Association of Phosphatidylinositol-Specific Phospholipase C with Calcium-Induced Biomineralization in the Coccolithophore Emiliania huxleyi
Abstract
1. Introduction
2. Materials and Methods
2.1. Algal Strain and Culture Conditions
2.2. Inorganic Phosphate Measurement
2.3. AP Activity Measurement
2.4. Determination of TIC and TOC
2.5. Cell Growth and Microscopic Observation
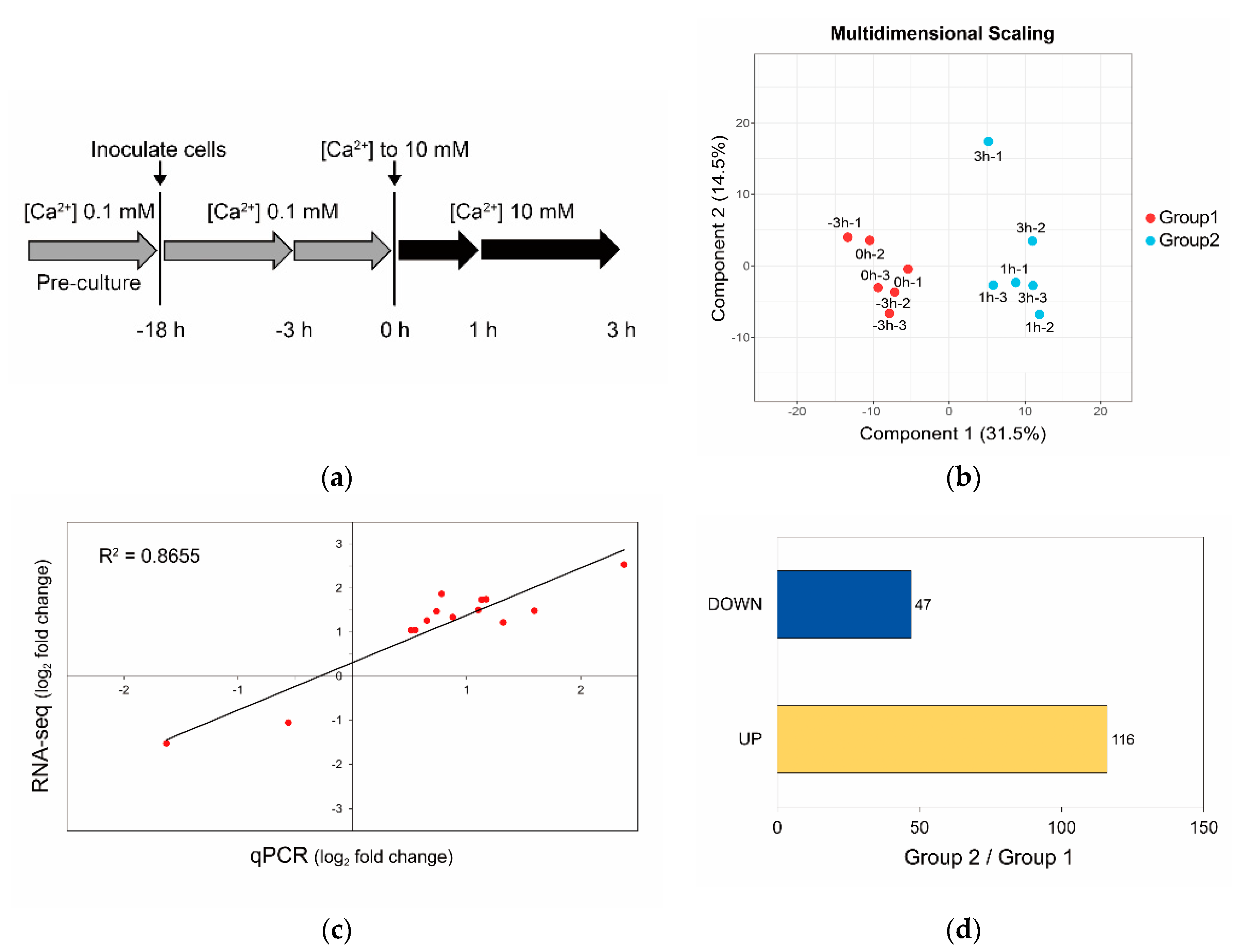
2.6. Sample Preparation
2.7. RNA-seq and Transcriptome Analysis
2.8. qPCR
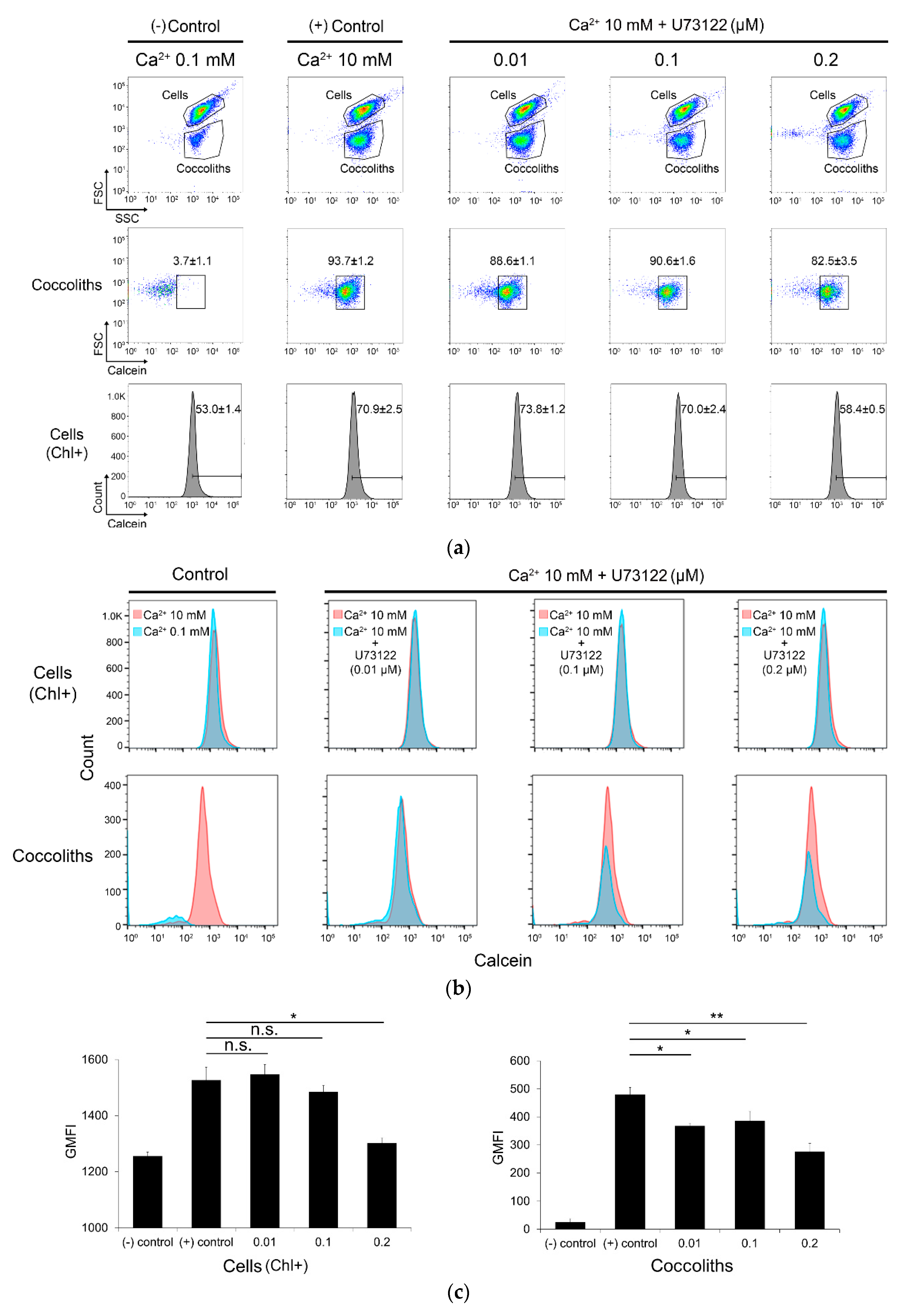
2.9. Calcein Staining and Flow Cytometry Analysis
3. Results
3.1. Coccolith Formation in E. huxleyi CCMP 371 under Phosphate-Replete Conditions
3.2. Morphological and Quantitative Monitoring of Calcium-Induced Coccolith Production
3.3. Calcium-Induced Biomineralization-Associated Genes
3.4. Inhibition of Coccolith Production by PI-PLC inhibitor U73122
4. Discussion
4.1. Physiological Regulation of Cell Culture for Providing Calcium-Induced Calcified Cells
4.2. Identification of Calcium-Induced Biomineralization-Associated Genes
4.3. Association of PKS with Calcium-Induced Biomineralization
4.4. Association of PI-PLC with Calcium-Induced Biomineralization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Westbroek, P.; Brown, C.W.; van Bleijswijk, J.; Brownlee, C.; Brummer, G.J.; Conte, M.; Egge, J.; Fernández, E.; Jordan, R.; Knappertsbusch, M. A model system approach to biological climate forcing. The example of Emiliania huxleyi. Glob. Planet. Chang. 1993, 8, 27–46. [Google Scholar]
- Monteiro, F.M.; Bach, L.T.; Brownlee, C.; Bown, P.; Rickaby, R.E.; Poulton, A.J.; Tyrrell, T.; Beaufort, L.; Dutkiewicz, S.; Gibbs, S. Why marine phytoplankton calcify. Sci. Adv. 2016, 2, e1501822. [Google Scholar] [CrossRef] [PubMed]
- Kondrik, D.; Kazakov, E.; Pozdnyakov, D. A synthetic satellite dataset of the spatio-temporal distributions of Emiliania huxleyi blooms and their impacts on Arctic and sub-Arctic marine environments (1998–2016). Earth Syst. Sci. Data 2019, 11, 119–128. [Google Scholar]
- Matson, P.G.; Washburn, L.; Fields, E.A.; Gotschalk, C.; Ladd, T.M.; Siegel, D.A.; Welch, Z.S.; Iglesias-Rodriguez, M.D. Formation, development, and propagation of a rare coastal coccolithophore bloom. J. Geophys. Res. Ocean. 2019, 124, 3298–3316. [Google Scholar] [CrossRef]
- Brown, C.W.; Yoder, J.A. Coccolithophorid blooms in the global ocean. J. Geophys. Res. Ocean. 1994, 99, 7467–7482. [Google Scholar] [CrossRef]
- Holligan, P.M.; Fernández, E.; Aiken, J.; Balch, W.M.; Boyd, P.; Burkill, P.H.; Finch, M.; Groom, S.B.; Malin, G.; Muller, K. A biogeochemical study of the coccolithophore, Emiliania huxleyi, in the North Atlantic. Glob. Biogeochem. Cycles 1993, 7, 879–900. [Google Scholar]
- Westbroek, P.; Young, J.; Linschooten, K. Coccolith production (biomineralization) in the marine alga Emiliania huxleyi. J. Eukaryot. Microbiol. 1989, 36, 368–373. [Google Scholar] [CrossRef]
- Paasche, E. A review of the coccolithophorid Emiliania huxleyi (Prymnesiophyceae), with particular reference to growth, coccolith formation, and calcification-photosynthesis interactions. Phycologia 2001, 40, 503–529. [Google Scholar]
- Marsh, M. Regulation of CaCO3 formation in coccolithophores. Comp. Biochem. Physiol. Part B Biochem. Mol. Biol. 2003, 136, 743–754. [Google Scholar]
- Taylor, A.R.; Brownlee, C.; Wheeler, G. Coccolithophore cell biology: Chalking up progress. Annu. Rev. Mar. Sci. 2017, 9, 283–310. [Google Scholar]
- Skeffington, A.W.; Scheffel, A. Exploiting algal mineralization for nanotechnology: Bringing coccoliths to the fore. Curr. Opin. Biotechnol. 2018, 49, 57–63. [Google Scholar] [CrossRef]
- Fukuda, S.-Y.; Suzuki, Y.; Shiraiwa, Y. Difference in physiological responses of growth, photosynthesis and calcification of the coccolithophore Emiliania huxleyi to acidification by acid and CO2 enrichment. Photosynth. Res. 2014, 121, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Satoh, M.; Iwamoto, K.; Suzuki, I.; Shiraiwa, Y. Cold stress stimulates intracellular calcification by the coccolithophore, Emiliania huxleyi (Haptophyceae) under phosphate-deficient conditions. Mar. Biotechnol. 2009, 11, 327–333. [Google Scholar] [CrossRef][Green Version]
- Bach, L.T.; Mackinder, L.C.; Schulz, K.G.; Wheeler, G.; Schroeder, D.C.; Brownlee, C.; Riebesell, U. Dissecting the impact of CO2 and pH on the mechanisms of photosynthesis and calcification in the coccolithophore Emiliania huxleyi. New Phytol. 2013, 199, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Benner, I.; Diner, R.E.; Lefebvre, S.C.; Li, D.; Komada, T.; Carpenter, E.J.; Stillman, J.H. Emiliania huxleyi increases calcification but not expression of calcification-related genes in long-term exposure to elevated temperature and pCO2. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2013, 368, 20130049. [Google Scholar] [CrossRef] [PubMed]
- Richier, S.; Fiorini, S.; Kerros, M.-E.; Von Dassow, P.; Gattuso, J.-P. Response of the calcifying coccolithophore Emiliania huxleyi to low pH/high pCO2: From physiology to molecular level. Mar. Biol. 2011, 158, 551–560. [Google Scholar] [CrossRef]
- Dyhrman, S.T.; Haley, S.T.; Birkeland, S.R.; Wurch, L.L.; Cipriano, M.J.; McArthur, A.G. Long serial analysis of gene expression for gene discovery and transcriptome profiling in the widespread marine coccolithophore Emiliania huxleyi. Appl. Environ. Microbiol. 2006, 72, 252–260. [Google Scholar] [CrossRef][Green Version]
- Nguyen, B.; Bowers, R.M.; Wahlund, T.M.; Read, B.A. Suppressive subtractive hybridization of and differences in gene expression content of calcifying and noncalcifying cultures of Emiliania huxleyi strain 1516. Appl. Environ. Microbiol. 2005, 71, 2564–2575. [Google Scholar] [CrossRef]
- Quinn, P.; Bowers, R.M.; Zhang, X.; Wahlund, T.M.; Fanelli, M.A.; Olszova, D.; Read, B.A. cDNA microarrays as a tool for identification of biomineralization proteins in the coccolithophorid Emiliania huxleyi (Haptophyta). Appl. Environ. Microbiol. 2006, 72, 5512–5526. [Google Scholar] [CrossRef]
- Wahlund, T.M.; Zhang, X.; Read, B.A. Expressed sequence tag profiles from calcifying and non-calcifying cultures of Emiliania huxleyi. Micropaleontology 2004, 50, 145–155. [Google Scholar] [CrossRef]
- von Dassow, P.; Ogata, H.; Probert, I.; Wincker, P.; Da Silva, C.; Audic, S.; Claverie, J.-M.; de Vargas, C. Transcriptome analysis of functional differentiation between haploid and diploid cells of Emiliania huxleyi, a globally significant photosynthetic calcifying cell. Genome Biol. 2009, 10, R114. [Google Scholar] [CrossRef] [PubMed]
- Mackinder, L.; Wheeler, G.; Schroeder, D.; von Dassow, P.; Riebesell, U.; Brownlee, C. Expression of biomineralization-related ion transport genes in Emiliania huxleyi. Environ. Microbiol. 2011, 13, 3250–3265. [Google Scholar] [CrossRef] [PubMed]
- Nam, O.; Shiraiwa, Y.; Jin, E. Calcium-related genes associated with intracellular calcification of Emiliania huxleyi (Haptophyta) CCMP 371. Algae 2018, 33, 181–189. [Google Scholar] [CrossRef]
- Nam, O.; Park, J.-M.; Lee, H.; Jin, E. De novo transcriptome profile of coccolithophorid alga Emiliania huxleyi CCMP371 at different calcium concentrations with proteome analysis. PLoS ONE 2019, 14, e0221938. [Google Scholar] [CrossRef]
- Mebarek, S.; Abousalham, A.; Magne, D.; Do, L.D.; Bandorowicz-Pikula, J.; Pikula, S.; Buchet, R. Phospholipases of mineralization competent cells and matrix vesicles: Roles in physiological and pathological mineralizations. Int. J. Mol. Sci. 2013, 14, 5036–5129. [Google Scholar] [CrossRef] [PubMed]
- Vines, C.M. Phospholipase C. In Calcium Signaling; Springer: Dordrecht, The Netherlands, 2012; pp. 235–254. [Google Scholar]
- Soto, A.R.; Zheng, H.; Shoemaker, D.; Rodriguez, J.; Read, B.A.; Wahlund, T.M. Identification and preliminary characterization of two cDNAs encoding unique carbonic anhydrases from the marine alga Emiliania huxleyi. Appl. Environ. Microbiol. 2006, 72, 5500–5511. [Google Scholar] [CrossRef]
- Guillard, R.R. Culture of phytoplankton for feeding marine invertebrates. In Culture of Marine Invertebrate Animals; Springer: Boston, MA, USA, 1975; pp. 29–60. [Google Scholar]
- Araie, H.; Sakamoto, K.; Suzuki, I.; Shiraiwa, Y. Characterization of the selenite uptake mechanism in the coccolithophore Emiliania huxleyi (Haptophyta). Plant Cell Physiol. 2011, 52, 1204–1210. [Google Scholar] [CrossRef]
- Murphy, J.; Riley, J.P. A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 1962, 27, 31–36. [Google Scholar] [CrossRef]
- Reichardt, W.; Overbeck, J.; Steubing, L. Free dissolved enzymes in lake waters. Nature 1967, 216, 1345–1347. [Google Scholar] [CrossRef]
- Pelusi, A.; Hanawa, Y.; Araie, H.; Suzuki, I.; Giordano, M.; Shiraiwa, Y. Rapid detection and quantification of haptophyte alkenones by Fourier transform infrared spectroscopy (FTIR). Algal Res. 2016, 19, 48–56. [Google Scholar] [CrossRef]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [PubMed]
- Fox, E.; Meyer, E.; Panasiak, N.; Taylor, A.R. Calcein staining as a tool to investigate coccolithophore calcification. Front. Mar. Sci. 2018, 5, 326. [Google Scholar] [CrossRef]
- Herfort, L.; Thake, B.; Roberts, J. Acquisition and use of bicarbonate by Emiliania huxleyi. New Phytol. 2002, 156, 427–436. [Google Scholar] [CrossRef]
- Herfort, L.; Loste, E.; Meldrum, F.; Thake, B. Structural and physiological effects of calcium and magnesium in Emiliania huxleyi (Lohmann) Hay and Mohler. J. Struct. Biol. 2004, 148, 307–314. [Google Scholar] [CrossRef]
- Trimborn, S.; Langer, G. Effect of varying calcium concentrations and light intensities on calcification and photosynthesis in Emiliania huxleyi. Limnol. Oceanogr. 2007, 52, 2285–2293. [Google Scholar] [CrossRef]
- Leonardos, N.; Read, B.; Thake, B.; Young, J.R. No mechanistic dependence of photosynthesis on calcification in the coccolithophorid Emiliania huxleyi (Haptophyta). J. Phycol. 2009, 45, 1046–1051. [Google Scholar] [CrossRef]
- Read, B.A.; Kegel, J.; Klute, M.J.; Kuo, A.; Lefebvre, S.C.; Maumus, F.; Mayer, C.; Miller, J.; Monier, A.; Salamov, A. Pan genome of the phytoplankton Emiliania underpins its global distribution. Nature 2013, 499, 209–213. [Google Scholar] [CrossRef]
- Paasche, E. Coccolith formation. Nature 1962, 193, 1094–1095. [Google Scholar] [CrossRef]
- Khosla, C.; Gokhale, R.S.; Jacobsen, J.R.; Cane, D.E. Tolerance and specificity of polyketide synthases. Annu. Rev. Biochem. 1999, 68, 219–253. [Google Scholar] [CrossRef] [PubMed]
- Hojo, M.; Omi, A.; Hamanaka, G.; Shindo, K.; Shimada, A.; Kondo, M.; Narita, T.; Kiyomoto, M.; Katsuyama, Y.; Ohnishi, Y. Unexpected link between polyketide synthase and calcium carbonate biomineralization. Zool. Lett. 2015, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Beeble, A.; Calestani, C. Expression pattern of polyketide synthase-2 during sea urchin development. Gene Expr. Patterns 2012, 12, 7–10. [Google Scholar] [CrossRef]
- John, U.; Beszteri, B.; Derelle, E.; Van de Peer, Y.; Read, B.; Moreau, H.; Cembella, A. Novel insights into evolution of protistan polyketide synthases through phylogenomic analysis. Protist 2008, 159, 21–30. [Google Scholar] [CrossRef]
- Rokitta, S.D.; de Nooijer, L.J.; Trimborn, S.; de Vargas, C.; Rost, B.; John, U. Transcriptome analyses reveal different gene expression patterns between the life-cycle stages of Emiliania huxleyi (Haptophyta) and reflect specialization to different ecological niches. J. Phycol. 2011, 47, 829–838. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.M.; Edwards, R.J.; Skipp, P.J.; O’Connor, C.D.; Iglesias-Rodriguez, M.D. Shotgun proteomic analysis of Emiliania huxleyi, a marine phytoplankton species of major biogeochemical importance. Mar. Biotechnol. 2011, 13, 496–504. [Google Scholar] [CrossRef]
- Heilmann, I. Phosphoinositide signaling in plant development. Development 2016, 143, 2044–2055. [Google Scholar] [CrossRef]
- Razzini, G.; Brancaccio, A.; Lemmon, M.A.; Guarnieri, S.; Falasca, M. The role of the pleckstrin homology domain in membrane targeting and activation of phospholipase Cβ1. J. Biol. Chem. 2000, 275, 14873–14881. [Google Scholar] [CrossRef][Green Version]
- Rebecchi, M.J.; Pentyala, S.N. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol. Rev. 2000, 80, 1291–1335. [Google Scholar] [CrossRef]
- Wing, M.R.; Houston, D.; Kelley, G.G.; Der, C.J.; Siderovski, D.P.; Harden, T.K. Activation of phospholipase C-ε by heterotrimeric G protein βγ-subunits. J. Biol. Chem. 2001, 276, 48257–48261. [Google Scholar] [CrossRef]
- Díaz Añel, A.M. Phospholipase C β3 is a key component in the Gβγ/PKCη/PKD-mediated regulation of trans-Golgi network to plasma membrane transport. Biochem. J. 2007, 406, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Brownlee, C.; Taylor, A. Calcification in coccolithophores: A cellular perspective. In Coccolithophores; Springer: Berlin/Heidelberg, Germany, 2004; pp. 31–49. [Google Scholar]
- Brownlee, C.; Wheeler, G.L.; Taylor, A.R. Coccolithophore biomineralization: New questions, new answers. Semin. Cell Dev. Biol. 2015, 52, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Young, J.R.; Davis, S.A.; Bown, P.R.; Mann, S. Coccolith ultrastructure and biomineralisation. J. Struct. Biol. 1999, 126, 195–215. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Yoshida, M.; Uji, T.; Saga, N.; Inoue, K.; Nagasawa, H. Stable nuclear transformation system for the Coccolithophorid alga Pleurochrysis carterae. Sci. Rep. 2016, 6, 22252. [Google Scholar] [CrossRef] [PubMed]
- Endo, H.; Hanawa, Y.; Araie, H.; Suzuki, I.; Shiraiwa, Y. Overexpression of Tisochrysis lutea Akd1 identifies a key cold-induced alkenone desaturase enzyme. Sci. Rep. 2018, 8, 11230. [Google Scholar] [CrossRef] [PubMed]
- Sviben, S.; Gal, A.; Hood, M.A.; Bertinetti, L.; Politi, Y.; Bennet, M.; Krishnamoorthy, P.; Schertel, A.; Wirth, R.; Sorrentino, A. A vacuole-like compartment concentrates a disordered calcium phase in a key coccolithophorid alga. Nat. Commun. 2016, 7, 11228. [Google Scholar] [CrossRef] [PubMed]
- Gal, A.; Sorrentino, A.; Kahil, K.; Pereiro, E.; Faivre, D.; Scheffel, A. Native-state imaging of calcifying and noncalcifying microalgae reveals similarities in their calcium storage organelles. Proc. Natl. Acad. Sci. USA 2018, 115, 11000–11005. [Google Scholar] [CrossRef]



| Term ID | Description | Count in Gene Set | FDR |
|---|---|---|---|
| PF14765 | Polyketide synthase dehydratase | 5 of 18 | 1.28 × 10−7 |
| PF00550 | Phosphopantetheine attachment site | 5 of 27 | 3.78 × 10−7 |
| PF00975 | Thioesterase domain | 4 of 19 | 5.50 × 10−6 |
| PF02801 | Beta-ketoacyl synthase, C-terminal domain | 4 of 19 | 5.50 × 10−6 |
| PF00109 | Beta-ketoacyl synthase, N-terminal domain | 4 of 24 | 7.57 × 10−6 |
| PF08659 | KR domain | 5 of 127 | 0.00016 |
| PF00127 | Copper binding proteins, plastocyanin/azurin family | 2 of 9 | 0.0034 |
| PF00063 | Myosin head (motor domain) | 3 of 57 | 0.0036 |
| PF08811 | Protein of unknown function (DUF1800) | 2 of 10 | 0.0036 |
| PF00388 | Phosphatidylinositol-specific phospholipase C, X domain | 2 of 19 | 0.009 |
| PF00169 | PH domain | 2 of 35 | 0.0255 |
| Term ID | Description | Count in Gene Set | FDR |
|---|---|---|---|
| IPR006162 | Phosphopantetheine attachment site | 5 of 19 | 2.35 × 10−7 |
| IPR013968 | Polyketide synthase, ketoreductase domain | 5 of 23 | 2.35 × 10−7 |
| IPR020806 | Polyketide synthase, phosphopantetheine-binding domain | 5 of 19 | 2.35 × 10−7 |
| IPR020807 | Polyketide synthase, dehydratase domain | 5 of 18 | 2.35 × 10−7 |
| IPR009081 | Phosphopantetheine binding ACP domain | 5 of 27 | 2.78 × 10−7 |
| IPR036736 | ACP-like superfamily | 5 of 28 | 2.78 × 10−7 |
| IPR020843 | Polyketide synthase, enoylreductase domain | 5 of 34 | 5.60 × 10−7 |
| IPR001031 | Thioesterase | 4 of 13 | 1.03 × 10−6 |
| IPR016039 | Thiolase-like | 5 of 40 | 1.03 × 10−6 |
| IPR014031 | Beta-ketoacyl synthase, C-terminal | 4 of 19 | 3.03 × 10−6 |
| IPR020841 | Polyketide synthase, beta-ketoacyl synthase domain | 4 of 19 | 3.03 × 10−6 |
| IPR014030 | Beta-ketoacyl synthase, N-terminal | 4 of 21 | 3.59 × 10−6 |
| IPR011032 | GroES-like superfamily | 5 of 59 | 3.83 × 10−6 |
| IPR008972 | Cupredoxin | 3 of 33 | 0.00083 |
| IPR016040 | NAD(P)-binding domain | 5 of 211 | 0.0013 |
| IPR018201 | Beta-ketoacyl synthase, active site | 2 of 6 | 0.0014 |
| IPR000923 | Blue (type 1) copper domain | 2 of 8 | 0.0021 |
| IPR013217 | Methyltransferase type 12 | 2 of 8 | 0.0021 |
| IPR001609 | Myosin head, motor domain | 3 of 53 | 0.0023 |
| IPR014917 | Protein of unknown function DUF1800 | 2 of 10 | 0.0026 |
| IPR036774 | ERV/ALR sulfhydryl oxidase domain superfamily | 2 of 12 | 0.0035 |
| IPR000909 | Phosphatidylinositol-specific phospholipase C, X domain | 2 of 19 | 0.0075 |
| IPR001711 | Phospholipase C, phosphatidylinositol-specific, Y domain | 2 of 23 | 0.0103 |
| IPR008979 | Galactose-binding-like domain superfamily | 3 of 103 | 0.0117 |
| IPR001192 | Phosphoinositide phospholipase C family | 2 of 27 | 0.0127 |
| IPR017946 | PLC-like phosphodiesterase, TIM beta/alpha-barrel domain superfamily | 2 of 30 | 0.0146 |
| IPR036961 | Kinesin motor domain superfamily | 3 of 115 | 0.0146 |
| IPR036291 | NAD(P)-binding domain superfamily | 5 of 471 | 0.0234 |
| IPR020845 | AMP-binding, conserved site | 2 of 44 | 0.0273 |
| IPR029058 | Alpha/Beta hydrolase fold | 5 of 527 | 0.0347 |
| IPR016035 | Acyl transferase/acyl hydrolase/lysophospholipase | 2 of 59 | 0.0442 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, O.; Suzuki, I.; Shiraiwa, Y.; Jin, E. Association of Phosphatidylinositol-Specific Phospholipase C with Calcium-Induced Biomineralization in the Coccolithophore Emiliania huxleyi. Microorganisms 2020, 8, 1389. https://doi.org/10.3390/microorganisms8091389
Nam O, Suzuki I, Shiraiwa Y, Jin E. Association of Phosphatidylinositol-Specific Phospholipase C with Calcium-Induced Biomineralization in the Coccolithophore Emiliania huxleyi. Microorganisms. 2020; 8(9):1389. https://doi.org/10.3390/microorganisms8091389
Chicago/Turabian StyleNam, Onyou, Iwane Suzuki, Yoshihiro Shiraiwa, and EonSeon Jin. 2020. "Association of Phosphatidylinositol-Specific Phospholipase C with Calcium-Induced Biomineralization in the Coccolithophore Emiliania huxleyi" Microorganisms 8, no. 9: 1389. https://doi.org/10.3390/microorganisms8091389
APA StyleNam, O., Suzuki, I., Shiraiwa, Y., & Jin, E. (2020). Association of Phosphatidylinositol-Specific Phospholipase C with Calcium-Induced Biomineralization in the Coccolithophore Emiliania huxleyi. Microorganisms, 8(9), 1389. https://doi.org/10.3390/microorganisms8091389





