Microbial Communities in Soils and Endosphere of Solanum tuberosum L. and their Response to Long-Term Fertilization
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design, Sample Collection and Processing
2.2. DNA Isolation
2.3. 16S rRNA Gene and ITS Region Amplicon Generation from Soil Materials
2.4. 16S rRNA Gene and ITS Region Amplicon Generation from Plant Materials
2.5. Amplicon Purification and Sequencing
2.6. Data Processing
2.7. Multivariate Statistical Analyses
3. Results
3.1. Analysis of Microbial Diversity in Soil and Potatoes
3.2. Microbial Community Structure in Soil and Tubers
3.3. Specific Effect of Fertilization on Microbial Community Structure
3.4. Genera Significantly Enriched in Fertilized Soil or Tubers
3.5. Transmission of Potential Human Pathogens
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Johnston-Monje, D.; Raizada, M.N. Plant and Endophyte Relationships. In Comprehensive Biotechnology; Elsevier: Amsterdam, The Netherlands, 2011; pp. 713–727. ISBN 978-0-08-088504-9. [Google Scholar]
- European Commission COM(2015) 614 European Commission. Closing the Loop—An EU Action Plan for the Circular Economy; European Commission: Brussels, Belgium, 2015; COM (2015) 614. [Google Scholar]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. Microbial Diversity in Soil: Selection of Microbial Populations by Plant and Soil Type and Implications for Disease Suppressiveness. Annu. Rev. Phytopathol. 2004, 42, 243–270. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Yang, T.; Friman, V.-P.; Xu, Y.; Shen, Q.; Jousset, A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun 2015, 6, 8413. [Google Scholar] [CrossRef] [PubMed]
- Treseder, K.K.; Balser, T.C.; Bradford, M.A.; Brodie, E.L.; Dubinsky, E.A.; Eviner, V.T.; Hofmockel, K.S.; Lennon, J.T.; Levine, U.Y.; MacGregor, B.J.; et al. Integrating microbial ecology into ecosystem models: Challenges and priorities. Biogeochemistry 2012, 109, 7–18. [Google Scholar] [CrossRef]
- Rousk, J.; Bengtson, P. Microbial regulation of global biogeochemical cycles. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Schulz, S.; Brankatschk, R.; Dümig, A.; Kögel-Knabner, I.; Schloter, M.; Zeyer, J. The role of microorganisms at different stages of ecosystem development for soil formation. Biogeosciences 2013, 10, 3983–3996. [Google Scholar] [CrossRef]
- Anderson, T.-H. Microbial eco-physiological indicators to asses soil quality. Agric. Ecosyst. Environ. 2003, 98, 285–293. [Google Scholar] [CrossRef]
- Herencia, J.F.; Ruiz-Porras, J.C.; Melero, S.; Garcia-Galavis, P.A.; Morillo, E.; Maqueda, C. Comparison between Organic and Mineral Fertilization for Soil Fertility Levels, Crop Macronutrient Concentrations, and Yield. Agron. J. 2007, 99, 973–983. [Google Scholar] [CrossRef]
- Zhang, Y.; Shen, H.; He, X.; Thomas, B.W.; Lupwayi, N.Z.; Hao, X.; Thomas, M.C.; Shi, X. Fertilization Shapes Bacterial Community Structure by Alteration of Soil pH. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Blanchet, G.; Gavazov, K.; Bragazza, L.; Sinaj, S. Responses of soil properties and crop yields to different inorganic and organic amendments in a Swiss conventional farming system. Agric. Ecosyst. Environ. 2016, 230, 116–126. [Google Scholar] [CrossRef]
- Geisseler, D.; Scow, K.M. Long-term effects of mineral fertilizers on soil microorganisms—A review. Soil Biol. Biochem. 2014, 75, 54–63. [Google Scholar] [CrossRef]
- Francioli, D.; Schulz, E.; Lentendu, G.; Wubet, T.; Buscot, F.; Reitz, T. Mineral vs. Organic Amendments: Microbial Community Structure, Activity and Abundance of Agriculturally Relevant Microbes Are Driven by Long-Term Fertilization Strategies. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Chaparro, J.M.; Sheflin, A.M.; Manter, D.K.; Vivanco, J.M. Manipulating the soil microbiome to increase soil health and plant fertility. Biol. Fertil. Soils 2012, 48, 489–499. [Google Scholar] [CrossRef]
- Papik, J.; Folkmanova, M.; Polivkova, M.; Suman, J.; Uhlik, O. The Invisible Life Inside Plants: Deciphering the Riddles of Endophytic Bacterial Diversity. Biotechnol. Adv. 2020, 107614. [Google Scholar] [CrossRef] [PubMed]
- Miliute, I.; Buzaite, O.; Baniulis, D.; Stanys, V. Bacterial endophytes in agricultural crops and their role in stress tolerance: A review. Zemdirb. Agric. 2015, 102, 465–478. [Google Scholar] [CrossRef]
- Guo, B.; Wang, Y.; Sun, X.; Tang, K. Bioactive natural products from endophytes: A review. Appl. Biochem. Microbiol. 2008, 44, 136–142. [Google Scholar] [CrossRef]
- Yadav, A. Exploring the Potential of Endophytes in Agriculture: A Minireview. APAR 2017, 6. [Google Scholar] [CrossRef]
- White, J.F.; Kingsley, K.L.; Zhang, Q.; Verma, R.; Obi, N.; Dvinskikh, S.; Elmore, M.T.; Verma, S.K.; Gond, S.K.; Kowalski, K.P. Review: Endophytic microbes and their potential applications in crop management. Pest. Manag. Sci. 2019, 75, 2558–2565. [Google Scholar] [CrossRef]
- Krell, V.; Unger, S.; Jakobs-Schoenwandt, D.; Patel, A.V. Endophytic Metarhizium brunneum mitigates nutrient deficits in potato and improves plant productivity and vitality. Fungal Ecology 2018, 34, 43–49. [Google Scholar] [CrossRef]
- Ikeda, S.; Sasaki, K.; Okubo, T.; Yamashita, A.; Terasawa, K.; Bao, Z.; Liu, D.; Watanabe, T.; Murase, J.; Asakawa, S.; et al. Low Nitrogen Fertilization Adapts Rice Root Microbiome to Low Nutrient Environment by Changing Biogeochemical Functions. Microb. Environ. 2014, 29, 50–59. [Google Scholar] [CrossRef]
- Redman, R.S.; Kim, Y.O.; Woodward, C.J.D.A.; Greer, C.; Espino, L.; Doty, S.L.; Rodriguez, R.J. Increased Fitness of Rice Plants to Abiotic Stress Via Habitat Adapted Symbiosis: A Strategy for Mitigating Impacts of Climate Change. PLoS ONE 2011, 6, e14823. [Google Scholar] [CrossRef]
- Buchholz, F.; Kostić, T.; Sessitsch, A.; Mitter, B. The potential of plant microbiota in reducing postharvest food loss. Microb. Biotechnol. 2018, 11, 971–975. [Google Scholar] [CrossRef] [PubMed]
- Seghers, D.; Wittebolle, L.; Top, E.M.; Verstraete, W.; Siciliano, S.D. Impact of Agricultural Practices on the Zea mays L. Endophytic Community. Appl. Environ Microbiol. 2004, 70, 1475–1482. [Google Scholar] [CrossRef] [PubMed]
- Pariona-Llanos, R.; Ibañez de Santi Ferrara, F.; Soto Gonzales, H.H.; Barbosa, H.R. Influence of organic fertilization on the number of culturable diazotrophic endophytic bacteria isolated from sugarcane. Eur. J. Soil Biol. 2010, 46, 387–393. [Google Scholar] [CrossRef]
- Yang, Q.; Zhang, H.; Guo, Y.; Tian, T. Influence of Chicken Manure Fertilization on Antibiotic-Resistant Bacteria in Soil and the Endophytic Bacteria of Pakchoi. Int. J. Environ. Res. Public Health 2016, 13, 662. [Google Scholar] [CrossRef] [PubMed]
- Manyi-Loh, C.; Mamphweli, S.; Meyer, E.; Makaka, G.; Simon, M.; Okoh, A. An Overview of the Control of Bacterial Pathogens in Cattle Manure. IJERPH 2016, 13, 843. [Google Scholar] [CrossRef]
- Stiborova, H.; Wolfram, J.; Demnerova, K.; Macek, T.; Uhlik, O. Bacterial community structure in treated sewage sludge with mesophilic and thermophilic anaerobic digestion. Folia Microbiol. 2015, 60, 531–539. [Google Scholar] [CrossRef]
- FAO. World Food and Agriculture—Statistical Pocketbook 2019; Food And Agriculture Organization of the United Nations: Rome, Italy, 2019; ISBN 978-92-5-131849-2. [Google Scholar]
- Lopez-Echartea, E.; Strejcek, M.; Mukherjee, S.; Uhlik, O.; Yrjälä, K. Bacterial succession in oil-contaminated soil under phytoremediation with poplars. Chemosphere 2020, 243, 125242. [Google Scholar] [CrossRef]
- Fraraccio, S.; Strejcek, M.; Dolinova, I.; Macek, T.; Uhlik, O. Secondary compound hypothesis revisited: Selected plant secondary metabolites promote bacterial degradation of cis-1,2-dichloroethylene (cDCE). Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Taylor, D.L.; Walters, W.A.; Lennon, N.J.; Bochicchio, J.; Krohn, A.; Caporaso, J.G.; Pennanen, T. Accurate Estimation of Fungal Diversity and Abundance through Improved Lineage-Specific Primers Optimized for Illumina Amplicon Sequencing. Appl. Environ. Microbiol. 2016, 82, 7217–7226. [Google Scholar] [CrossRef]
- R Core Team R. A Language and Environment for Statistical Computing in R Foundation for Statistical Computing; R Core Team R: Vienna, Austria, 2017. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Callahan, B.J. Silva Taxonomic Training Data Formatted for DADA2 (Silva version 132) [Data set]; Zenodo: Callahan, Benjamin, 2018. [Google Scholar] [CrossRef]
- UNITE Community. UNITE General FASTA Release for Fungi 2. Version 18.11.2018. UNITE Community, 2019. Available online: https://unite.ut.ee/repository.php (accessed on 04 September 2020).
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. vegan: Community Ecology Package. R package version 2.5-6. 2019. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 04 September 2020).
- Callahan, B.J.; Sankaran, K.; Fukuyama, J.A.; McMurdie, P.J.; Holmes, S.P. Bioconductor workflow for microbiome data analysis: From raw reads to community analyses. F1000Res 2016, 5, 1492. [Google Scholar] [CrossRef] [PubMed]
- Legendre, P.; Gallagher, E.D.G. Ecologically Meaningful Transformations for Ordination of Species Data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; Balakrishnan, N., Colton, T., Everitt, B., Piegorsch, W., Ruggeri, F., Teugels, J.L., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2014; pp. 1–15. ISBN 978-1-118-44511-2. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001, 26, 32–46. [Google Scholar]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST + : Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal Biodiversity and Their Role in Soil Health. Front. Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef]
- Rashid, M.I.; Mujawar, L.H.; Shahzad, T.; Almeelbi, T.; Ismail, I.M.I.; Oves, M. Bacteria and fungi can contribute to nutrients bioavailability and aggregate formation in degraded soils. Microbiol. Res. 2016, 183, 26–41. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.-P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef]
- Wang, Z.; Liu, Y.; Zhao, L.; Zhang, W.; Liu, L. Change of soil microbial community under long-term fertilization in a reclaimed sandy agricultural ecosystem. PeerJ 2019, 7, e6497. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zhou, A.; Wen, K.; Liu, Z.; Liu, W.; Wang, A.; Yue, X. Upgrading VFAs bioproduction from waste activated sludge via co-fermentation with soy sauce residue. Front. Environ. Sci. Eng. 2019, 13, 3. [Google Scholar] [CrossRef]
- Pang, L.; Ge, L.; Yang, P.; He, H.; Zhang, H. Degradation of organophosphate esters in sewage sludge: Effects of aerobic/anaerobic treatments and bacterial community compositions. Bioresour. Technol. 2018, 255, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Shao, M.-F.; Ye, L. 454 Pyrosequencing reveals bacterial diversity of activated sludge from 14 sewage treatment plants. ISME J. 2012, 6, 1137. [Google Scholar] [CrossRef] [PubMed]
- Adams, G.O.; Fufeyin, P.T.; Okoro, S.E.; Ehinomen, I. Bioremediation, Biostimulation and Bioaugmention: A Review. Int. J. Environ. Bioremediation Biodegrad. 2015, 3, 28–39. [Google Scholar] [CrossRef]
- Hemmat-Jou, M.H.; Safari-Sinegani, A.A.; Mirzaie-Asl, A.; Tahmourespour, A. Analysis of microbial communities in heavy metals-contaminated soils using the metagenomic approach. Ecotoxicology 2018, 27, 1281–1291. [Google Scholar] [CrossRef]
- Lopez-Echartea, E.; Strejcek, M.; Mateju, V.; Vosahlova, S.; Kyclt, R.; Demnerova, K.; Uhlik, O. Bioremediation of chlorophenol-contaminated sawmill soil using pilot-scale bioreactors under consecutive anaerobic-aerobic conditions. Chemosphere 2019, 227, 670–680. [Google Scholar] [CrossRef]
- Khillare, P.S.; Sattawan, V.K.; Jyethi, D.S. Profile of polycyclic aromatic hydrocarbons in digested sewage sludge. Environ. Technol. 2018, 1–10. [Google Scholar] [CrossRef]
- Stiborova, H.; Kolar, M.; Vrkoslavova, J.; Pulkrabova, J.; Hajslova, J.; Demnerova, K.; Uhlik, O. Linking toxicity profiles to pollutants in sludge and sediments. J. Hazard. Mater. 2017, 321, 672–680. [Google Scholar] [CrossRef]
- Stiborova, H.; Strejcek, M.; Musilova, L.; Demnerova, K.; Uhlik, O. Diversity and phylogenetic composition of bacterial communities and their association with anthropogenic pollutants in sewage sludge. Chemosphere 2020, 238, 124629. [Google Scholar] [CrossRef]
- Maropola, M.K.A.; Ramond, J.-B.; Trindade, M. Impact of metagenomic DNA extraction procedures on the identifiable endophytic bacterial diversity in Sorghum bicolor (L. Moench). J. Microbiol. Methods 2015, 112, 104–117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Yuen, G.Y. The Role of Chitinase Production by Stenotrophomonas maltophilia Strain C3 in Biological Control of Bipolaris sorokiniana. Phytopathology 2000, 90, 384–389. [Google Scholar] [CrossRef] [PubMed]
- Sommermann, L.; Geistlinger, J.; Wibberg, D.; Deubel, A.; Zwanzig, J.; Babin, D.; Schlüter, A.; Schellenberg, I. Fungal community profiles in agricultural soils of a long-term field trial under different tillage, fertilization and crop rotation conditions analyzed by high-throughput ITS-amplicon sequencing. PLoS ONE 2018, 13, e0195345. [Google Scholar] [CrossRef]
- Berruti, A.; Lumini, E.; Balestrini, R.; Bianciotto, V. Arbuscular Mycorrhizal Fungi as Natural Biofertilizers: Let’s Benefit from Past Successes. Front. Microbiol. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Miransari, M. Soil microbes and plant fertilization. Appl. Microbiol. Biotechnol. 2011, 92, 875–885. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wu, X.; Li, G.; Qin, P. Interactions between arbuscular mycorrhizal fungi and phosphate-solubilizing fungus (Mortierella sp.) and their effects on Kostelelzkya virginica growth and enzyme activities of rhizosphere and bulk soils at different salinities. Biol. Fertil. Soils 2011, 47, 543. [Google Scholar] [CrossRef]
- Liu, K.; Ding, H.; Yu, Y.; Chen, B. A Cold-Adapted Chitinase-Producing Bacterium from Antarctica and Its Potential in Biocontrol of Plant Pathogenic Fungi. Marine Drugs 2019, 17, 695. [Google Scholar] [CrossRef] [PubMed]
- Mishra, P.; Kshirsagar, P.R.; Nilegaonkar, S.S.; Singh, S.K. Statistical optimization of medium components for production of extracellular chitinase by Basidiobolus ranarum: A novel biocontrol agent against plant pathogenic fungi. J. Basic Microbiol. 2012, 52, 539–548. [Google Scholar] [CrossRef] [PubMed]
- Hallen, H.E.; Watling, R.; Adams, G.C. Taxonomy and toxicity of Conocybe lactea and related species. Mycol. Res. 2003, 107, 969–979. [Google Scholar] [CrossRef]
- Gu, H.; Lou, J.; Wang, H.; Yang, Y.; Wu, L.; Wu, J.; Xu, J. Biodegradation, Biosorption of Phenanthrene and Its Trans-Membrane Transport by Massilia sp. WF1 and Phanerochaete chrysosporium. Front. Microbiol. 2016, 7, 38. [Google Scholar] [CrossRef]
- Wemheuer, F.; Kaiser, K.; Karlovsky, P.; Daniel, R.; Vidal, S.; Wemheuer, B. Bacterial endophyte communities of three agricultural important grass species differ in their response towards management regimes. Sci. Rep. 2017, 7, 40914. [Google Scholar] [CrossRef] [PubMed]
- Nakasaki, K.; Saito, M.; Suzuki, N. Coprinellus curtus (Hitoyo-take) prevents diseases of vegetables caused by pathogenic fungi. FEMS Microbiol. Lett. 2007, 275, 286–291. [Google Scholar] [CrossRef] [PubMed]
- Duczek, L.J. Biological control of common root rot in barley by Idriella bolleyi. Can. J. Plant Pathol. 1997, 19, 402–405. [Google Scholar] [CrossRef]
- Lascaris, D.; Deacon, J.W. Colonization of wheat roots from seed-applied spores of Idriella (Microdochium) bolleyi: A biocontrol agent of take-all. Biocontrol. Sci. Technol. 1991, 1, 229–240. [Google Scholar] [CrossRef]
- Roux, J.; Wingfield, M. Ceratocystis species: Emerging pathogens of non-native plantation Eucalyptus and Acacia species. South. For. J. For. Sci. 2009, 71, 115–120. [Google Scholar] [CrossRef]
- Tóth, G.; Montanarella, L.; Stolbovoy, V.; Máté, F.; Bódis, K.; Jones, A.; Panagos, P.; Van Liedekerke, M.; European Commission; Joint Research Centre; et al. Soils of the European Union; Publications Office: Luxembourg, 2008; ISBN 978-92-79-09530-6. [Google Scholar]
- Yao, H.; He, Z.; Wilson, M.J.; Campbell, C.D. Microbial Biomass and Community Structure in a Sequence of Soils with Increasing Fertility and Changing Land Use. Microb. Ecol. 2000, 40, 223–237. [Google Scholar] [CrossRef]
- Afzal, I.; Shinwari, Z.K.; Sikandar, S.; Shahzad, S. Plant beneficial endophytic bacteria: Mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 2019, 221, 36–49. [Google Scholar] [CrossRef]
- Gadhave, K.R.; Devlin, P.F.; Ebertz, A.; Ross, A.; Gange, A.C. Soil Inoculation with Bacillus spp. Modifies Root Endophytic Bacterial Diversity, Evenness, and Community Composition in a Context-Specific Manner. Microb. Ecol. 2018, 76, 741–750. [Google Scholar] [CrossRef]
- Fang, K.; Bao, Z.-S.-N.; Chen, L.; Zhou, J.; Yang, Z.-P.; Dong, X.-F.; Zhang, H.-B. Growth-promoting characteristics of potential nitrogen-fixing bacteria in the root of an invasive plant Ageratina adenophora. PeerJ 2019, 7, e7099. [Google Scholar] [CrossRef]
- Ramakrishna, W.; Yadav, R.; Li, K. Plant growth promoting bacteria in agriculture: Two sides of a coin. Appl. Soil Ecol. 2019, 138, 10–18. [Google Scholar] [CrossRef]
- Grall, S.; Manceau, C. Colonization of Vitis vinifera by a Green Fluorescence Protein-Labeled, gfp-Marked Strain of Xylophilus ampelinus, the Causal Agent of Bacterial Necrosis of Grapevine. AEM 2003, 69, 1904–1912. [Google Scholar] [CrossRef] [PubMed]
- Adegoke, A.A.; Awolusi, O.O.; Stenström, T.A. Organic Fertilizers: Public Health Intricacies. In Organic Fertilizers—From Basic Concepts to Applied Outcomes; Larramendy, M.L., Soloneski, S., Eds.; InTech: Rijeka, Croatia, 2016; ISBN 978-953-51-2449-8. [Google Scholar]
- Sharma, M.; Reynnells, R. Importance of Soil Amendments: Survival of Bacterial Pathogens in Manure and Compost Used as Organic Fertilizers. In Preharvest Food Safety; Thakur, K., Ed.; American Society of Microbiology: Washington, DC, USA, 2018; pp. 159–175. ISBN 978-1-55581-707-7. [Google Scholar]
- Solomon, E.B.; Yaron, S.; Matthews, K.R. Transmission of Escherichia coli O157:H7 from Contaminated Manure and Irrigation Water to Lettuce Plant Tissue and Its Subsequent Internalization. AEM 2002, 68, 397–400. [Google Scholar] [CrossRef]
- Szczech, M.; Kowalska, B.; Smolińska, U.; Maciorowski, R.; Oskiera, M.; Michalska, A. Microbial quality of organic and conventional vegetables from Polish farms. Int. J. Food Microbiol. 2018, 286, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Kljujev, I.; Raicevic, V.; Jovicic-Petrovic, J.; Vujovic, B.; Mirkovic, M.; Rothballer, M. Listeria monocytogenes—Danger for health safety vegetable production. Microb. Pathog. 2018, 120, 23–31. [Google Scholar] [CrossRef] [PubMed]
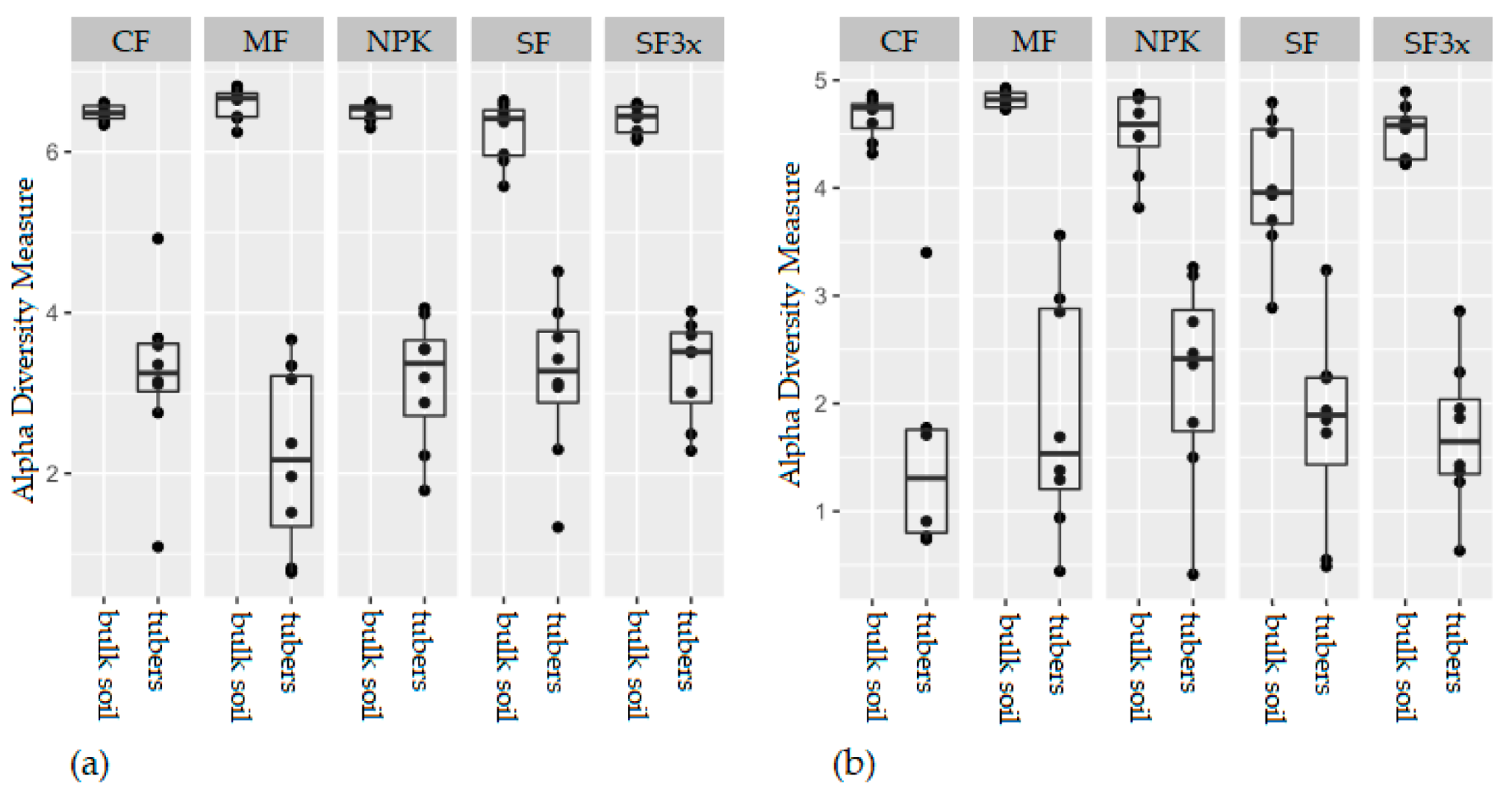
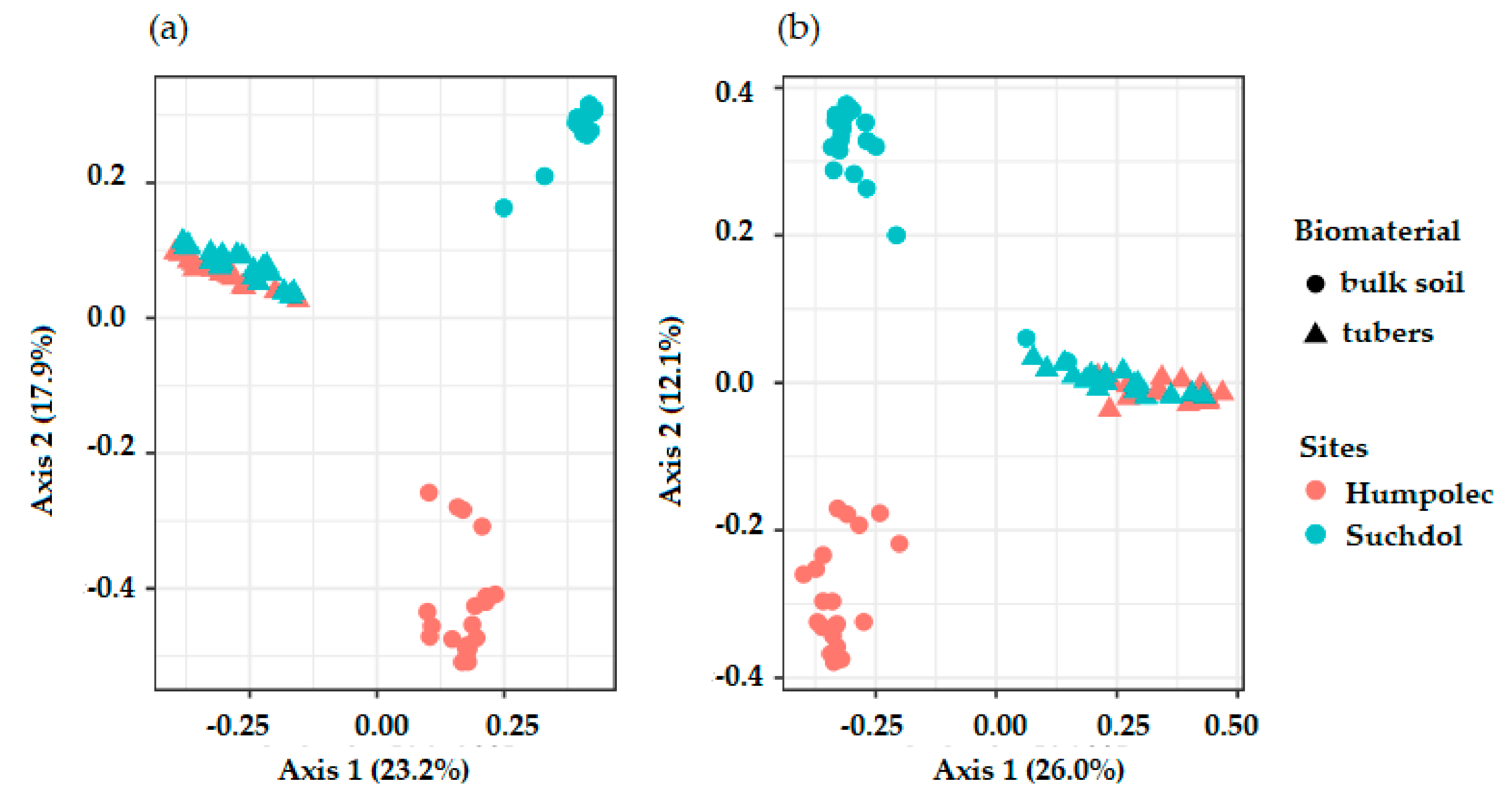
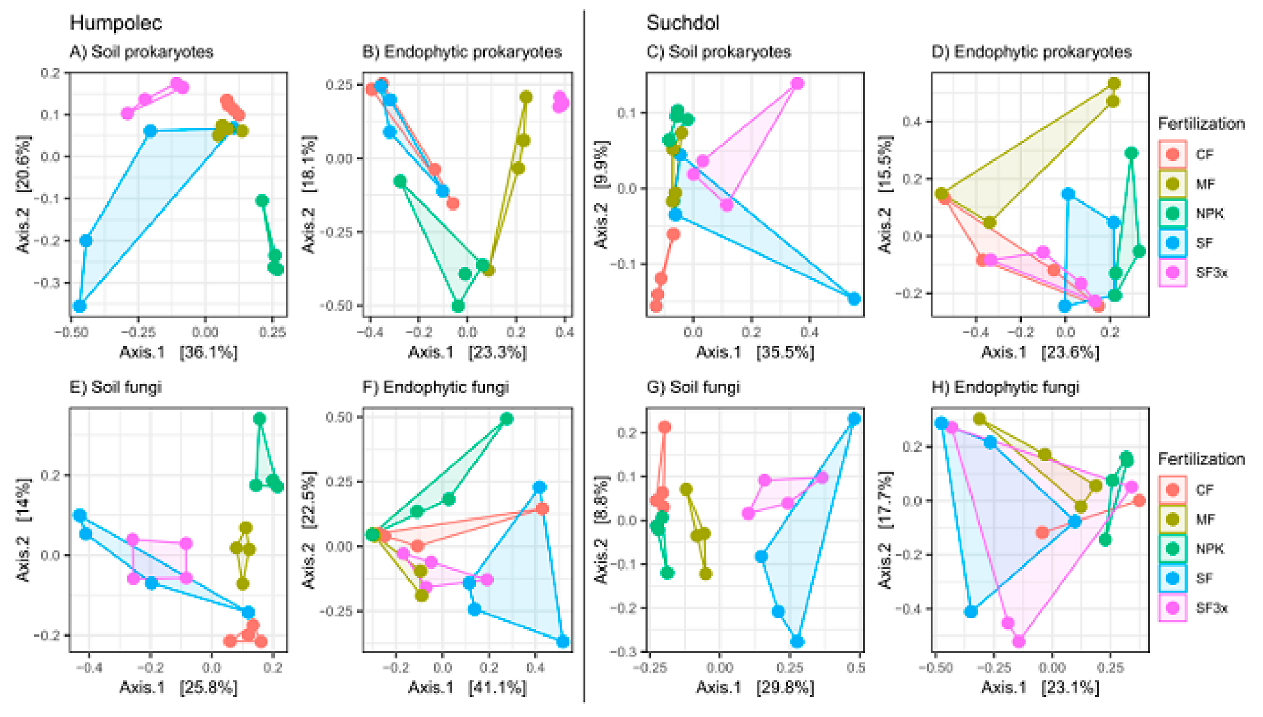
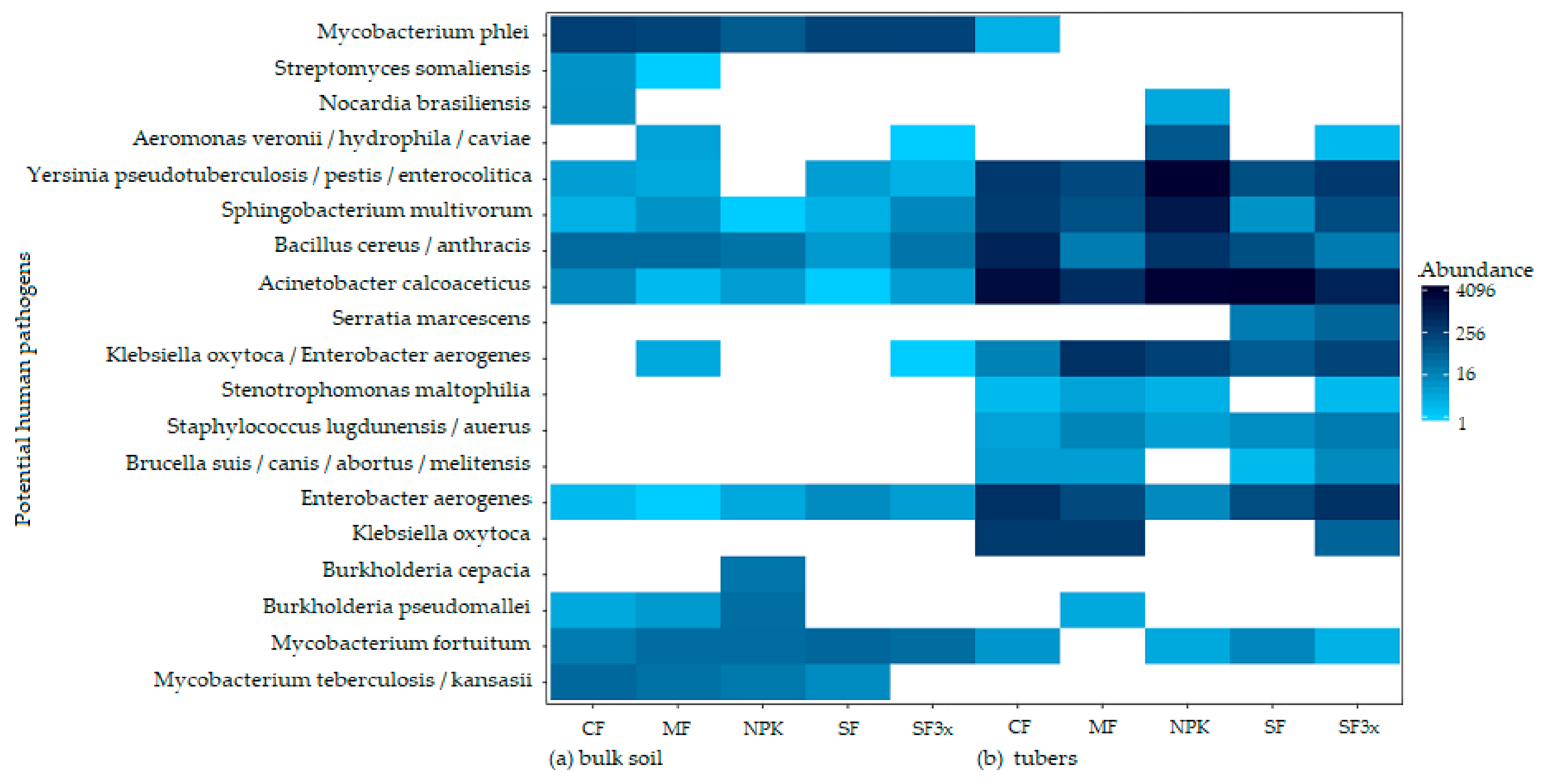
| Site 1: Humpolec | Site 2: Prague-Suchdol | |
|---|---|---|
| GPS | 49°33′16″ N, 15°21′2″ E | 50°7′40″ N, 14°22′33″ E |
| Elevation [m] | 525 | 286 |
| CEC 1 [mmol(+)/kg] | 90 | 262 |
| Cox [%] | 1.24 | 1.76 |
| pH | 5.27 ± 0.5 | 7.8 ± 0.4 |
| Bulk density [g/cm3] | 1.40 | 1.43 |
| Clay [%] | 5.84 | 2.18 |
| Silt [%] | 43.55 | 71.8 |
| Sand [%] | 50.61 | 26.03 |
| Soil type (WRB 2006) | Cambisol | Chernozem |
| NRCS 2 USDA | silty loam | silty loam |
| Bulk Soil | Tubers | |||||||
|---|---|---|---|---|---|---|---|---|
| Prokaryotes | Fungi | Prokaryotes | Fungi | |||||
| R2 | Padj | R2 | Padj | R2 | Padj | R2 | Padj | |
| Site characteristics | 57% | 0.001 * | 39% | 0.001 * | 10% | 0.001 * | 7% | 0.001 * |
| Fertilization | 11% | 0.002 * | 14% | 0.003 * | 14% | 0.001 * | 11% | 0.087 |
| Fertilization X Site char. | 8% | 0.004 * | 11% | 0.007 * | 18% | 0.001 * | 19% | 0.001 * |
| (A) Significantly Enriched Bacteria in Fertilized Soil | ||
|---|---|---|
| Fertilization | Total | Genera |
| MF SF SF3x | 2 | Arenimonas, Herminiimonas |
| NPK SF SF3x | 2 | Nitrosospira, Rhodanobacter |
| SF SF3x | 12 | Turicibacter, Ferruginibacter, Hydrogenophaga, Flavobacterium, Stenotrophomonas, Actinomadura, Devosia, Sphingomonas, Chitinophaga, Clostridium sensu stricto 1, Chryseolinea, Aminobacter |
| NPK SF | 1 | Granulicella |
| SF | 3 | Candidatus_Caldatribacterium, Pseudoxanthomonas, Coprothermobacter |
| (B) Significantly Enriched Fungi in Fertilized Soil | ||
| Fertilization | Total | Genera |
| MF NPK SF SF3x | 2 | Apodus, Operculomyces |
| MF NPK SF | 2 | Funneliformis, Diversispora |
| MF SF | 1 | Basidiobolus |
| SF | 1 | Mortierella |
| NPK | 1 | Conocybe |
| (C) Significantly Enriched Bacteria in Tubers Planted in Fertilized Soil | ||
| Fertilization | Total | Genera |
| MF SF3x | 1 | Xylophilus |
| NPK SF3x | 1 | Duganella |
| NPK | 1 | Acinetobacter |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kracmarova, M.; Karpiskova, J.; Uhlik, O.; Strejcek, M.; Szakova, J.; Balik, J.; Demnerova, K.; Stiborova, H. Microbial Communities in Soils and Endosphere of Solanum tuberosum L. and their Response to Long-Term Fertilization. Microorganisms 2020, 8, 1377. https://doi.org/10.3390/microorganisms8091377
Kracmarova M, Karpiskova J, Uhlik O, Strejcek M, Szakova J, Balik J, Demnerova K, Stiborova H. Microbial Communities in Soils and Endosphere of Solanum tuberosum L. and their Response to Long-Term Fertilization. Microorganisms. 2020; 8(9):1377. https://doi.org/10.3390/microorganisms8091377
Chicago/Turabian StyleKracmarova, Martina, Jana Karpiskova, Ondrej Uhlik, Michal Strejcek, Jirina Szakova, Jiri Balik, Katerina Demnerova, and Hana Stiborova. 2020. "Microbial Communities in Soils and Endosphere of Solanum tuberosum L. and their Response to Long-Term Fertilization" Microorganisms 8, no. 9: 1377. https://doi.org/10.3390/microorganisms8091377
APA StyleKracmarova, M., Karpiskova, J., Uhlik, O., Strejcek, M., Szakova, J., Balik, J., Demnerova, K., & Stiborova, H. (2020). Microbial Communities in Soils and Endosphere of Solanum tuberosum L. and their Response to Long-Term Fertilization. Microorganisms, 8(9), 1377. https://doi.org/10.3390/microorganisms8091377







