M.A.; Yang, J. Impact of salicylic acid and PGPR on the drought tolerance and phytoremediation potential of Helianthus annus. Front. Microbiol. 2018, 9, 2507. [Google Scholar] [CrossRef] [Green Version]
- Pirhadi, M.; Enayatizamir, N.; Motamedi, H.; Sorkheh, K. Screening of salt tolerant sugarcane endophytic bacteria with potassium and zinc for their solubilizing and antifungal activity. Biosci. Biotechnol. Res. Commun. 2016, 93, 530–538. [Google Scholar] [CrossRef]
- Tindall, B.J.; Rosselló-Móra, R.; Busse, H.J.; Ludwig, W.; Kämpfer, P. Notes on the characterization of prokaryote strains for taxonomic purposes. Int. J. Syst. Evol. 2010, 60, 249–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, S.; Pandey, S. ACC deaminase producing bacteria with multifarious plant growth promoting traits alleviates salinity stress in French bean (Phaseolus vulgaris) plants. Front. Microbiol. 2019, 10, 1506. [Google Scholar] [CrossRef]
- Misra, S.; Chauhan, P.S. ACC deaminase-producing rhizosphere competent Bacillus spp. mitigate salt stress and promote Zea mays growth by modulating ethylene metabolism. 3 Biotech 2020, 10, 1–14. [Google Scholar] [CrossRef]
- Passari, A.K.; Chandra, P.; Mishra, V.K.; Leo, V.V.; Gupta, V.K.; Kumar, B.; Singh, B.P. Detection of biosynthetic gene and phytohormone production by endophytic actinobacteria associated with Solanum lycopersicum and their plant-growth-promoting effect. Res. Microbiol. 2016, 167, 692–705. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Hao, T.; Chen, S. Colonization and maize growth promotion induced by phosphate solubilizing bacterial isolates. Int. J. Mol. Sci. 2017, 18, 1253. [Google Scholar] [CrossRef] [Green Version]
- Chakdar, H.; Dastager, S.G.; Khire, J.M.; Rane, D.; Dharne, M.S. Characterization of mineral phosphate solubilizing and plant growth promoting bacteria from termite soil of arid region. 3 Biotech 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Singh, R.; Jha, P.N. Alleviation of salinity-induced damage on wheat plant by an ACC deaminase-producing halophilic bacterium Serratia sp. SL-12 isolated from a Salt lake. Symbiosis 2016, 69, 101–111. [Google Scholar] [CrossRef]
- Singh, P.; Singh, R.K.; Li, H.B.; Guo, D.J.; Sharma, A.; Lakshmanan, P.; Malviya, M.K.; Song, X.P.; Solanki, M.K.; Verma, K.K.; et al. Diazotrophic bacteria Pantoea dispersa and Enterobacter asburiae promote sugarcane growth by inducing nitrogen uptake and defense-related gene expression. Front. Microbiol. 2021, 11, 600417. [Google Scholar] [CrossRef]
- Dey, R.; Pal, K.K.; Bhatt, D.M.; Chauhan, S.M. Growth promotion and yield enhancement of peanut (Arachis hypogaea L.) by application of plant growth-promoting rhizobacteria. Microbiol. Res. 2004, 159, 371–394. [Google Scholar] [CrossRef]
- Kumari, S.; Vaishnav, A.; Jain, S.; Varma, A.; Choudhary, D.K. Bacterial-mediated induction of systemic tolerance to salinity with expression of stress alleviating enzymes in soybean Glycine max L. Merrill. J. Plant Growth Regul. 2015, 343, 558–573. [Google Scholar] [CrossRef]
- Qurashi, A.W.; Sabri, A.N. Osmoadaptation and plant growth promotion by salt tolerant bacteria under salt stress. Afr. J. Microbiol. Res. 2011, 521, 3546–3554. [Google Scholar]
- Sharma, A.; Kashyap, P.L.; Srivastava, A.K.; Bansal, Y.K.; Kaushik, R. Isolation and characterization of salt-tolerant bacilli from chickpea (Cicer arietinum L.) rhizosphere for plant growth promotion and biocontrol traits. Eur. J. Plant Pathol. 2019, 153, 787–800. [Google Scholar] [CrossRef]
- Guo, D.J.; Singh, R.K.; Singh, P.; Li, D.P.; Sharma, A.; Xing, Y.X.; Song, X.P.; Yang, L.T.; Li, Y.R. Complete genome sequence of Enterobacter roggenkampii ED5, a nitrogen fixing plant growth promoting endophytic bacterium with biocontrol and stress tolerance properties, isolated from sugarcane root. Front. Microbiol. 2020, 11, 2270. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, D.P.; Saikia, R. Genetic diversity of plant growth promoting rhizobacteria isolated from rhizosphere soil of wheat under saline condition. Curr. Microbiol. 2009, 59, 489–496. [Google Scholar] [CrossRef]
- Singh, R.K.; Singh, P.; Li, H.B.; Song, Q.Q.; Guo, D.J.; Solanki, M.K.; Verma, K.K.; Malviya, M.K.; Song, X.P.; Lakshmanan, P.; et al. Diversity of nitrogen-fixing rhizobacteria associated with sugarcane: A comprehensive study of plant-microbe interactions for growth enhancement in Saccharum spp. BMC Plant Biol. 2020, 20, 1–21. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Anee, T.I.; Khan, M.I.R.; Fujita, M. Silicon-mediated regulation of antioxidant defense and glyoxalase systems confers drought stress tolerance in Brassica napus L. S. Afr. J. Bot. 2018, 115, 50–57. [Google Scholar] [CrossRef]
- El-Tayeb, M.A. Response of barley grains to the interactive e. ect of salinity and salicylic acid. Plant Growth Regul. 2005, 45, 215–224. [Google Scholar] [CrossRef]
- Al-Garni, S.M.; Khan, M.M.A.; Bahieldin, A. Plant growth-promoting bacteria and silicon fertilizer enhance plant growth and salinity tolerance in Coriandrum Sativum. J. Plant Interact. 2019, 14, 386–396. [Google Scholar] [CrossRef] [Green Version]
- Jangpromma, N.; Songsri, P.; Thammasirirak, S.; Jaisil, P. Rapid assessment of chlorophyll content in sugarcane using a SPAD chlorophyll meter across different water stress conditions. Asian J. Plant Sci. 2010, 96, 368–374. [Google Scholar] [CrossRef] [Green Version]
- Kousar, B.; Bano, A.; Khan, N. PGPR modulation of secondary metabolites in tomato infested with Spodoptera litura. Agronomy 2020, 10, 778. [Google Scholar] [CrossRef]
- Singh, R.P.; Jha, P.N. A halotolerant bacterium Bacillus licheniformis HSW-16 augments induced systemic tolerance to salt stress in wheat plant (Triticum aestivum). Front. Plant Sci. 2016, 7, 1890. [Google Scholar] [CrossRef]
- Qureshi, M.I.; Abdin, M.Z.; Ahmad, J.; Iqbal, M. Effect of long-term salinity on cellular antioxidants, compatible solute and fatty acid profile of Sweet Annie (Artemisia annua L.). Phytochem 2013, 95, 215–223. [Google Scholar] [CrossRef]
- Weisany, W.; Sohrabi, Y.; Heidari, G.; Siosemardeh, A.; Ghassemi-Golezani, K. Changes in antioxidant enzymes activity and plant performance by salinity stress and zinc application in soybean Glycine max L. Plant Omics 2012, 52, 60–67. [Google Scholar]
- Choudhary, D.K. Plant growth-promotion (PGP) activities and molecular characterization of rhizobacterial strains isolated from soybean (Glycine max L. Merril) plants against charcoal rot pathogen, Macrophomina phaseolina. Biotechnol. Lett. 2011, 33, 2287–2295. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, A.; Kumari, S.; Jain, S.; Varma, A.; Tuteja, N.; Choudhary, D.K. PGPR-mediated expression of salt tolerance gene in soybean through volatiles under sodium nitroprusside. J. Basic Microbiol. 2016, 5611, 1274–1288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Fan, C.; Wang, Y.; Xia, Y.; Xiao, W.; Cui, X. Salt-tolerant and plant-growth-promoting bacteria isolated from high-yield paddy soil. Can. J. Microbiol. 2018, 64, 968–978. [Google Scholar] [CrossRef]
- Jamali, H.; Sharma, A.; Kushwaha, P.; Roohi, P.L.K.; Srivastava, A.K. Exploitation of multifarious abiotic stresses, antagonistic activity and plant growth promoting attributes of Bacillus amyloliquefaciens AH53 for sustainable agriculture production. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 751–763. [Google Scholar] [CrossRef]
- Bokhari, A.; Essack, M.; Lafi, F.F.; Andres-Barrao, C.; Jalal, R.; Alamoudi, S.; Saad, M.M. Bioprospecting desert plant Bacillus endophytic strains for their potential to enhance plant stress tolerance. Sci. Rep. 2019, 91, 1–13. [Google Scholar] [CrossRef]
- Sharma, A.; Singh, P.; Kumar, S.; Kashyap, P.L.; Srivastava, A.K.; Chakdar, H.; Singh, R.N.; Kaushik, R.K.; Saxena, A.K.; Sharma, A.K. Deciphering diversity of salt-tolerant bacilli from saline soils of eastern Indo-gangetic plains of India. Geomicrobiol. J. 2015, 32, 170–180. [Google Scholar] [CrossRef]
- Khan, N.; Ali, S.; Shahid, M.A.; Mustafa, A.; Sayyed, R.Z.; Curá, J.A. Insights into the Interactions among Roots, Rhizosphere, and Rhizobacteria for Improving Plant Growth and Tolerance to Abiotic Stresses: A Review. Cells 2021, 10, 1551. [Google Scholar] [CrossRef]
- Shultana, R.; Kee Zuan, A.T.; Yusop, M.R.; Saud, H.M. Characterization of salt-tolerant plant growth-promoting rhizobacteria and the effect on growth and yield of saline-affected rice. PLoS ONE 2020, 159, e0238537. [Google Scholar] [CrossRef]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 2020, 140682. [Google Scholar] [CrossRef]
- Ha-Tran, D.M.; Nguyen, T.T.M.; Hung, S.H.; Huang, E.; Huang, C.C. Roles of plant growth-promoting rhizobacteria PGPR) in stimulating salinity stress defense in plants: A Review. Int. J. Mol. Sci. 2021, 226, 3154. [Google Scholar] [CrossRef] [PubMed]
- Grover, M.; Bodhankar, S.; Sharma, A.; Sharma, P.; SINGH, J.; Nain, L. PGPR mediated alterations in root traits: Way towards sustainable crop production. Front. Sustain. Food Syst. 2020, 4, 287. [Google Scholar]
- Dimkpa, C.; Weinand, T.; Asch, F. Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 2009, 3212, 1682–1694. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.R. Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 2014, 1691, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Hashem, A.; Abd_Allah, E.F.; Alqarawi, A.A.; Al-Huqail, A.A.; Wirth, S.; Egamberdieva, D. The interaction between arbuscular mycorrhizal fungi and endophytic bacteria enhances plant growth of Acacia gerrardii under salt stress. Front. Microbiol. 2016, 7, 1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shultana, R.; Tan Kee Zuan, A.; Yusop, M.R.; Mohd Saud, H.; Ayanda, A.F. Effect of salt-tolerant bacterial inoculations on rice seedlings differing in salt-tolerance under saline soil conditions. Agronomy 2020, 107, 1030. [Google Scholar] [CrossRef]
- Khan, N.; Bano, A. Growth and Yield of Field Crops Grown under Drought Stress Condition is Influenced by the Application of PGPR. In Field Crops: Sustainable Management by PGPR; Springer: Cham, Switzerland, 2019; pp. 337–349. [Google Scholar]
- Sarropoulou, V.; Dimassi-Theriou, K.; Therios, I. In vitro plant regeneration from leaf explants of the cherry rootstocks CAB-6P, Gisela 6, and MxM 14 using sodium nitroprusside. In Vitro Cell Dev. Biol. Plant. 2014, 50, 226–234. [Google Scholar] [CrossRef]
- Chen, Z.; Zhao, X.; Hu, Z.; Leng, P. Nitric oxide modulating ion balance in Hylotelephium erythrostictum roots subjected to NaCl stress based on the analysis of transcriptome, fluorescence, and ion fluxes. Sci. Rep. 2019, 91, 1–12. [Google Scholar] [CrossRef]
- Panahi, B. Effects of osmotic and salt stresses on water relation parameters of pistachio seedlings. Plant Ecophysiol. 2009, 1, 1–8. [Google Scholar]
- Khan, M.N.; Siddiqui, M.H.; Mohammad, F.; Naeem, M. Interactive role of nitric oxide and calcium chloride in enhancing tolerance to salt stress. Nitric Oxide 2012, 27, 210–218. [Google Scholar] [CrossRef] [PubMed]
- Mahmood, S.; Daur, I.; Al-Solaimani, S.G.; Ahmad, S.; Madkour, M.H.; Yasir, M.; Ali, Z. Plant growth promoting rhizobacteria and silicon synergistically enhance salinity tolerance of mung bean. Front. Plant Sci. 2016, 7, 876. [Google Scholar] [CrossRef] [Green Version]
- Ilyas, N.; Mazhar, R.; Yasmin, H.; Khan, W.; Iqbal, S.; Enshasy, H.E.; Dailin, D.J. Rhizobacteria isolated from saline soil induce systemic tolerance in wheat (Triticum aestivum L.) against salinity stress. Agronomy 2020, 10, 989. [Google Scholar] [CrossRef]
- Romero-Munar, A.; Baraza, E.; Gulías, J.; Cabot, C. Arbuscular mycorrhizal fungi confer salt tolerance in giant reed (Arundo donax L.) plants grown under low phosphorus by reducing leaf Na+ concentration and improving phosphorus use efficiency. Front. Plant Sci. 2019, 10, 843. [Google Scholar] [CrossRef] [Green Version]
- Abdelaziz, M.E.; Abdelsattar, M.; Abdeldaym, E.A.; Atia, M.A.; Mahmoud, A.W.M.; Saad, M.M.; Hirt, H. Piriformospora indica alters Na+/K+ homeostasis, antioxidant enzymes and LeNHX1 expression of greenhouse tomato grown under salt stress. Sci. Hortic. 2019, 256, 108532. [Google Scholar] [CrossRef]
- Zhao, L.; Zhang, F.; Guo, J.; Yang, Y.; Li, B.; Zhang, L. Nitric oxide functions as a signal in salt resistance in the calluses from two ecotypes of reed. Plant Physiol. 2004, 134, 849–857. [Google Scholar] [CrossRef] [Green Version]
- Barnawal, D.; Bharti, N.; Pandey, S.S.; Pandey, A.; Chanotiya, C.S.; Kalra, A. Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plant. 2017, 161, 502–514. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.; Ali, S.; Tariq, H.; Latif, S.; Yasmin, H.; Mehmood, A.; Shahid, M.A. Water Conservation and Plant Survival Strategies of Rhizobacteria under Drought Stress. Agronomy 2020, 10, 1683. [Google Scholar] [CrossRef]
- Dodd, I.C.; Zinovkina, N.Y.; Safronova, V.I.; Belimov, A.A. Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 2010, 157, 361–379. [Google Scholar] [CrossRef]
- Bürling, K.; Cerovic, Z.G.; Cornic, G.; Ducruet, J.M.; Noga, G.; Hunsche, M. Fluorescence-based sensing of drought-induced stress in the vegetative phase of four contrasting wheat genotypes. Environ. Exp. Bot. 2013, 89, 51–59. [Google Scholar] [CrossRef]
- Verma, K.K.; Song, X.P.; Zeng, Y.; Guo, D.J.; Singh, M.; Rajput, V.D.; Malviya, M.K.; Wei, K.J.; Sharma, A.; Li, D.P.; et al. Foliar application of silicon boosts growth, photosynthetic leaf gas exchange, antioxidative response and resistance to limited water irrigation in sugarcane (Saccharum officinarum L.). Plant Physiol. Biochem. 2021, 166, 582–592. [Google Scholar] [CrossRef]
- Mohamed, I.A.; Shalby, N.; Bai, C.; Qin, M.; Agami, R.A.; Jie, K.; Wang, B.; Zhou, G. Stomatal and photosynthetic traits are associated with investigating sodium chloride tolerance of Brassica napus L. cultivars. Plants 2020, 9, 62. [Google Scholar] [CrossRef] [Green Version]
- Kanwal, H.; Ashraf, M.; Shahbaz, M. Assessment of salt tolerance of some newly developed and candidate wheat (Triticum aestivum L.) cultivars using gas exchange and chlorophyll fluorescence attributes. Pak. J. Bot. 2011, 43, 2693–2699. [Google Scholar]
- Yasin, N.A.; Khan, W.U.; Ahmad, S.R.; Ali, A.; Ahmad, A.; Akram, W. Imperative roles of salt-tolerantplant growth-promoting rhizobacteria and kinetin in improving salt tolerance and growth of black gram (Phaseolus mungo). Environ. Sci. Pollut. Res. 2018, 25, 4491–4505. [Google Scholar] [CrossRef]
- Yasmeen, T.; Ahmad, A.; Arif, M.S.; Mubin, M.; Rehman, K.; Shahzad, S.M.; Iqbal, S.; Rizwan, M.; Ali, S.; Alyemeni, M.N.; et al. Biofilm forming rhizobacteria enhance growth and salt tolerance in sunflower plants by stimulating antioxidant enzymes activity. Plant Physiol. Biochem. 2020, 156, 242–256. [Google Scholar] [CrossRef]
- Ansari, F.A.; Ahmad, I.; Pichtel, J. Growth stimulation and alleviation of salinity stress to wheat by the biofilm forming Bacillus pumilus strain FAB10. Appl. Soil Ecol. 2019, 143, 45–54. [Google Scholar] [CrossRef]
- Cechin, I.; Cardoso, G.S.; Fumis, T.D.F.; Corniani, N. Nitric oxide reduces oxidative damage induced by water stress in sunflower plants. Bragantia 2015, 74, 200–206. [Google Scholar] [CrossRef] [Green Version]
- Wani, K.I.; Naeem, M.; Castroverde, C.D.M.; Kalaji, H.M.; Albaqami, M.; Aftab, T. Molecular Mechanisms of Nitric Oxide (NO) Signaling and Reactive Oxygen Species (ROS) Homeostasis during Abiotic Stresses in Plants. Int. J. Mol. Sci. 2021, 22, 9656. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, D.P. Effect of salt-tolerant plant growth-promoting rhizobacteria on wheat plants and soil health in a saline environment. Plant Biol. 2015, 17, 288–293. [Google Scholar] [CrossRef]
- Ashraf, M.F.M.R.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Tuna, A.L.; Kaya, C.; Dikilitas, M.; Higgs, D. The combined effects of gibberellic acid and salinity on some antioxidant enzyme activities, plant growth parameters and nutritional status in maize plants. Environ. Exp. Bot. 2008, 621, 1–9. [Google Scholar] [CrossRef]
- Shukla, P.S.; Agarwal, P.K.; Jha, B. Improved salinity tolerance of (Arachis hypogaea L.) by the interaction of salt-tolerantplant-growth-promoting rhizobacteria. J. Plant Grow. Regul. 2012, 312, 195–206. [Google Scholar] [CrossRef]
- Han, Q.Q.; Lü, X.P.; Bai, J.P.; Qiao, Y.; Paré, P.W.; Wang, S.M.; Zhang, J.L.; Wu, Y.N.; Pang, X.P.; Xu, W.B.; et al. Beneficial soil bacterium Bacillus subtilis (GB03) augments salt tolerance of white clover. Front. Plant Sci. 2014, 5, 525. [Google Scholar] [CrossRef]
- Hakim, S.; Naqqash, T.; Nawaz, M.S.; Laraib, I.; Siddique, M.J.; Zia, R.; Mirza, M.S.; Imran, A. Rhizosphere Engineering with Plant Growth-Promoting Microorganisms for Agriculture and Ecological Sustainability. Front. Sustain. Food Syst. 2021, 5, 16. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Islam, F.; Yasmeen, T.; Arif, M.S.; Ali, S.; Ali, B.; Hameed, S.; Zhou, W. Plant growth promoting bacteria confer salt tolerance in Vigna radiata by up-regulating antioxidant defense and biological soil fertility. Plant. Growth. Regul. 2016, 80, 23–36. [Google Scholar] [CrossRef]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 2020, 11, 552969. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Saxena, A.K.; Singh, D.P. Impact of PGPR inoculation on growth and antioxidant status of wheat under saline conditions. Plant Biol. 2012, 14, 605–611. [Google Scholar] [CrossRef]
- Abd_Allah, E.F.; Alqarawi, A.A.; Hashem, A.; Radhakrishnan, R.; Al-Huqail, A.A.; Al-Otibi, F.O.N.; Malik, J.A.; Alharbi, R.I.; Egamberdieva, D. Endophytic bacterium Bacillus subtilis (BERA 71) improves salt tolerance in chickpea plants by regulating the plant defense mechanisms. J. Plant. Interact. 2018, 13, 37–44. [Google Scholar] [CrossRef] [Green Version]
- Naseem, H.; Ahsan, M.; Shahid, M.A.; Khan, N. Exopolysaccharides producing rhizobacteria and their role in plant growth and drought tolerance. J. Basic Microbiol. 2018, 58, 1009–1022. [Google Scholar] [CrossRef]
- Agami, R.A.; Medani, R.A.; Abd El-Mola, I.A.; Taha, R.S. Exogenous application with plant growth promoting rhizobacteria (PGPR) or proline induces stress tolerance in basil plants (Ocimum basilicum L.) exposed to water stress. Int. J. Environ. Agric. Res. 2016, 2, 78. [Google Scholar]
- Pissolato, M.D.; Silveira, N.M.; Prataviera, P.J.C.; Machado, E.C.; Seabra, A.B.; Pelegrino, M.T.; Sodek, L.; Ribeiro, R.V. Enhanced nitric oxide synthesis through nitrate supply improves drought tolerance of sugarcane plants. Front. Plant. Sci. 2020, 11, 970. [Google Scholar] [CrossRef]
- Moran, J.F.; Becana, M.; Iturbe-Ormaetxe, I.; Frechilla, S.; Klucas, R.V.; Aparicio-Tejo, P. Drought induces oxidative stress in pea plants. Planta 1994, 194, 346–352. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, S.; Prasad, V.; Chauhan, P.S.; Lata, C. Bacillus amyloliquefaciens confers tolerance to various abiotic stresses and modulates plant response to phytohormones through osmoprotection and gene expression regulation in rice. Front. Plant. Sci. 2017, 8, 1510. [Google Scholar] [CrossRef] [Green Version]
- Augustine, S.M.; Narayan, J.A.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Overexpression of EaDREB2 and pyramiding of EaDREB2 with the pea DNA helicase gene (PDH45) enhance drought and salinity tolerance in sugarcane (Saccharum spp. hybrid). Plant Cell. Rep. 2015, 34, 247–263. [Google Scholar] [CrossRef]
- Hu, L.; Huang, Z.; Liu, S.; Fu, J. Growth response and gene expression in antioxidant-related enzymes in two bermudagrass genotypes differing in salt tolerance. J. Am. Soc. Hortic. Sci. 2012, 137, 134–143. [Google Scholar] [CrossRef]
| Salinity Level (% NaCl) | IAA Production (μg/mL) | ACC Deaminase | P-Solubilization Index (PSI) | Siderophore Production | HCN Production | Ammonia Production | EPS Production |
|---|---|---|---|---|---|---|---|
| 0% | 196.48 ± 1.12 | +++ | +++ | +++ | ++ | +++ | +++ |
| 5% | 122.13 ± 1.52 | ++ | ++ | ++ | ++ | ++ | +++ |
| 10% | 74.62 ± 1.70 | ++ | + | + | + | ++ | ++ |
| Treatment | RWC (%) | EL (%) | Na+ (mg/g DW) | K+ (mg/g DW) | Proline (mg/g FW) | TSS (mg/g FW) | STI |
|---|---|---|---|---|---|---|---|
| T-1 | 63.30 ± 1.19 d | 39.41 ± 0.74 c | 4.12 ± 0.14 e | 8.10 ± 0.24 f | 10.62 ± 0.44 f | 123.28 ± 1.17 e | - |
| T-2 | 75.17 ± 1.43 b | 32.73 ± 0.61 d | 3.85 ± 0.13 e | 9.72 ± 0.25 e | 11.54 ± 0.48 f | 136.16 ± 0.90 c | - |
| T-3 | 79.13 ± 1.50 b | 31.05 ± 0.58 d | 3.23 ± 0.10 f | 10.05 ± 0.12 e | 12.62 ± 0.52 ef | 141.41 ± 0.52 b | - |
| T-4 | 88.03 ± 1.67 a | 27.24 ± 0.51 e | 2.84 ± 0.11 f | 11.19 ± 0.17 d | 13.80 ± 0.58 e | 150.19 ± 0.88 a | - |
| T-5 | 36.60 ± 0.68 f | 60.95 ± 1.14 a | 12.69 ± 0.25 a | 4.93 ± 0.13 g | 18.19 ± 0.42 d | 90.32 ± 1.04 g | 0.7 ± 0.04 d |
| T-6 | 57.37 ± 1.09 e | 42.38 ± 0.79 b | 10.41 ± 0.21 b | 12.39 ± 0.14 c | 23.39 ± 0.51 c | 118.36 ± 0.84 f | 2.1 ± 0.12 c |
| T-7 | 62.30 ± 1.19 d | 37.25 ± 0.70 c | 8.12 ± 0.16 c | 14.83 ± 0.17 b | 30.20 ± 0.65 b | 129.29 ± 0.89 d | 2.5 ± 0.14 b |
| T-8 | 70.23 ± 1.33 c | 32.13 ± 0.60 d | 6.23 ± 0.12 d | 16.28 ±0.18 a | 36.65 ± 0.91 a | 136.40 ± 0.96 c | 3.1 ± 0.18 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, A.; Singh, R.K.; Singh, P.; Vaishnav, A.; Guo, D.-J.; Verma, K.K.; Li, D.-P.; Song, X.-P.; Malviya, M.K.; Khan, N.; et al. Insights into the Bacterial and Nitric Oxide-Induced Salt Tolerance in Sugarcane and Their Growth-Promoting Abilities. Microorganisms 2021, 9, 2203. https://doi.org/10.3390/microorganisms9112203
Sharma A, Singh RK, Singh P, Vaishnav A, Guo D-J, Verma KK, Li D-P, Song X-P, Malviya MK, Khan N, et al. Insights into the Bacterial and Nitric Oxide-Induced Salt Tolerance in Sugarcane and Their Growth-Promoting Abilities. Microorganisms. 2021; 9(11):2203. https://doi.org/10.3390/microorganisms9112203
Chicago/Turabian StyleSharma, Anjney, Rajesh Kumar Singh, Pratiksha Singh, Anukool Vaishnav, Dao-Jun Guo, Krishan K. Verma, Dong-Ping Li, Xiu-Peng Song, Mukesh Kumar Malviya, Naeem Khan, and et al. 2021. "Insights into the Bacterial and Nitric Oxide-Induced Salt Tolerance in Sugarcane and Their Growth-Promoting Abilities" Microorganisms 9, no. 11: 2203. https://doi.org/10.3390/microorganisms9112203
APA StyleSharma, A., Singh, R. K., Singh, P., Vaishnav, A., Guo, D.-J., Verma, K. K., Li, D.-P., Song, X.-P., Malviya, M. K., Khan, N., Lakshmanan, P., & Li, Y.-R. (2021). Insights into the Bacterial and Nitric Oxide-Induced Salt Tolerance in Sugarcane and Their Growth-Promoting Abilities. Microorganisms, 9(11), 2203. https://doi.org/10.3390/microorganisms9112203











