Can Human Handling Increase the Presence of Multidrug Resistance (MDR) in Salmonella spp. Isolated from Food Sources?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Salmonella spp. Strains
2.2. Antibiotic Susceptibility Assay Using the Disc Diffusion Method
2.3. MIC Determination for MDR Strains
2.4. Detection of Antibiotic Resistance Genes in MDR Strains
2.5. Statistical Analysis
3. Results
3.1. Antibiotic Susceptibility Assay Using the Disc Diffusion Method
3.2. MIC Determination
3.3. Antibiotic Resistance Gene Detection
3.4. Statistical Analyses
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Food Safety Authority. European Centre for Disease Prevention and Control the European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2017/2018. EFSA J. 2020, 18, e06007. [Google Scholar] [CrossRef] [Green Version]
- Foti, M.; Mascetti, A.; Fisichella, V.; Fulco, E.; Orlandella, B.M.; Lo Piccolo, F. Antibiotic Resistance Assessment in Bacteria Isolated in Migratory Passeriformes Transiting through the Metaponto Territory (Basilicata, Italy). Avian Res. 2017, 8, 26. [Google Scholar] [CrossRef] [Green Version]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-Resistant, Extensively Drug-Resistant and Pandrug-Resistant Bacteria: An International Expert Proposal for Interim Standard Definitions for Acquired Resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tangcharoensathien, V.; Sattayawutthipong, W.; Kanjanapimai, S.; Kanpravidth, W.; Brown, R.; Sommanustweechai, A. Antimicrobial Resistance: From Global Agenda to National Strategic Plan, Thailand. Bull. World Health Organ. 2017, 95, 599–603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poli, G. Microbiologia e Immunologia Veterinaria; UTET: Torino, Italy, 2001; ISBN 978-88-02-04989-2. [Google Scholar]
- Habrun, B.; Listes, E.; Spicic, S.; Cvetnic, Z.; Lukacevic, D.; Jemersic, L.; Lojkic, M.; Kompes, G. An Outbreak of Salmonella Abortusovis Abortions in Sheep in South Croatia. J. Vet. Med. Ser. B 2006, 53, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Gambino, D.; Vicari, D.; Vitale, M.; Schirò, G.; Mira, F.; Giglia, M.L.; Riccardi, A.; Gentile, A.; Giardina, S.; Carrozzo, A.; et al. Study on Bacteria Isolates and Antimicrobial Resistance in Wildlife in Sicily, Southern Italy. Microorganisms 2021, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Shrivastava, S.; Shrivastava, P.; Ramasamy, J. World Health Organization Releases Global Priority List of Antibiotic-Resistant Bacteria to Guide Research, Discovery, and Development of New Antibiotics. J. Med. Soc. 2018, 32, 76. [Google Scholar] [CrossRef]
- Franco, A.; Leekitcharoenphon, P.; Feltrin, F.; Alba, P.; Cordaro, G.; Iurescia, M.; Tolli, R.; D’Incau, M.; Staffolani, M.; Di Giannatale, E.; et al. Emergence of a Clonal Lineage of Multidrug-Resistant ESBL-Producing Salmonella Infantis Transmitted from Broilers and Broiler Meat to Humans in Italy between 2011 and 2014. PLoS ONE 2015, 10, e0144802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility: Supplement M100, 31st ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2021; ISBN 978-1-68440-105-5. [Google Scholar]
- Gambino, D.; Sciortino, S.; Migliore, S.; Galuppo, L.; Puleio, R.; Dara, S.; Vicari, D.; Seminara, S.; Gargano, V. Preliminary Results on the Prevalence of Salmonella Spp. in Marine Animals Stranded in Sicilian Coasts: Antibiotic Susceptibility Profile and ARGs Detection in the Isolated Strains. Pathogens 2021, 10, 930. [Google Scholar] [CrossRef] [PubMed]
- Dashti, A.A.; Jadaon, M.M.; Abdulsamad, A.M.; Dashit, H.M. Heat Treatment of Bacteria: A Simple Method of DNA Extraction for Molecular Techniques. Kwait Med. J. 2009, 41, 117–122. [Google Scholar]
- Lynne, A.M.; Rhodes-Clark, B.S.; Bliven, K.; Zhao, S.; Foley, S.L. Antimicrobial Resistance Genes Associated with Salmonella Enterica Serovar Newport Isolates from Food Animals. AAC 2008, 52, 353–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stangroom, J. Chi-Square Test Calculator. Available online: https://www.socscistatistics.com/tests/chisquare2/default2.aspx (accessed on 12 February 2021).
- European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2017. EFSA J. 2018, 16, e05500. [Google Scholar] [CrossRef]
- Vitale, M.; Galluzzo, P.; Buffa, P.G.; Carlino, E.; Spezia, O.; Alduina, R. Comparison of Antibiotic Resistance Profile and Biofilm Production of Staphylococcus Aureus Isolates Derived from Human Specimens and Animal-Derived Samples. Antibiotics 2019, 8, 97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abruzzo, N. Improvement of Reproductive Performances with a Combined Strategy (Sementusa®) in Sheep Farms in Sicily. Large Anim. Rev. 2014, 20, 209–2013. [Google Scholar]
- Guardabassi, L. Pet Animals as Reservoirs of Antimicrobial-Resistant Bacteria: Review. J. Antimicrob. Chemother. 2004, 54, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-T.; Tsao, S.-M.; Hsueh, P.-R. Clinical Outcomes of Tigecycline Alone or in Combination with Other Antimicrobial Agents for the Treatment of Patients with Healthcare-Associated Multidrug-Resistant Acinetobacter Baumannii Infections. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1211–1220. [Google Scholar] [CrossRef] [PubMed]
- Mąka, Ł.; Popowska, M. Antimicrobial Resistance of Salmonella Spp. Isolated from Food. Rocz. Panstw. Zakl. Hig. 2016, 67, 343–358. [Google Scholar] [PubMed]
- Ferri, M.; Ranucci, E.; Romagnoli, P.; Giaccone, V. Antimicrobial Resistance: A Global Emerging Threat to Public Health Systems. Crit. Rev. Food Sci. Nutr. 2017, 57, 2857–2876. [Google Scholar] [CrossRef] [PubMed]
- European Food Safety Authority. European Centre for Disease Prevention and Control The European Union Summary Report on Antimicrobial Resistance in Zoonotic and Indicator Bacteria from Humans, Animals and Food in 2018/2019. EFS2 2021, 19, e06490. [Google Scholar] [CrossRef]
| Target Name | Resistance Mechanism | Amplicon Size (bp) | Reference |
|---|---|---|---|
| blaTEM | β-Lactamases | 858 | [12] |
| blaCTXM | β-Lactamases | 112 | [7] |
| blaSHV | β-Lactamases | 807 | [7] |
| blaOXA | β-Lactamases | 590 | [7] |
| tet(A) | Efflux | 210 | [7] |
| tet(B) | Efflux | 659 | [13] |
| sulI | Dihydropteroate synthase inhibitor | 316 | [13] |
| sulII | Dihydropteroate synthase inhibitor | 191 | [7] |
| sulIII | Dihydropteroate synthase inhibitor | 799 | [13] |
| Antibiotics Classes | Antibiotics | Strains | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ID. 17 | ID. 36 | ID. 49 | ID. 50 | ID. 51 | ID. 52 | ID. 53 | ID. 57 | ID. 58 | ||
| Aminoglycosides | GEN | ≥8 | ≤0.5 | ≤0.5 | ≥1 | ≤0.5 | ≤0.5 | ≥1 | ≤0.5 | ≤0.5 |
| Beta lactams | AMP | ≥64 | >64 | >64 | >64 | >64 | >64 | ≥4 | ≤1 | ≥2 |
| TAZ | ≥0.5 | ≤0.5 | ≤0.5 | ≥4 | ≥4 | ≥4 | ≤0.5 | ≤0.5 | ≥1 | |
| FOT | ≤0.25 | ≤0.25 | ≤0.25 | >4 | >4 | >4 | ≤0.25 | ≤0.25 | ≥0.5 | |
| MERO | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | ≤0.03 | |
| Phenicoles | CHL | ≤8 | ≥16 | ≤8 | ≤8 | ≤8 | ≥16 | ≤8 | ≤8 | ≤8 |
| Polymixine | COL | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≤1 | ≥4 | ≤1 | ≤1 |
| Quinolones | NAL | ≥8 | >128 | ≥16 | >128 | >128 | >128 | >128 | >128 | >128 |
| CIP | ≥0.03 | ≥0.25 | ≥0.5 | ≥0.12 | ≥0.25 | ≥0.25 | ≥0.5 | ≥1 | ≥0.25 | |
| Sulfonamides | SMX | ≥64 | >1024 | >1024 | >1024 | >1024 | >1024 | >1024 | >1024 | >1024 |
| TMP | ≤0.25 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | >32 | |
| Tetracyclines | TET | >64 | >64 | >64 | ≥64 | >64 | >64 | >64 | >64 | >64 |
| TGC | ≥1 | ≥2 | ≥2 | ≥1 | ≥1 | ≥2 | ≥1 | ≥2 | ≥1 | |
| Macrolides | AZI | ≥8 | ≥8 | ≥8 | ≥16 | ≥4 | ≥4 | ≥16 | ≥8 | ≥8 |
| Strains | Source | Serotype | Phenotypic Resistance | Resistance Genes |
|---|---|---|---|---|
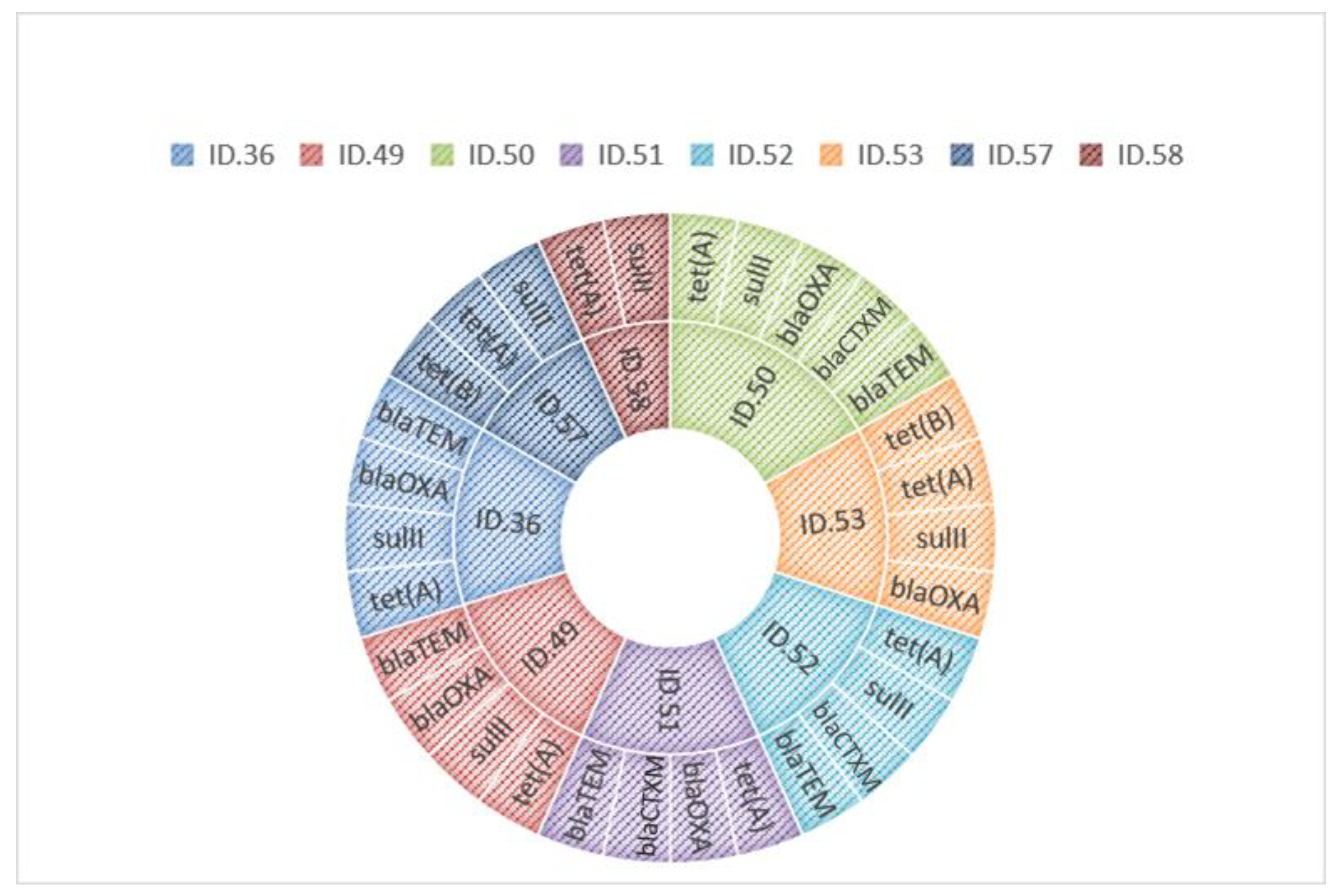
| ID. 36 | Carduelis carduelis | S. enterica subsp. Infantis | AMP, SMX, TET, TGC | blaTEM, blaOXA, sulII, tet(A) |
| ID. 49 | Turkey meat | S. enterica subsp. Newport | AMP, SMX, TET, TGC | blaTEM, blaOXA, sulII, tet(A) |
| ID. 50 | Bovine meat | S. enterica subsp. Infantis | AMP, FOT, SMX, TET, TGC | blaTEM,blaCTXM, blaOXA, sulII, tet(A) |
| ID. 51 | Bovine meat | S. enterica subsp. Infantis | AMP, FOT, TET, TGC | blaTEM,blaCTXM, blaOXA, tet(A) |
| ID. 52 | Poultry meat | S. enterica subsp. Infantis | AMP, FOT, SMX, TET, TGC | blaTEM,blaCTXM, sulII, tet(A) |
| ID. 53 | Poultry meat | S. enterica subsp. Infantis | SMX, TET, TGC | blaOXA, sulII, tet(A), tet(B) |
| ID. 57 | Poultry meat | S. enterica subsp. Infantis | SMX, TET, TGC | sulII, tet(A), tet(B) |
| ID. 58 | Poultry meat | S. enterica subsp. Infantis | SMX, TET, TGC | sulII, tet(A) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gargano, V.; Gambino, D.; Migliore, S.; Vitale, M.; Sciortino, S.; Costa, A.; Vicari, D. Can Human Handling Increase the Presence of Multidrug Resistance (MDR) in Salmonella spp. Isolated from Food Sources? Microorganisms 2021, 9, 2018. https://doi.org/10.3390/microorganisms9102018
Gargano V, Gambino D, Migliore S, Vitale M, Sciortino S, Costa A, Vicari D. Can Human Handling Increase the Presence of Multidrug Resistance (MDR) in Salmonella spp. Isolated from Food Sources? Microorganisms. 2021; 9(10):2018. https://doi.org/10.3390/microorganisms9102018
Chicago/Turabian StyleGargano, Valeria, Delia Gambino, Sergio Migliore, Maria Vitale, Sonia Sciortino, Antonella Costa, and Domenico Vicari. 2021. "Can Human Handling Increase the Presence of Multidrug Resistance (MDR) in Salmonella spp. Isolated from Food Sources?" Microorganisms 9, no. 10: 2018. https://doi.org/10.3390/microorganisms9102018
APA StyleGargano, V., Gambino, D., Migliore, S., Vitale, M., Sciortino, S., Costa, A., & Vicari, D. (2021). Can Human Handling Increase the Presence of Multidrug Resistance (MDR) in Salmonella spp. Isolated from Food Sources? Microorganisms, 9(10), 2018. https://doi.org/10.3390/microorganisms9102018









