Is Gut Microbiota Dysbiosis a Predictor of Increased Susceptibility to Poor Outcome of COVID-19 Patients? An Update
Abstract
1. The “Gut Microbiota Hypothesis” in Poor Outcomes of COVID-19 Patients
2. What Is the Evidence So Far that Links Gut Microbiota Composition to COVID-19 Severity?
3. Concluding Remarks and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Ma, W.T.; Pang, M.; Fan, Q.L.; Hua, J.L. The Commensal Microbiota and Viral Infection: A Comprehensive Review. Front. Immunol. 2019, 10, 1551. [Google Scholar] [CrossRef] [PubMed]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Guinane, C.M.; Cotter, P.D. Role of the gut microbiota in health and chronic gastrointestinal disease: Understanding a hidden metabolic organ. Ther. Adv. Gastroenterol. 2013, 6, 295–308. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.K.; Cabral, C.; Kumar, R.; Ganguly, R.; Rana, H.K.; Gupta, A.; Lauro, M.R.; Carbone, C.; Reis, F.; Pandey, A.K. Beneficial Effects of Dietary Polyphenols on Gut Microbiota and Strategies to Improve Delivery Efficiency. Nutrients 2019, 11, 2216. [Google Scholar] [CrossRef]
- Wang, B.; Yao, M.; Lv, L.; Ling, Z.; Li, L. The Human Microbiota in Health and Disease. Engineering 2017, 3, 71–82. [Google Scholar] [CrossRef]
- Carding, S.; Verbeke, K.; Vipond, D.T.; Corfe, B.M.; Owen, L.J. Dysbiosis of the gut microbiota in disease. Microb. Ecol. Health Dis. 2015, 26, 26191. [Google Scholar] [CrossRef]
- Fernandes, R.; Viana, S.D.; Nunes, S.; Reis, F. Diabetic gut microbiota dysbiosis as an inflammaging and immunosenescence condition that fosters progression of retinopathy and nephropathy. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1876–1897. [Google Scholar] [CrossRef]
- Hasan, N.; Yang, H. Factors affecting the composition of the gut microbiota, and its modulation. PeerJ 2019, 7, e7502. [Google Scholar] [CrossRef]
- Moran-Ramos, S.; Lopez-Contreras, B.E.; Villarruel-Vazquez, R.; Ocampo-Medina, E.; Macias-Kauffer, L.; Martinez-Medina, J.N.; Villamil-Ramirez, H.; León-Mimila, P.; Del Rio-Navarro, B.E.; Ibarra-Gonzalez, I.; et al. Environmental and intrinsic factors shaping gut microbiota composition and diversity and its relation to metabolic health in children and early adolescents: A population-based study. Gut Microbes 2020, 11, 900–917. [Google Scholar] [CrossRef]
- Groves, H.T.; Higham, S.L.; Moffatt, M.F.; Cox, M.J.; Tregoning, J.S. Respiratory Viral Infection Alters the Gut Microbiota by Inducing Inappetence. mBio 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Hanada, S.; Pirzadeh, M.; Carver, K.Y.; Deng, J.C. Respiratory Viral Infection-Induced Microbiome Alterations and Secondary Bacterial Pneumonia. Front. Immunol. 2018, 9, 2640. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Hensley, C.; Mahsoub, H.M.; Ramesh, A.K.; Zhou, P. Microbiota in viral infection and disease in humans and farm animals. Prog. Mol. Biol. Transl. Sci. 2020, 171, 15–60. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, M.; Kleine-Weber, H.; Krüger, N.; Müller, M.; Drosten, C.; Pöhlmann, S. The novel coronavirus 2019 (2019-nCoV) uses the SARS-coronavirus receptor ACE2 and the cellular protease TMPRSS2 for entry into target cells. bioRxiv 2020. [Google Scholar] [CrossRef]
- Wu, T.; Zuo, Z.; Kang, S.; Jiang, L.; Luo, X.; Xia, Z.; Liu, J.; Xiao, X.; Ye, M.; Deng, M. Multi-organ Dysfunction in Patients with COVID-19: A Systematic Review and Meta-analysis. Aging Dis. 2020, 11, 874–894. [Google Scholar] [CrossRef]
- COVID, Team CDC, and Response Team. Severe Outcomes Among Patients with Coronavirus Disease 2019 (COVID-19)-United States, February 12-March 16, 2020. Morb. Mortal. Wkly. Rep. 2020, 69, 343–346. [Google Scholar] [CrossRef]
- Roncon, L.; Zuin, M.; Rigatelli, G.; Zuliani, G. Diabetic patients with COVID-19 infection are at higher risk of ICU admission and poor short-term outcome. J. Clin. Virol. 2020, 127, 104354. [Google Scholar] [CrossRef]
- Siordia, J.A., Jr. Epidemiology and clinical features of COVID-19: A review of current literature. J. Clin. Virol. 2020, 127, 104357. [Google Scholar] [CrossRef]
- Du, M.; Cai, G.; Chen, F.; Christiani, D.C.; Zhang, Z.; Wang, M. Multiomics Evaluation of Gastrointestinal and Other Clinical Characteristics of COVID-19. Gastroenterology 2020, 158, 2298–2301.e2297. [Google Scholar] [CrossRef]
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009. [Google Scholar] [CrossRef]
- Villapol, S. Gastrointestinal symptoms associated with COVID-19: Impact on the gut microbiome. Transl. Res. 2020, 226, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Tu, L. Implications of gastrointestinal manifestations of COVID-19. Lancet Gastroenterol. Hepatol. 2020, 5, 629–630. [Google Scholar] [CrossRef]
- Hanrahan, T.P.; Lubel, J.S.; Garg, M. Lessons From COVID-19, ACE2, and Intestinal Inflammation: Could a Virus Trigger Chronic Intestinal Inflammation? Clin. Gastroenterol. Hepatol. 2020, 19, 206. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y.; Wu, W.; Chen, S.; Gu, J.W.; Li, X.L.; Song, H.J.; Du, F.; Wang, G.; Zhong, C.Q.; Wang, X.Y.; et al. Digestive system involvement of novel coronavirus infection: Prevention and control infection from a gastroenterology perspective. J. Dig. Dis. 2020, 21, 199–204. [Google Scholar] [CrossRef]
- Penninger, J.M.; Grant, M.B.; Sung, J.J.Y. The Role of Angiotensin Converting Enzyme 2 in Modulating Gut Microbiota, Intestinal Inflammation, and Coronavirus Infection. Gastroenterology 2020. ahead of print. [Google Scholar] [CrossRef]
- Xu, J.; Chu, M.; Zhong, F.; Tan, X.; Tang, G.; Mai, J.; Lai, N.; Guan, C.; Liang, Y.; Liao, G. Digestive symptoms of COVID-19 and expression of ACE2 in digestive tract organs. Cell Death Discov. 2020, 6, 76. [Google Scholar] [CrossRef]
- Kuba, K.; Imai, Y.; Penninger, J.M. Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ. J. 2013, 77, 301–308. [Google Scholar] [CrossRef]
- Perlot, T.; Penninger, J.M. ACE2—From the renin–angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013, 15, 866–873. [Google Scholar] [CrossRef]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef]
- Kotfis, K.; Skonieczna-Żydecka, K. COVID-19: Gastrointestinal symptoms and potential sources of SARS-CoV-2 transmission. Anaesthesiol. Intensive Ther. 2020, 52, 171–172. [Google Scholar] [CrossRef]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the Gut Microbiota in Patients with COVID-19 or H1N1 Influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Wang, J.; Li, F.; Shi, Y. Main Clinical Features of COVID-19 and Potential Prognostic and Therapeutic Value of the Microbiota in SARS-CoV-2 Infections. Front. Microbiol. 2020, 11, 1302. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.; Tso, E.; Yeoh, Y.K.; Chen, Z.; Boon, S.; Chan, F.K.L.; Chan, P.; et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2020, gutjnl-2020-322294. [Google Scholar] [CrossRef] [PubMed]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e948. [Google Scholar] [CrossRef] [PubMed]
- Viana, S.D.; Nunes, S.; Reis, F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities-Role of gut microbiota dysbiosis. Ageing Res. Rev. 2020, 62, 101123. [Google Scholar] [CrossRef]
- Busnelli, M.; Manzini, S.; Chiesa, G. The Gut Microbiota Affects Host Pathophysiology as an Endocrine Organ: A Focus on Cardiovascular Disease. Nutrients 2019, 12, 79. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Rodriguez, E.; Egea-Zorrilla, A.; Plaza-Díaz, J.; Aragón-Vela, J.; Muñoz-Quezada, S.; Tercedor-Sánchez, L.; Abadia-Molina, F. The Gut Microbiota and Its Implication in the Development of Atherosclerosis and Related Cardiovascular Diseases. Nutrients 2020, 12, 605. [Google Scholar] [CrossRef] [PubMed]
- Channappanavar, R.; Perlman, S. Pathogenic human coronavirus infections: Causes and consequences of cytokine storm and immunopathology. Semin. Immunopathol. 2017, 39, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Liu, J.; Zhang, D.; Xu, Z.; Ji, J.; Wen, C. Cytokine Storm in COVID-19: The Current Evidence and Treatment Strategies. Front. Immunol. 2020, 11, 1708. [Google Scholar] [CrossRef]
- Fuellen, G.; Liesenfeld, O.; Kowald, A.; Barrantes, I.; Bastian, M.; Simm, A.; Jansen, L.; Tietz-Latza, A.; Quandt, D.; Franceschi, C.; et al. The preventive strategy for pandemics in the elderly is to collect in advance samples & data to counteract chronic inflammation (inflammaging). Ageing Res. Rev. 2020, 62, 101091. [Google Scholar] [CrossRef] [PubMed]
- Hojyo, S.; Uchida, M.; Tanaka, K.; Hasebe, R.; Tanaka, Y.; Murakami, M.; Hirano, T. How COVID-19 induces cytokine storm with high mortality. Inflamm. Regen. 2020, 40, 37. [Google Scholar] [CrossRef] [PubMed]
- Calder, P.C. Nutrition, immunity and COVID-19. BMJ Nutr. Prev. Health 2020, 3, 74–92. [Google Scholar] [CrossRef]
- Lazar, V.; Ditu, L.-M.; Pircalabioru, G.G.; Gheorghe, I.; Curutiu, C.; Holban, A.M.; Picu, A.; Petcu, L.; Chifiriuc, M.C. Aspects of Gut Microbiota and Immune System Interactions in Infectious Diseases, Immunopathology, and Cancer. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef]
- Schirmer, M.; Smeekens, S.P.; Vlamakis, H.; Jaeger, M.; Oosting, M.; Franzosa, E.A.; Ter Horst, R.; Jansen, T.; Jacobs, L.; Bonder, M.J.; et al. Linking the Human Gut Microbiome to Inflammatory Cytokine Production Capacity. Cell 2016, 167, 1125–1136.e1128. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, C.; Viana, S.D.; Reis, F. Gut Microbiota Dysbiosis-Immune Hyperresponse-Inflammation Triad in Coronavirus Disease 2019 (COVID-19): Impact of Pharmacological and Nutraceutical Approaches. Microorganisms 2020, 8, 1514. [Google Scholar] [CrossRef] [PubMed]
- Kaźmierczak-Siedlecka, K.; Vitale, E.; Makarewicz, W. COVID-19-gastrointestinal and gut microbiota-related aspects. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 10853–10859. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Aolymat, I.; Al-Holy, M.; Ayyash, M.; Abu Ghoush, M.; Al-Nabulsi, A.A.; Osaili, T.; Apostolopoulos, V.; Liu, S.Q.; Shah, N.P. The potential application of probiotics and prebiotics for the prevention and treatment of COVID-19. NPJ Sci. Food 2020, 4, 17. [Google Scholar] [CrossRef]
- Kalantar-Zadeh, K.; Ward, S.A.; Kalantar-Zadeh, K.; El-Omar, E.M. Considering the Effects of Microbiome and Diet on SARS-CoV-2 Infection: Nanotechnology Roles. ACS Nano 2020, 14, 5179–5182. [Google Scholar] [CrossRef]
- Kataoka, K. The intestinal microbiota and its role in human health and disease. J. Med. Investig. JMI 2016, 63, 27–37. [Google Scholar] [CrossRef]
- Wu, H.J.; Wu, E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes 2012, 3, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef] [PubMed]
- Catanzaro, M.; Fagiani, F.; Racchi, M.; Corsini, E.; Govoni, S.; Lanni, C. Immune response in COVID-19: Addressing a pharmacological challenge by targeting pathways triggered by SARS-CoV-2. Signal Transduct. Target. Ther. 2020, 5, 84. [Google Scholar] [CrossRef]
- Chowdhury, M.A.; Hossain, N.; Kashem, M.A.; Shahid, M.A.; Alam, A. Immune response in COVID-19: A review. J. Infect. Public Health 2020, 13, 1619–1629. [Google Scholar] [CrossRef]
- Kalfaoglu, B.; Almeida-Santos, J.; Tye, C.A.; Satou, Y.; Ono, M. T-Cell Hyperactivation and Paralysis in Severe COVID-19 Infection Revealed by Single-Cell Analysis. Front. Immunol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Tang, J.; Ye, C.; Dong, L. The immunology of COVID-19: Is immune modulation an option for treatment? Lancet Rheumatol. 2020, 2, e428–e436. [Google Scholar] [CrossRef]
- Fathi, N.; Rezaei, N. Lymphopenia in COVID-19: Therapeutic opportunities. Cell Biol. Int. 2020, 44, 1792–1797. [Google Scholar] [CrossRef]
- Huang, I.; Pranata, R. Lymphopenia in severe coronavirus disease-2019 (COVID-19): Systematic review and meta-analysis. J. Intensive Care 2020, 8, 36. [Google Scholar] [CrossRef]
- Tan, L.; Wang, Q.; Zhang, D.; Ding, J.; Huang, Q.; Tang, Y.-Q.; Wang, Q.; Miao, H. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal Transduct. Target. Ther. 2020, 5, 33. [Google Scholar] [CrossRef]
- Tavakolpour, S.; Rakhshandehroo, T.; Wei, E.X.; Rashidian, M. Lymphopenia during the COVID-19 infection: What it shows and what can be learned. Immunol. Lett. 2020, 225, 31–32. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef] [PubMed]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef]
- Swiecki, M.; Colonna, M. Type I interferons: Diversity of sources, production pathways and effects on immune responses. Curr. Opin. Virol. 2011, 1, 463–475. [Google Scholar] [CrossRef]
- Britton, G.J.; Chen-Liaw, A.; Cossarini, F.; Livanos, A.E.; Spindler, M.P.; Plitt, T.; Eggers, J.; Mogno, I.; Gonzalez-Reiche, A.; Siu, S.; et al. SARS-CoV-2-specific IgA and limited inflammatory cytokines are present in the stool of select patients with acute COVID-19. medRxiv 2020. preprint. [Google Scholar] [CrossRef]
- Liu, Z.; Long, W.; Tu, M.; Chen, S.; Huang, Y.; Wang, S.; Zhou, W.; Chen, D.; Zhou, L.; Wang, M.; et al. Lymphocyte subset (CD4+, CD8+) counts reflect the severity of infection and predict the clinical outcomes in patients with COVID-19. J. Infect. 2020, 81, 318–356. [Google Scholar] [CrossRef]
- Hou, H.; Zhang, B.; Huang, H.; Luo, Y.; Wu, S.; Tang, G.; Liu, W.; Mao, L.; Mao, L.; Wang, F.; et al. Using IL-2R/lymphocytes for predicting the clinical progression of patients with COVID-19. Clin. Exp. Immunol. 2020, 201, 76–84. [Google Scholar] [CrossRef]
- Dhar, D.; Mohanty, A. Gut microbiota and Covid-19- possible link and implications. Virus Res. 2020, 285, 198018. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e911. [Google Scholar] [CrossRef]
- Fontaine, S.S.; Kohl, K.D. Optimal integration between host physiology and functions of the gut microbiome. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190594. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the normal gut microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Martin, A.M.; Sun, E.W.; Rogers, G.B.; Keating, D.J. The Influence of the Gut Microbiome on Host Metabolism Through the Regulation of Gut Hormone Release. Front. Physiol. 2019, 10, 428. [Google Scholar] [CrossRef] [PubMed]
- Budden, K.F.; Gellatly, S.L.; Wood, D.L.; Cooper, M.A.; Morrison, M.; Hugenholtz, P.; Hansbro, P.M. Emerging pathogenic links between microbiota and the gut-lung axis. Nat. Rev. Microbiol. 2017, 15, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Dumas, A.; Bernard, L.; Poquet, Y.; Lugo-Villarino, G.; Neyrolles, O. The role of the lung microbiota and the gut-lung axis in respiratory infectious diseases. Cell. Microbiol. 2018, 20, e12966. [Google Scholar] [CrossRef]
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The Gut-Lung Axis in Health and Respiratory Diseases: A Place for Inter-Organ and Inter-Kingdom Crosstalks. Front. Cell. Infect. Microbiol. 2020, 10, 9. [Google Scholar] [CrossRef]
- Gibson, P.G.; Qin, L.; Puah, S.H. COVID-19 acute respiratory distress syndrome (ARDS): Clinical features and differences from typical pre-COVID-19 ARDS. Med. J. Aust. 2020, 213, 54–56.e51. [Google Scholar] [CrossRef]
- Grasselli, G.; Tonetti, T.; Protti, A.; Langer, T.; Girardis, M.; Bellani, G.; Laffey, J.; Carrafiello, G.; Carsana, L.; Rizzuto, C.; et al. Pathophysiology of COVID-19-associated acute respiratory distress syndrome: A multicentre prospective observational study. Lancet Respir. Med. 2020, 8, 1201–1208. [Google Scholar] [CrossRef]
- van der Lelie, D.; Taghavi, S. COVID-19 and the Gut Microbiome: More than a Gut Feeling. mSystems 2020, 5. [Google Scholar] [CrossRef]
- Dickson, R.P.; Schultz, M.J.; van der Poll, T.; Schouten, L.R.; Falkowski, N.R.; Luth, J.E.; Sjoding, M.W.; Brown, C.A.; Chanderraj, R.; Huffnagle, G.B.; et al. Lung Microbiota Predict Clinical Outcomes in Critically Ill Patients. Am. J. Respir. Crit. Care Med. 2020, 201, 555–563. [Google Scholar] [CrossRef]
- Gou, W.; Fu, Y.; Yue, L.; Chen, G.-d.; Cai, X.; Shuai, M.; Xu, F.; Yi, X.; Chen, H.; Zhu, Y.J.; et al. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv 2020. [Google Scholar] [CrossRef]
- Zhang, W.; Li, L.; Liu, J.; Chen, L.; Zhou, F.; Jin, T.; Jiang, L.; Li, X.; Yang, M.; Wang, H. The characteristics and predictive role of lymphocyte subsets in COVID-19 patients. Int. J. Infect. Dis. 2020, 99, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Nie, H.X.; Hu, K.; Wu, X.J.; Zhang, Y.T.; Wang, M.M.; Wang, T.; Zheng, Z.S.; Li, X.C.; Zeng, S.L. Abnormal immunity of non-survivors with COVID-19: Predictors for mortality. Infect. Dis. Poverty 2020, 9, 108. [Google Scholar] [CrossRef] [PubMed]
- Morrow, L.E.; Kollef, M.H.; Casale, T.B. Probiotic prophylaxis of ventilator-associated pneumonia: A blinded, randomized, controlled trial. Am. J. Respir. Crit. Care Med. 2010, 182, 1058–1064. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Wang, C.T.; Zhang, F.S.; Qi, F.; Wang, S.F.; Ma, S.; Wu, T.J.; Tian, H.; Tian, Z.T.; Zhang, S.L.; et al. Effect of probiotics on the incidence of ventilator-associated pneumonia in critically ill patients: A randomized controlled multicenter trial. Intensive Care Med. 2016, 42, 1018–1028. [Google Scholar] [CrossRef]
- Ceccarelli, G.; Scagnolari, C.; Pugliese, F.; Mastroianni, C.M.; d’Ettorre, G. Probiotics and COVID-19. Lancet Gastroenterol. Hepatol. 2020, 5, 721–722. [Google Scholar] [CrossRef]
- Mak, J.W.Y.; Chan, F.K.L.; Ng, S.C. Probiotics and COVID-19: One size does not fit all. Lancet Gastroenterol. Hepatol. 2020, 5, 644–645. [Google Scholar] [CrossRef]
| Study Sample | Major Findings Regarding Gut Microbiota, Inflammation and Immune System Changes | Ref. |
|---|---|---|
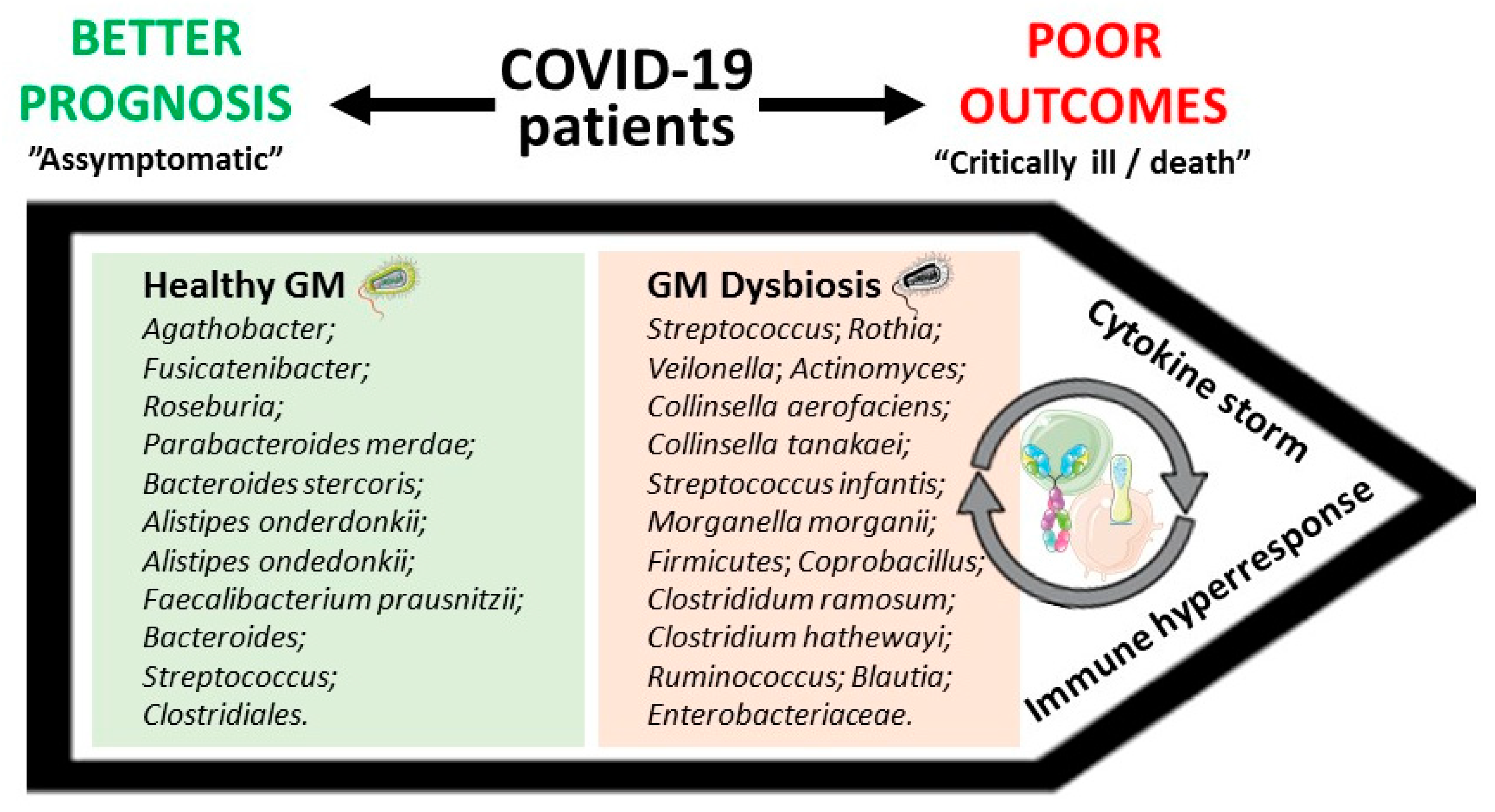
| 30 COVID-19 patients, 24 H1N1 patients, and 30 matched controls. | (i) Compared with healthy controls, COVID-19 patients displayed a depletion of Agathobacter, Fusicatenibacter, Roseburia, and Ruminocaccaceae UCG-013, whose abundancy was negatively correlated, mainly, with the levels of CRP, PCT, and D-dimer; (ii) CRP and D-dimer levels were positively correlated with bacteria abundance in COVID-19 cases, including Streptococcus, Rothia, Veilonella, and Actinomyces. | [31] |
| 15 hospitalized COVID-19 patients, of which 7 had stool positivity for SARS-CoV-2. | (i) Faecal samples with high SARS-CoV-2 infectivity had higher abundances of Collinsella aerofaciens, Collinsella tanakaei, Streptococcus infantis, and Morganella morganii, and higher functional capacity for nucleotide de novo biosynthesis, amino acid biosynthesis, and glycolysis; (ii) faecal samples with signature of low-to-none SARS-CoV-2 infectivity had higher abundances of SCFAs-producing bacteria, Parabacteroides merdae, Bacteroides stercoris, Alistipes onderdonkii, and Lachnospiraceae bacterium 1_1_57FAA. | [34] |
| 15 COVID-19 patients, 6 patients with pneumonia, and 15 healthy controls. | (i) In a sample of seven antibiotic-naïve patients, 23 bacterial taxa were discovered to correlate with disease severity; (ii) the main bacteria positively correlated with disease severity belonged to the phylum Firmicutes, Coprobacillus genus, and Clostrididum ramosum and Clostridium hathewayi species; (iii) the main bacteria negatively correlated with disease severity were Alistipes ondedonkii and Faecalibacterium prausnitzii. | [35] |
| COVID-19 patients (moderate vs. severe). | (i) Versus moderate patients, CD4+ T-cells from severe COVID-19 patients expressed higher levels of the AP-1 genes fos, fosb, and jun; the activation marker MKI67 (Ki67); Th2-related genes il4r and maf; and chemokines including CCL2, CCL3, CCL4, CCL7, CCL8, and CXCL8; (ii) CD4+ T-cells in severe patients expressed higher levels of immunoregulatory genes including immune checkpoints (ctla4, havcr2 [tim-3], and lgals3 [galectin-3]) as well as the Tregs and T-cell activation marker IL2RA (CD25); (iii) CD4+ T-cells from severe patients showed decreased expression of interferon-induced genes (ifit1, ifit2, ifit3, and ifitm1) and genes related to downstream pathways. | [55] |
| 12 COVID-19 death cases (moderate vs. severe). | (i) Among all parameters studied, blood lymphocyte percentage (LYM%) showed the most significant and consistent trend, suggesting it as an indicator of disease progression; (ii) LYM% of severe patients decreased initially and then increased to >10% until being discharged; (iii) LYM% of moderate patients fluctuated very little after disease onset and was >20% when discharged. | [59] |
| 522 COVID-19 patients, 43 of which were admitted in the ICU; vs. 40 healthy controls. | (i) In the severe and critical disease patients, as well as the perished ones, total T cells, CD4+, and CD8+ T cells were significantly lower than in the mild/moderate patients; (ii) total T cells, CD8+ T, or CD4+ T cells lower than 800, 300, and 400/μL, respectively, were negatively correlated with patient survival; (iii) T cell numbers were negatively correlated with serum IL-6, IL-10, and TNF-α levels, which were significantly higher in ICU patients than in non-ICU ones; (iv) increasing PD-1 and Tim-3 expression on T cells in patients progressed to symptomatic stages. | [61] |
| 39 COVID-19 patients. | (i) The more serious the disease and the worse the prognosis, the lower the T cell, CD4+ T cell, and CD8+ T cell counts on admission. | [66] |
| 452 COVID-19 patients: 166 non-severe; 286 severe. | (i) With increased severity of illness, leukocytes, neutrophils, infection biomarkers (CRP, PCT, and ferritin) and cytokines (IL-2R, IL-6, IL-8, IL-10, and TNF-α) were significantly increased, while lymphocytes were significantly decreased; (ii) the ratio of IL-2R to lymphocytes was found to be remarkably increased in severe and critical patients. IL-2R/lymphocytes ratio was superior vs. other markers for the identification of critical illness COVID-19; (iii) cytokine profile and IL-2R/lymphocytes were significantly decreased in recovered patients. | [67] |
| 91 critically ill patients, within 24 h of their ICU admission. | (i) ARDS patients display an increased number of Lachnospiraceae and Enterobacteriaceae, which predicted fewer ventilator-free days; (ii) enrichment of the lungs with gut-associated bacteria was positively correlated with high levels of inflammatory markers, particularly TNF-α; (iii) increased Lachnospiraceae was a strong predictor of reduced survival in ARDS patients. | [79] |
| 31 COVID-19 patients (18 non-severe cases and 13 severe cases) and 990 non-COVID-19 individuals. | (i) Each 10% increment in PRS was associated with a 57% higher risk of progressing to severe phases, as indicated by proteomic biomarkers SAA1, SAA2, SAA4, SERPINA3, C6, and CFB; (ii) higher PRS significantly correlated with increased serum levels of all the studied inflammatory markers among older healthy individuals (>58 years); (iii) in the same individuals, Bacteroides genus, Streptococcus genus, and Clostridiales order were negatively correlated with cytokines (IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12p70, IL-13, TNF-α, and IFN-γ), whereas Ruminococcus genus, Blautia genus, and Lactobacillus genus showed positive associations. | [80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ferreira, C.; Viana, S.D.; Reis, F. Is Gut Microbiota Dysbiosis a Predictor of Increased Susceptibility to Poor Outcome of COVID-19 Patients? An Update. Microorganisms 2021, 9, 53. https://doi.org/10.3390/microorganisms9010053
Ferreira C, Viana SD, Reis F. Is Gut Microbiota Dysbiosis a Predictor of Increased Susceptibility to Poor Outcome of COVID-19 Patients? An Update. Microorganisms. 2021; 9(1):53. https://doi.org/10.3390/microorganisms9010053
Chicago/Turabian StyleFerreira, Carolina, Sofia D. Viana, and Flávio Reis. 2021. "Is Gut Microbiota Dysbiosis a Predictor of Increased Susceptibility to Poor Outcome of COVID-19 Patients? An Update" Microorganisms 9, no. 1: 53. https://doi.org/10.3390/microorganisms9010053
APA StyleFerreira, C., Viana, S. D., & Reis, F. (2021). Is Gut Microbiota Dysbiosis a Predictor of Increased Susceptibility to Poor Outcome of COVID-19 Patients? An Update. Microorganisms, 9(1), 53. https://doi.org/10.3390/microorganisms9010053







