Abstract
Persistent infections with the human pathogen Helicobacter pylori (H. pylori) have been closely associated with the induction and progression of a wide range of gastric disorders, including acute and chronic gastritis, ulceration in the stomach and duodenum, mucosa-associated lymphoid tissue (MALT) lymphoma, and gastric adenocarcinoma. The pathogenesis of H. pylori is determined by a complicated network of manifold mechanisms of pathogen–host interactions, which involves a coordinated interplay of H. pylori pathogenicity and virulence factors with host cells. While these molecular and cellular mechanisms have been intensively investigated to date, the knowledge about outer membrane vesicles (OMVs) derived from H. pylori and their implication in bacterial pathogenesis is not well developed. In this review, we summarize the current knowledge on H. pylori-derived OMVs.
1. Background
Helicobacter pylori (H. pylori) is a Gram-negative human pathogen that specifically colonizes the epithelial lining in the human stomach. Persistent infections with H. pylori are invariably associated with a mild chronic inflammation of the gastric mucosa, which often remains asymptomatic [1]. However, a significant proportion of infected individuals develop clinically relevant symptoms of gastritis, gastric and duodenal ulcers, mucosa-associated lymphoid tissue (MALT), or gastric cancer [2,3]. Consequently, H. pylori was classified as the first bacterial group-I carcinogen by the World Health Organization in 1994 [4]. It is estimated that in connection with environmental influences, genetic predispositions of the host or microbiota, H. pylori represents the primary cause of more than 90% of gastric cancer cases [5,6]. H. pylori-dependent gastric adenocarcinomas develop through a series of sequential transformations of the gastric mucosa, which is known as the ’Correa cascade´ [7]. This process is initiated by chronic gastritis, leading to atrophic gastritis, intestinal metaplasia, dysplasia, and finally to carcinoma [8]. Eradication of H. pylori with antibiotics has been demonstrated to be an effective strategy for preventing cancer development and progression [9,10].
H. pylori developed a number of survival stratagems that allow bacterial persistence and immune evasion despite severe pathogenesis. In this context, H. pylori expresses a wide range of niche and virulence factors (reviewed in [11,12,13]), which are implicated in a complex molecular network that controls the induction and progression of H. pylori-dependent disorders [14]. Outer membrane vesicles (OMVs) provide an emerging mechanism to interfere with host cell physiology. Membrane vesicles are ubiquitously produced among organisms from all domains of life, including prokaryotes, eukaryotes and archaea [15]. Mammalian cells secrete extracellular vesicles (EVs), derived from both the plasma membrane (microvesicles or ectosomes) and from intracellular endosome-derived multivesicular bodies (exosomes), exhibiting manifold functions in health and disease [16]. Membrane vesicles from Gram-negative bacteria are termed OMVs to emphasize their origin from the outer membrane [17]. Functionally, OMVs are implicated in intermicrobial communication through exchanges of genetic material, chemical compounds and signaling molecules [18]. Nevertheless, several publications also suggest that OMVs play a direct role in bacterial pathogenesis [19,20,21,22,23]. In this review, we will summarize the current state of research on H. pylori-derived OMVs.
2. Physicochemical Characteristics of H. pylori-Derived OMVs
The physical interaction between H. pylori and gastric epithelial cells has been intensively investigated over the last 30 years, and has revealed a complex network of manifold cellular and molecular mechanisms, which are coordinated in time and place [12,24,25]. H. pylori-derived OMVs add a new aspect to the pathogenesis of H. pylori. OMVs of H. pylori were analyzed for the first time in 1997 by dual silver staining [26]. Since then, H. pylori OMVs have been studied both in vivo and in vitro in gastric biopsies [19,27,28], in gastric juice samples of infected individuals [29], and in culture media of H. pylori [19,27,28,30,31,32], especially during the late stationary growth phase and during biofilm formation [30,33,34,35,36]. A standard methodology in the EV field is Nanoparticle Tracking Analysis (NTA), which uses a light diffraction-based detection of particles diffusing through an observation volume, and sizing based on translational diffusion analysis of the Brownian motion, allowing the detection of particles >50 nm. An unbiased size determination is currently only possible by electron microscopy (EM) or atomic force microscopy (AFM) [37]. Generally, bacterial OMVs are thought to be relatively stable structures. Long-term storage has not been studied extensively, but one study reported that OMVs from H. pylori J99 or NCTC 11637 (ATCC 43504) stored at 4 °C overnight or at −20 °C did not differ in their macroscopic structure, resembling a deformed football [38]. Usually, OMVs from different H. pylori strains range between 20 and 300 nm in size, but some studies reported diameters of 400 nm to 500 nm (Table 1). Size analyses also suggest that OMV diameters change with bacterial growth phase. Vesicles isolated from bacteria grown to early logarithmic phases were larger compared to vesicles derived from bacteria in the stationary phase [39]. OMVs in this study were examined by NTA, and therefore it is possible that vesicles even smaller than 50 nm are produced but not detected. Whether the observed OMV size differences are specific for different H. pylori strains, and/or are the result of differing culture and assay conditions, isolation methods and vesicle analysis methods, needs to be investigated in the future. A systematic and technically unbiased evaluation of H. pylori vesicle sizes might, however, be particularly relevant to advancing our understanding of OMV function, since it has been implied that OMV size influences host cell entry and protein content [40]. In addition to the size, the number of OMVs released by H. pylori also depends on bacterial growth and environmental conditions. The release of H. pylori OMVs is inversely related to the cell growth, shifting from low quantities of OMVs released during the logarithmic growth phase to high levels produced during the stationary phase. This process is also correlated with the shape transitions of the bacterium from spiral to curved, doughnut-shaped and finally coccoid morphologies [41]. The number of OMVs secreted also changes with culturing conditions or environmental factors, which the bacteria encounter during the infection process, as well as bacterial mutations. A direct comparison of H. pylori 251 strain and an isogenic mutant exhibiting a deletion in the “colicin”-tolerant (tol) B (HP1126) locus revealed a more than 600-fold increase in OMV concentrations, while the protein composition characterized by LC/MS-proteomics was largely overlapping [42]. In H. pylori, the HP1126 locus is not well understood, but in Escherichia coli (E. coli), TolB is a periplasmic protein interacting with outer membrane proteins (OMPs) and peptidoglycan-associated lipoprotein (Pal), which was originally identified to play a role in the import of a number of colicins [43]. Disruptions in tol-pal clusters can result in defects in the growth, motility and virulence of E. coli, as well as of other Gram-negative bacteria [44]. Hence future studies are necessary to investigate the impact of HP1226 on OMV biogenesis in H. pylori hyper-vesiculating strains.

Table 1.
Size of H. pylori OMVs.
3. H. pylori OMVs Promote Biofilm Formation
Biofilms are surface-associated communities of bacteria that are embedded in a hydrated matrix of extracellular polymeric substances. Biofilm formation helps bacterial cells to avoid antimicrobials and protects them from being killed by the host’s immune system, thus enabling chronic infections. The bacteria in biofilms are phenotypically distinct from their planktonic counterparts. Not much is known about H. pylori’s ability to develop biofilms, its structure and composition, or the genes associated with this mode of growth [45]. However, several studies have demonstrated the ability of H. pylori to produce a biofilm in vitro [30,33,34,35,46]. In addition, there are also some reports on biofilm formation during the colonization of the human gastric mucosa with H. pylori. In SEM analyses of biopsy specimens, the presence of dense layers resembles biofilm-associated bacteria at the mucosal surface of H. pylori-positive patients [47,48].
In vitro studies of H. pylori biofilm formation indicate an involvement of OMVs in this process; however, it is still unclear which molecular and cellular mechanisms are involved. One study reported that the biofilm formation ability of H. pylori was dependent on OMVs mediating direct cell-to-cell contacts between the bacteria. The addition of OMVs, from a clinical isolate (TK1402) with strong biofilm-forming activity, to H. pylori cultures significantly enhanced biofilm formation ability [35]. An unidentified 22 kD protein [33] and an omp20 (HP0912, AlpB) protein in OMVs were described as important factors in biofilm formation [34]. Another study has shown that H. pylori ATCC 43629/NCTC 11639 OMVs isolated from a 48 h biofilm contained fourfold extracellular DNA (eDNA), compared to OMVs isolated from planktonic cultures of the same strain, indicating that OMVs may prevent eDNA degradation, which could additionally support or modulate biofilm formation or play a role in the intercellular exchange of genetic material. H. pylori (ATCC 43629/NCTC 11639) OMVs isolated directly from biofilms (bOMVs) showed a broader size range between 10 nm and 2.5 µm, with a major peak at 681.1 nm (59%) and two minor populations at 1760 nm (29.9%) and 2557 nm (11%) [30]. The enzyme carbonic anhydrase was also identified, and it was suggested that carbonic anhydrase and dsDNA released into the medium could stabilize the biofilm formation [36]. The change in the content of isolated OMVs during the development of biofilms was also observed in other H. pylori strains [33]. Despite individual reports investigating H. pylori biofilms, a significant role for biofilms in H. pylori survival and pathogenesis has yet to be determined.
4. The Content of H. pylori OMVs
In several reports, the content of isolated OMVs was analyzed, and large sets of lipids, nucleic acids, peptidoglycan and proteins were identified (Table 2). Dual silver staining revealed an increased ratio of carbohydrate to protein in OMVs compared to whole bacterial cells [26,27]. Protein composition as well as the carbohydrate to protein ratio of OMVs differ between bacterial strains [26]. Differences in the size and density of OMVs are also reflected in differences in vesicle composition. The high density vesicles contain higher protein to lipid ratios. Furthermore, differences in environmental conditions affect the content of vesicles [41] and the bacterial growth stage influences the abundance of key virulence determinants in OMVs [39].
4.1. Lipids, Peptidoglycan, and Nucleic Acids in H. pylori OMVs
Analysis of the membrane lipids provides information about OMV biogenesis since the inner membrane (IM) differs from outer membrane (OM) and OMVs in phospholipid content. Consistent with the biogenesis of OMVs, the phospholipid composition of OMVs closely resembles that of the OM [41]. IM contains lower amounts of phosphatidylglycerol and lyso-phosphatidyletanolamine, compared to OM and OMVs membrane fractions. In turn, phosphatidylcholine was solely detected in OM and OMVs. Phosphatidylethanolamine and cardiolipin were detected as major phospholipids in whole cell extracts (WCE), OM and IM, and purified OMVs. Significant amounts of cholesterol were also detected in WCE as well as in OMVs. Using 13C, 1H NMR correlation spectra analysis, it was estimated that the cholesterol content is about 10% of total phospholipids present in both WCE and OMVs [41], which is generally lower than the cholesterol fraction reported for mammalian EVs [49]. H. pylori lipopolysaccharide (LPS) is enriched in OMVs compared to OM [26,50], and is influenced by environmental conditions. LPS levels in H. pylori and OMVs are reduced in iron-limited conditions, probably due to the decreases in energy supply indicated by lower ATP levels [50].
In addition to dsDNA, several studies have shown the presence of small RNAs in H. pylori OMVs, and suggested a functional role upon their transfer into host cells [51,52]. More than a half million unique small non-coding (snc) RNA sequences were identified by RNA seq of OMVs purified from the supernatants of H. pylori strain J99, and RNAse protection assays for selected sncRNAs suggested an encapsulation within the OMV lumen [53]—or at least a lower susceptibility to digestion due to OMV binding and protection by a protein corona on the OMV surface, as has been suggested for small RNAs in mammalian EVs [54,55]. The uptake of sncRNAs into AGS cells was suggested by quantitative RT- PCR and microscopy using double labeling with a lipid dye (DiI) and an unspecific RNA intercalating dye. Of note, the lipid labeling of mammalian EVs can easily give rise to a number of artefacts, including micelle formation, unspecific labeling of other particles within the heterogeneous samples and the secondary redistribution of the dye, and it is conceivable that similar issues also apply for OMVs. Nevertheless, this study reports a functional effect of the H. pylori OMV on LPS-stimulated interleukin-8 (IL-8) secretion by gastric epithelial AGS cells, and has convincingly attributed it to two candidate small RNAs identified in the OMV transcriptome, and the authors hypothesized that these small RNAs might target the human IL-8 mRNA directly [53]. While more studies are required, these data suggest that small RNAs within H. pylori OMVs might play a fundamental role in directly tuning the host immune response.
4.2. The Protein Content of H. pylori OMVs
H. pylori OMVs exhibit a large heterogeneity in their protein composition. Interestingly, in contrast to the lipid composition, the proteome of OMVs significantly differs from whole cells and periplasm, suggesting a regulated sorting mechanism as is well known for mammalian EVs [38,41]. OMVs are predominantly enriched in uncharacterized proteins, flagellar basal body proteins [39] or outer membrane proteins (OMPs) (Table 2) [41]. More than 500 different H. pylori proteins were identified using mass spectroscopy analyses. However, most available reports show conflicting data, most likely due to the unstandardized sample preparation and analysis protocols. Nevertheless, the heterogeneity of OMV proteins is influenced by strain type, different culture conditions and bacterial growth phase [23]. Consistently identified targets include the proteins involved in cellular processing and signaling (especially cell motility and cell wall, membrane or envelope biogenesis), metabolism (energy production and conversion), and several well-known H. pylori virulence factors. Proteins identified in OMVs and considered as virulence factors (Table 2) belong to the group of proteins implicated in adherence, motility, acid resistance, immune evasion and modulation, and protein secretion of proteins (Figure 1).
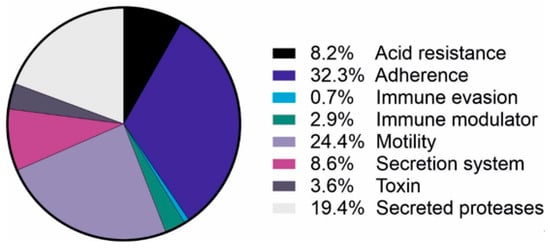
Figure 1.
Virulence factors detected in H. pylori-derived OMVs. Identified virulence factors of H. pylori by mass spectroscopy [38,39,40,41,42,45] and their role in bacterial pathogenesis.
Uptake of H. pylori OMVs: Delivery of Virulence Factors into Host Cells?
It has been reported that orally administered OMVs remain inside the stomach of mice for at least 24 h and can enter gastric epithelial cells [29]. Additionally, OMVs attach to, and internalize into, primary human antrum cells [31]. Clathrin-dependent and clathrin-independent mechanisms are described for OMV internalization, resulting in a highly efficient and rapid uptake in gastric epithelial AGS cells [56,57] (Figure 2). It has been suggested that caveolin mediates the entry of smaller (20–100 nm) OMVs into AGS cells. Larger OMVs might enter AGS cells via macropinocytosis, caveolin and clathrin-dependent endocytosis without preference [40]. By EM imaging, OMVs were found within the endosomes of human gastric mucosa cells [19,32]. Internalized OMVs colocalize with markers for early endosomes [57,58] and lysosomes, suggesting transport through the endocytic pathway [57] (Figure 2). This is particularly intriguing since it converges with the uptake and trafficking routes of mammalian exosomes within their recipient cells [59]. Once internalized, OMVs are relatively stable at least for 12 h within AGS [29,60], or 72 h within MKN28 cells [32]. Two independent studies reported that the uptake of H. pylori OMVs into AGS cells was significantly decreased upon the cell membrane depletion of cholesterol and the disruption of cholesterol-rich lipid rafts [20,57]. OMVs from H. pylori strain 60190 did not affect AGS cells’ viability but did influence proliferation; it was indicated that at low doses, OMVs slightly increased [22], and at higher doses, OMVs reduced proliferation significantly [22,60].
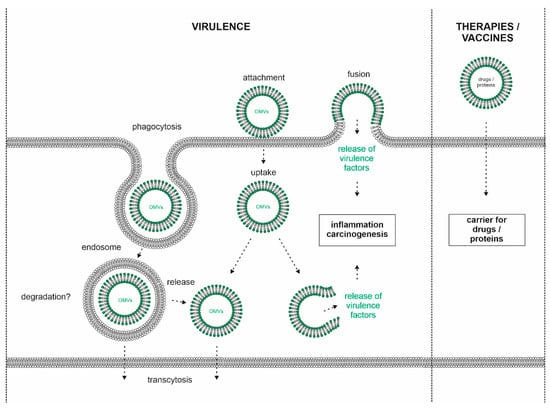
Figure 2.
Model of OMV interaction with host cells. H. pylori releases OMVs, which attach to the cell surface of gastric epithelial cells. Clathrin-dependent and -independent uptake of OMVs could occur via phagocytosis leading to the formation of endosomes and/or fusion of the bacterial OMV membrane with the host´s membrane. The fate of internalized OMVs is unclear, but the release of OMVs at the basolateral membrane (transcytosis) or into the cytoplasm was hypothesized. OMV-delivered virulence factors might be released into the cytosol where they can interfere with host cell signaling leading to inflammation and carcinogenesis (left panel). Technical applications of H. pylori OMVs as nanocarriers for drugs or proteins (right panel).
OMV proteins involved in H. pylori motility comprise a relatively large group (Table 2, Figure 1) and include proteins involved in flagella assembly [61]. H. pylori P12 and 18943 OMVs contained catalase (KatA), which is implicated in the protection of bacteria from extracellular ROS in respiratory burst reactions [62]. Among virulence factors, several known and putative adhesins were detected in OMVs. This group of OMV proteins include HopZ and HorB proteins, the blood group antigen binding adhesin (BabA, BabB), sialic acid-binding adhesion (SabA, SabB), H. pylori adhesin A (HpaA), and adherence-associated lipoproteins (AlpA, AlpB) (Table 2). The finding of adhesins on OMV surfaces is important since they could mediate the binding and uptake of OMVs into gastric epithelial cells [41].
Urease is important for buffering the acidic environment for the acid-sensitive pathogen, and thus is crucial for successful colonization of the human stomach [63]. Weak signals for urease were detected for OMVs of H. pylori 189 and 249 in Western blot analyses [20]. Further, through mass spectroscopy analysis, the urease subunit β was identified in H. pylori 7.13 OMVs [45], and both subunits of the active enzyme complex (α and β) were found in H. pylori CCUG 17874 [41], NCTC 11637 and J99 [38], 251 OMVs [40], and 26695, independently of growth phase [39]. Urease accessory proteins (UreH, UreG, UreF) were also identified by mass spectroscopy in H. pylori OMVs (Table 2). Additional well-known virulence factors of H. pylori (e.g., vacuolating cytotoxin A (VacA), cytotoxin-associated gene A (CagA), high temperature requirement A (HtrA), etc.) have been repeatedly identified in OMVs, suggesting that OMVs are involved in the delivery of virulence factors when taken up by host cells.
VacA was identified in OMVs by mass spectroscopy [39,40,41,42,45] and Western blot analysis [27,29,32,38,41,64,65] from many different H. pylori strains [19,27,29,32,38,39,40,41,42,45,65,66,67]. VacA is pleiotropic pathogenic factor of H. pylori, which is secreted into the bacterial environment. A prominent VacA-mediated phenotype is the excessive formation of massive cell vacuoles harboring both late endosomal and lysosomal markers [68]. Moreover, VacA induces autophagosome formation [69], can induce apoptosis via mitochondrial damage [70], and might also weaken the epithelial barrier by impeding tight junction integrity [71]. VacA was found on the surface of OMVs [19,27,41]. Ultrastructural immunolocalization of VacA in H. pylori 60190 OMVs isolated from bacteria culture and in biopsies from infected patients, confirming that this protein is membrane-associated during the growth of H. pylori in vitro and in vivo [19,27]. It was further suggested that the uptake of H. pylori 60190 OMVs by AGS cells was enhanced by VacA and reduced by LPS [56]. OMVs isolated from the H. pylori 60190 strain and its ∆VacA isogenic strain do not differ in morphology and quantity [65]. H. pylori 60190 OMVs have been described to induce the vacuolization of HEp-2 cells [27,72] and AGS cells [22], suggesting that VacA is functionally active on OMVs. Even though the presence of OMVs in host cells was not further analyzed in these studies, OMVs were extensively washed to exclude the presence of free soluble VacA, indicating that OMVs might transfer functionally active VacA into the recipient cells. However, the quantification of extracellular VacA revealed that H. pylori OMV VacA accounted for about 25% of total VacA, while the remaining was free-soluble VacA. The ratio was similar for both 60190 and CCUG 17874 strains used [73]. Ultrastructural immunocytochemistry confirmed the release of VacA by H. pylori both as free-soluble and as OMV-associated toxin; both forms of VacA are taken up by MKN28 cells and are localized inside vacuoles [19,32,73]. On the contrary, when strains 60190 and SS1 were compared, the amount of OMV VacA differed substantially. A significant difference was also seen in the OMV-mediated vacuolization of RK13 rabbit kidney cells, where OMVs from 60190 induced vacuolization in about 49%, and SS1 OMVs induced it in only 13% of cells [65]. H. pylori 60190 and CCUG 17874 OMV-associated VacA poorly vacuolate in MKN28 and HeLa cells [73]. Along with the observation that OMV internalization requires cholesterol [20,57], it is interesting that the disruption of lipid rafts by methyl-β-cyclodextrin (MβCD) decreased vacuolization in cells treated with VacA-containing OMVs, pointing to the essentiality of cholesterol in VacA function [56]. It has been discussed that environmental factors can influence VacA levels in OMVs. Iron limitation affects H. pylori 60190 growth, and as OMV numbers and OMV protein content increase, VacA decreases to levels similar to those detected for an isogenic VacA deletion mutant [72]. A significant increase in the number of cells with micronuclei was observed in AGS cells upon incubation with OMV from H. pylori strain 60190. This process was accompanied by the redistribution of lysosomal iron and a large decrease in cellular glutathione (GSH), both dependent on VacA [60]. The question whether VacA-containing OMVs impact cell survival is not fully answered. Both OMVs from VacA wildtype and a truncated, non-secretable VacA H. pylori strain induced a decrease in AGS viability through apoptosis. These OMVs induced caspase-8, -3 and -9 within the host cells, but apoptosis was mitochondria-independent as cytochrome c release was not detected [66]. This is in contrast with reports of soluble VacA, which leads to cytochrome c release [74].

Table 2.
Proteins detected in H. pylori OMVs.
Table 2.
Proteins detected in H. pylori OMVs.
| Virulence Factors—Acid Resistance | |||||
|---|---|---|---|---|---|
| Gene/Chromosome (NC_000915) | Function/Description | Virulence Factors | Confirmed by Mass Spectroscopy (Reference) | Confirmed by Western Blotting (Reference) | |
| ureH/HP0067 | urease accessory proteins, form a complex that acts as a GTP-hydrolysis-dependent molecular chaperone, activating the urease | Urease | [41] | ||
| ureG/HP0068 | [39] | ||||
| ureF/HP0069 | [39,41] | ||||
| ureB/HP0072 | urease subunit beta | neutralizes the gastric acidity, NH3 damages the gastric epithelium | [38,39,40,41,42,45] | [20] | |
| ureA/HP0073 | urease subunit alpha | [38,39,40,41,42] | [20] | ||
| Virulence factors—Adherence | |||||
| hopZ/HP0009 | Outer membrane protein | HopZ | [38,40,42,45] | ||
| horB/HP0127 | Outer membrane protein 4 | HorB | [38,39,40,41,45] | ||
| babA/hopS/HP0317/HP1243 | Outer membrane protein 9/28, binds with the epithelial cell receptor Leb, mediates bacterial attachment and colonization | Blood group antigen binding adhesins | [39,40,41,42] | [41] | |
| babB/hopT/HP0896 | Outer membrane protein 19, babA paralog | [38,39,40,41,42] | |||
| sabB/hopO/HP0722 | sabA homologue | Sialic acid binding adhesins | [40,41,42] | ||
| sabA/hopP/HP0725 | binds with sialyl-Lex antigen, mediates bacterial attachment and colonization | [38,40,41,42] | [41] | ||
| hpaA/HP0797 | neuraminyllactose-binding hemagglutinin | H. pylori adhesin A | [38,39,40,41,42] | [38] | |
| alpA/hopC/HP0912 | Outer membrane protein 20 | adherence-associated lipoprotein | [38,39,40,41,42] | ||
| alpB/hopB/HP0913 | Outer membrane protein 21 | [39,40,41,42] | [34,41] | ||
| Virulence factors—Immune evasion | |||||
| futA/HP0379 | alpha-(1,3)-fucosyltransferases, LPS oligosaccharide biosynthesis | Lipopoly-saccharide Lewis antigens | [39] | ||
| futB/HP0651 | [41] | ||||
| Virulence factors—Immune modulator | |||||
| napA/HP0243 | DNA protection during starvation, activates neutrophils, mast cells and monocytes | Neutrophil-activating protein (HP-NAP) | [39,40,42] | [38] | |
| oipA/hopH/HP0638 | Outer membrane protein 13, bacterial adherence to the gastric epithelium, damages mucosal layer, induces apoptosis and IL-8 expression | Outer inflammatory protein | [38,39,41,42] | [38,41] | |
| Virulence factors—Motility | |||||
| flaB/HP0115 | minor flagellin subunit, polymerizes to form the filaments of bacterial flagella | helps bacteria to minimize contact to acidic environment | Flagella | [39,41,42] | |
| flgI/HP0246 | P-ring protein, assembles around the rod | [39,41,42] | |||
| flgL/HP0295 | Hook-associated protein 3 | [39,41,42] | |||
| flgH/HP0325 | L-ring protein, assembles around the rod | [39,45] | |||
| flaA/HP0601 | predominant flagellin subunit, polymerizes to form the filaments of bacterial flagella | [38,39,40,41] | |||
| flaG/HP0751 | polar flagellin, rotor/switch protein | [39,42] | |||
| fliD/HP0752 | filament-capping protein, flagellin folding chaperone, required for the morphogenesis and for the elongation of the flagellar filament | [39,41,42] | |||
| flgE_1/HP0870 | hook protein, links the flagellar filament to the drive apparatus in the basal body | [39,40,41] | |||
| flgD/HP0907 | basal body rod modification protein, required for flagellar hook formation, scaffolding protein | [39,40,41,42] | |||
| flgE_2/HP0908 | flagellar basal body protein | [39,41] | |||
| flgG_1/HP1092 | flagellar basal body protein | [39,42] | |||
| flgK/HP1119 | first hook-filament junction protein | [39,41,42] | |||
| flgA/HP1477 | involved in the assembly process of the P-ring formation | [40,41] | |||
| fliE/HP1557 | MS-ring rod junction protein, | [39,40,42] | |||
| flgC/HP1558 | rod protein | [39] | |||
| flgB/HP1559 | rod protein of flagellar basal body | [39,42] | |||
| flgG_2/HP1585 | distal rod protein | [39,42] | |||
| Virulence factors—Secretion system (Proteins Required for Cag T4SS Activity) | |||||
| cag1/HP0520 | membrane protein in T4SS, associated with IL-8 expression induction and CagA delivery to host cells | Cag PAI type IV secretion system | [41] | ||
| cag3/HP0522 | defined localization in T4SS apparatus | [39] | |||
| cagX/HP0528 | defined localization in T4SS apparatus | [39] | [41] | ||
| cagT/HP532 | defined localization in T4SS apparatus, core complex protein in T4SS, helps in the translocation of CagA | [39,41] | [41] | ||
| cagM/HP0537 | defined localization in T4SS apparatus | [39,41] | [41] | ||
| cagN/HP0538 | localization in T4SS apparatus is not yet defined | [39] | [41] | ||
| cagF/HP0543 | localization in T4SS apparatus is not yet defined, CagA chaperone | [41] | |||
| cagD/HP0545 | localization in T4SS apparatus is not yet defined | [39,42] | |||
| cagA/HP0547 | Scaffold/hub protein, oncoprotein, becomes phosphorylated in the host cells, causes cellular proliferation and elongation, induces IL-8 expression | Secreted T4SS effector cytotoxin-associated gene A | [38,39,41,42,45] | [29,38,41] | |
| Virulence factors—Toxin | |||||
| vacA/HP0887 | induces vacuolization of epithelial cells and endoplasmic reticulum stress, causes cell vacuolization, necrosis and apoptosis, enhances activation of autophagy and increased cellular death | Vacuolating cytotoxin | [39,40,41,42,45] | [27,29,32,38,41,65,66] | |
| Other | |||||
| katA/HP0875 | antioxidant enzyme, neutralization of H2O2 and NaClO | [36,38,39,40,41] | enzyme activity determined in [62] | ||
| hcpD/HP0160 | beta-lactamase | Putative solenoid proteins | [39,40,41] | ||
| hcpA/HP0211 | beta-lactamase, cysteine-rich 28 kD protein | [39,40,42] | |||
| hcpE/HP0235 | beta-lactamase | [39,40,41,42] | |||
| hcpC/HP1098 | beta-lactamase, cysteine-rich protein C | [38,39,40,42] | |||
| tolB/HP1126 | periplasmic protein interacting with outer membrane proteins (OMPs) | [38,39,40,41,42] | |||
| csd3/HP0506 | conserved hypothetical secreted protein | Secreted proteases | [39,41] | ||
| ymxG/HP0657 | processing protease | [38,39,41,42] | |||
| pqqE/HP1012 | metalloendopeptidase | [38,39,40,41,42,45] | |||
| htrA/HP1018-9 | acts as protease, degrades misfolded proteins and tight junction protein enabling delivery of CagA | [38,39,40,41,42] | |||
| HP1037 | metal ion binding aminopeptidase | [39,41] | |||
| ggt/HP1118 | transpeptidation and amino acid synthesis, enhances cell apoptosis, inhibits cellular proliferation | [38,39,40,41,42] | |||
| ctpB/HP1350 | serine-type endopeptidase | [38,39,40,41] | |||
Several uncharacterized proteases (HP0506, HP0657, HP1012, HP1037, HP1350) secreted by H. pylori were identified in OMVs (Table 2). Further, the activity of unidentified gelatinolytic proteases (76 kD and 96 kD) was observed in OMVs isolated from H. pylori 60190 cultured under iron-limiting conditions [72]. Among the proteases, the chaperone and serine protease high temperature requirement A (HtrA) has also been detected. HtrA was identified by mass spectroscopy and Western blot analysis in H. pylori CCUG 17874 OMVs, and its relative amount was higher than in the OM fraction [41]. HtrA was also identified in OMVs from H. pylori J99 and NCTC 11637 [38], H. pylori 251 and its Δtol or Δpal deletion mutants [40,42]. In recent years, HtrA has been intensively investigated as a secreted H. pylori factor that disrupts adherens junctions by targeting the cell adhesion protein and tumor suppressor E-cadherin of gastric epithelial cells [75]. Recently, occludin and claudin-8 were identified as additional substrates for HtrA, suggesting that tight junction complexes can be opened by HtrA as well [76]. Functionally, HtrA enables H. pylori to transmigrate across the epithelial layer, allowing the direct interaction with integrin-β1 at basolateral membranes so as to inject CagA into the polarized gastric epithelial cells [75,76]. While the HtrA substrate E-cadherin is cleaved within its extracellular domain, it was an interesting finding that the occludin and claudin-8 cleavage sites were identified in their intracellular domains [76]. Therefore, the concept of OMV-delivered and released virulence factors of H. pylori would explain how HtrA can target occludin and claudin-8 intracellularly.
Another important factor in H. pylori-mediated pathogenesis is γ-glutamyltransferase (GGT), which has been identified in OMVs originating from H. pylori CCUG 17874 [41], H. pylori J99 and NCTC 11637 [38], and H. pylori 251 [40,42]. The presence of GGT was also confirmed in OMVs isolated from H. suis [77]. GGT is a secreted enzyme that converts glutamine into glutamate and ammonia, and converts glutathione into glutamate and cysteinyl glycine. H. pylori GGT and its catalyzed products have been implicated in cell cycle arrest and apoptosis. Further, H. pylori GGT can mediate immune tolerance by inhibiting T cell proliferation and modulating dendritic cell activation to give rise to regulatory T cell responses [24]. It is intriguing to speculate that OMVs contribute to the transport of the immunomodulatory H. pylori factors (e.g., VacA and GGT) across the epithelium and directly interact with immune cells.
One of the best characterized virulence factors is the bacterial effector and oncoprotein CagA, which is translocated via a specialized type-IV secretion system (T4SS) into the cytoplasm of infected host cells [13,24,78]. Translocated CagA has a molecular weight of 135 kD, and is rapidly tyrosine phosphorylated in glutamate-proline-isoleucine-tyrosine-alanine (EPIYA) motifs in its carboxy-terminal part by host cell kinases of the c-Src and c-Abl families [79,80,81,82,83]. Both phospho- and non-phospho-CagA interact with distinct signaling proteins, which alters signaling and cell functions [84]. The observed changes include inflammatory phenotypes, altered proliferation, de-differentiation and elevated migratory capacity. Although in individual reports CagA was not detected in the OMV fraction from cagPAI-positive strains [27], there is a number of publications reporting CagA in OMVs as identified by mass spectroscopy [38,39,41,42,45]. Analyzing CagA in H. pylori by STED (Stimulated Emission Depletion) microscopy revealed CagA localized in speckles resembling OMVs (Figure 3), underlining the previous reports. The CagA level seemed to be similar in H. pylori CCUG 17874 OMVs and OM fractions, and it is associated with the surface of H. pylori vesicles. The presence of CagA was also confirmed by Western blotting in OMVs of H. pylori HP99 [29], H. pylori CCUG 17874 [41] and H. pylori NCTC 11637 [38]. CagA was detectable in serum-derived exosomes, leading to the hypothesis that those exosomes serve as nanocarriers for virulence factors and could be a causative mechanism through which H. pylori infection promotes extragastric diseases [85] (Figure 2). A possible function of OMV-delivered CagA might be indicated in a recent study using OMVs from cagA-positive and cagA-negative H. pylori. OMVs from cagA-positive bacteria induced an enhanced HO-1 expression in dendritic cells via the activation of nuclear factor kappa B (NF-kB) and nuclear factor erythroid 2-related factor 2 (Nrf2) [86]. H. pylori OMVs containing CagA were found to localize to the cell junctions, and were implicated in zonula occludens 1 (ZO-1) redistribution [87]. While the T4SS-translocated CagA drastically changes the motogenic response and cellular morphology of infected gastric epithelial cells, the incubation of AGS cells with these vesicles did not induce changes in morphology [56], which is in contrast to serum-derived OMVs [85]. Since CagA-mediated cell elongation is strictly dependent on CagA tyrosine phosphorylation [82], detailed analyses of OMV-delivered CagA and the activity of CagA kinases are necessary.
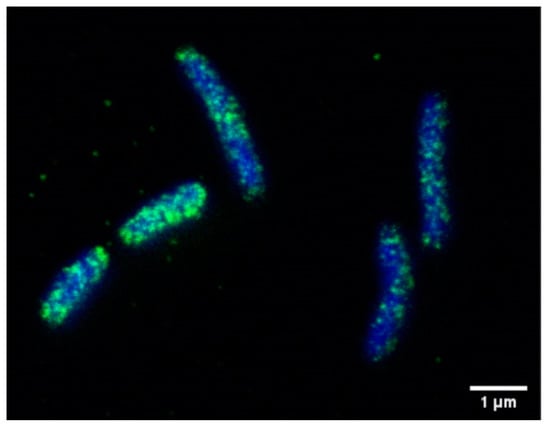
Figure 3.
Localization of CagA in H. pylori. CagA (green) was detected using an anti-CagA antibody and analyzed by STED microscopy. Bacterial DNA was stained using DAPI (blue). Bar, 1 µM.
4.3. Do H. pylori OMVs Modulate Host Immune Regulation?
Although the content of H. pylori-derived OMVs has been extensively analyzed, the functional consequences of OMVs’ interactions with host cells or H. pylori bacterial cells are largely unknown. Importantly, the gastric epithelial layer also recruits and activates cells’ immune system by the production and release of pro-inflammatory cytokines. As such, it is an interesting finding that H. pylori-derived OMVs induce IL-8 secretion by gastric epithelial AGS cells [40]. Corresponding to this observation, H. pylori NCTC 11637 OMVs induced the secretion of IL-8 from AGS cells, colonic epithelial T84 cells and duodenal explants [38]. OMVs induced IL-8 in a dose-dependent manner, which was shown to be independent of LPS [29]. Furthermore, IL-8 secretion was more pronounced using OMVs from cagPAI-negative Tx-30a compared to cagPAI-positive H. pylori strain 60190 [22]. This is in line with infection experiments investigating IL-8 secretion, which is clearly dependent on a functional T4SS, but independent of CagA translocation [88]. The sncRNAs identified in H. pylori strain J99 OMVs reduced IL-8 cytokine secretion [53], whereas it will be an interesting question to assess whether this might be mediated through a direct targeting of the IL-8 mRNA by the sncRNAs, and potential induction of RNA silencing. The fact that IL-8 is released in response to OMVs is interesting since it was further shown that OMVs also contain peptidoglycan (PG). Previously, PG has been identified to be translocated into H. pylori-infected cells via the T4SS, where it binds to the nucleotide-binding oligomerization domain-containing 1 (Nod1) protein and transactivates NF-κB-dependent pro-inflammatory genes [89]. Correspondingly, OMV derived PG has been described as an important factor in NF-κB-mediated IL-8 and CXCL2 (chemokine C-X-C motif ligand 2, macrophage inflammatory protein 2-alpha (MIP2-alpha)) induction in human and murine gastric cells, independent of the presence of proteins [20]. These data support the hypothesis that PG can induce NF-kB via OMVs independent of the T4SS. Recently, the H. pylori metabolite ADP heptose has been characterized as the key T4SS effector for the regulation of innate immune pathways [90,91]. Whether ADP heptose is also present in OMVs has not been analyzed yet. However, H. pylori OMVs can also induce NOD1 signaling, as shown by receptor-interacting protein (RIP2)-dependent autophagy prior to the release of IL-8 [58]. In summary, these data suggest that OMVs can induce the expression and release of proinflammatory cytokines and chemokines attracting immune cells to the place of infection.
Since molecules involved in immune evasion are also present in H. pylori OMVs, it was speculated that OMVs directly interfere with immune cell functions. As the fucosyltransferases (FutA and FutB) involved in lipopolysaccharide Lewis antigens modification were identified, as well were immune modulators neutrophil-activating protein (NapA) and outer inflammatory protein (OipA) (Table 2) [38,39,40,41,42], it would be interesting to investigate the biodistribution of OMVs and possible functional consequences of these factors for the immune response. A general effect of H. pylori-derived OMVs was also investigated on eosinophils isolated from the peripheral blood of human volunteers. H. pylori 60190 OMVs containing VacA led to cytoplasmic degranulation, increased CD11 and intracellular adhesion molecule (ICAM-1) expression, and to the release of the eosinophil cationic protein (ECP), which is a cytotoxic protein contributing to eosinophilic inflammation [92]. These OMVs were also used to study the response of peripheral blood mononuclear cells (PBMCs), whereby they strongly stimulated the release of IL-10 and IL-6, as well as a proliferative response, independent of VacA. OMVs were also tested on the Jurkat T cell line, where it caused pro- and anti-inflammatory responses, but also a loss of metabolic activity and apoptosis, which was promoted by VacA [65]. This observation is in contrast with a recent report describing that OMV treatment resulted in significant inhibition of Jurkat T cell proliferation, but did not induce apoptosis. In monocytes, H. pylori 60190 OMVs induced IL-10 and COX-2 expression [93]. OMVs isolated from H. pylori HP99 culture were shown to induce in vivo production of IL-6 in mouse macrophages, which was LPS-independent [29]. H. pylori 60190 OMVs induced expression of heme oxygenase-1 (HO-1) in bone marrow-derived mouse dendritic cells. HO-1 is also implicated in inflammation and oxidative damage. A such the authors speculated that upregulated HO-1 expression protects cells from immunopathogenesis or stress damage [86].
The effect of OMVs on the adaptive immune response was also investigated in vivo. The intragastric administration of H. pylori 60190 OMVs led to a production of specific IgG in mice [94,95]. Intragastric immunization was NOD1-dependent but independent of TLR signaling [20]. Furthermore, cytokine levels like IFN-γ, IL-17 and IL-4 were elevated in mouse spleens [29]. Immunization with H. pylori 60190 OMVs induced antibody reactivity against an 18 kD protein [26,94], which was later recognized as a specific anti-lipoprotein 20 (Lpp20, HP1456) response. Interestingly, animals transplanted with hybridoma tumors producing antibodies against H. pylori Lpp20 showed lower numbers of H. pylori colonizing the stomach, suggesting Lpp20 as a vaccine candidate [95]. In fact, mouse immunization studies with OMVs or recombinant Lpp20 revealed significantly lower levels of colonization in immunized animals [96].
5. Conclusions
Knowledge about the fate and function of OMVs in H. pylori pathogenesis is still at its beginning, but accumulating data point to its functional role in bacterial pathogenesis through attachment to, and possible uptake into, host cells (Figure 2). In this context, reliable and unbiased methods are required to analyze OMV internalization and the fate of OMVs and their cargo inside the host cells. It is an attractive hypothesis that OMVs are transported through the epithelial monolayer and are released at the basal membrane, which could explain the delivery of virulence factors to extragastric body sites (Figure 2). Additionally, the idea that the OMV content is released into the host cell´s cytosol is a fascinating novel concept concerning how H. pylori might interfere with host cell function, and needs to be investigated in future.
On the basis of their emerging pathophysiological functions in transporting functional macromolecules across biological barriers and kingdoms, both into other enteric bacteria as well as into cells and tissues of the mammalian host, it is intriguing to speculate about the potential biomedical applications of H. pylori OMVs. Beyond targeting the epithelium, the biodistribution of H. pylori OMVs in the human body has not been systematically documented to date. Due to the induction of mucosal barrier instability and their small size, it is conceivable that OMVs cannot only penetrate deeper layers of the gastric mucosa but potentially also enter the circulation. It was shown that CagA is present in vesicles derived from the serum of patients infected with cagA positive strains [85], suggesting that either OMVs themselves or secondary exosomes secreted from mucosal cells targeted by OMVs might enter the blood stream. If this holds true, this would open up a new avenue for vesicle-based diagnostics and biomarkers of H. pylori infection. Secondly, like extracellular vesicles from mammalian sources, OMVs are also of potential interest as a biological drug delivery vehicle due to their effectiveness in penetrating biological barriers, entering their recipient cells in a targeted manner to functionally deliver biological macromolecules to distant sites while evading (or modulating) the immune system (reviewed in [97]) (Figure 2). The ease of bioengineering the vesicles by genetic modification of the bacterial cells, as well as the relative ease of production at scale, makes OMVs a potentially attractive drug delivery platform. Their immunogenicity, as well as their similarity to their bacterial mother cells, might however make them of particular interest for vaccine development [61]. In fact, a Neisseria meningitidis OMV-containing vaccine (Bexsero) was approved in 2013 for meningitis B by the FDA and EMA, and further OMV based vaccines are currently in development for other pathogenic bacteria [17,98,99,100]. Vaccine development for H. pylori infection has historically been challenging. A recently published immunization study with oral administration of H. pylori OMVs in mice, however, has shown strong protective effects against H. pylori infection via a Th2 driven immune response [45], indicating that OMV-based H. pylori vaccines for clinical testing could eventually appear on the horizon. Finally, it has been proposed that OMVs of pathogenic bacteria might be used as biological antibiotics against other infectious pathogens; OMVs from Borrelia [101], E. coli [102,103], Actinomyces [104] or Helicobacter [51] have been used to deliver cytotoxic proteins or peptides, resulting in lysis and cell death, and it has been proposed that this activity is targeted to other bacteria rather than the host. This fascinating strategy of using one pathogenic microorganism to not only cross-regulate its host but also produce its own ‘predatory microvesicle artillery’ [97] to defend itself against other pathogenic species competing for the same niche might open up a fundamentally new principle for next generation antibiotics.
Author Contributions
Conceptualization, M.J., G.P., S.W. Writing—Original Draft Preparation, M.J., G.P., N.M.-K., S.W. Writing—Review and Editing, M.J., G.P., N.M.-K., S.W. Funding Acquisition, S.W. All authors have read and agreed to the published version of the manuscript.
Funding
Open Access Funding by the Austrian Science Fund (FWF). The work of SW was supported by the grants W_01213, P_31507, and I_4360 from the Austrian Science Fund (FWF).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Piazuelo, M.B.; Epplein, M.; Correa, P. Gastric Cancer: An Infectious Disease. Infect. Dis. Clin. N. Am. 2010, 24, 853–869. [Google Scholar] [CrossRef]
- Waskito, L.A.; Salama, N.R.; Yamaoka, Y. Pathogenesis of Helicobacter pylori infection. Helicobacter 2018, 23, e12516. [Google Scholar] [CrossRef]
- Venerito, M.; Vasapolli, R.; Rokkas, T.; Malfertheiner, P. Gastric cancer: Epidemiology, prevention, and therapy. Helicobacter 2018, 23, e12518. [Google Scholar] [CrossRef]
- Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon, 7–14 June 1994. IARC Monogr. Eval. Carcinog. Risks Hum. 1994, 61, 1–241.
- Parkin, D.M. The global health burden of infection-associated cancers in the year 2002. Int. J. Cancer 2006, 118, 3030–3044. [Google Scholar] [CrossRef]
- Moss, S.F. The Clinical Evidence Linking Helicobacter pylori to Gastric Cancer. Cell. Mol. Gastroenterol. Hepatol. 2017, 3, 183–191. [Google Scholar] [CrossRef]
- Correa, P.; Piazuelo, M.B. The gastric precancerous cascade. J. Dig. Dis. 2011, 13, 2–9. [Google Scholar] [CrossRef]
- Correa, P. Human gastric carcinogenesis: A multistep and multifactorial process—First American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992, 52, 6735–6740. [Google Scholar]
- Choi, I.J.; Kook, M.-C.; Kim, Y.-I.; Cho, S.-J.; Lee, J.Y.; Kim, C.G.; Park, B.; Nam, B.H. Helicobacter pylori Therapy for the Prevention of Metachronous Gastric Cancer. N. Engl. J. Med. 2018, 378, 1085–1095. [Google Scholar] [CrossRef]
- Ford, A.C.; Forman, D.; Hunt, R.H.; Yuan, Y.; Moayyedi, P. Helicobacter pylori eradication therapy to prevent gastric cancer in healthy asymptomatic infected individuals: Systematic review and meta-analysis of randomised controlled trials. BMJ 2014, 348, g3174. [Google Scholar] [CrossRef]
- Šterbenc, A.; Jarc, E.; Poljak, M.; Homan, M. Helicobacter pylori virulence genes. World J. Gastroenterol. 2019, 25, 4870–4884. [Google Scholar] [CrossRef]
- Posselt, G.; Backert, S.; Wessler, S. The functional interplay of Helicobacter pylori factors with gastric epithelial cells induces a multi-step process in pathogenesis. Cell Commun. Signal. 2013, 11, 77. [Google Scholar] [CrossRef]
- Backert, S.; Clyne, M. Pathogenesis of Helicobacter pylori Infection. Helicobacter 2011, 16, 19–25. [Google Scholar] [CrossRef]
- Zhang, X.-Y.; Zhang, P.-Y.; Aboul-Soud, M.A. From inflammation to gastric cancer: Role of Helicobacter pylori. Oncol. Lett. 2016, 13, 543–548. [Google Scholar] [CrossRef]
- Deatherage, B.L.; Cookson, B.T. Membrane Vesicle Release in Bacteria, Eukaryotes, and Archaea: A Conserved yet Underappreciated Aspect of Microbial Life. Infect. Immun. 2012, 80, 1948–1957. [Google Scholar] [CrossRef]
- Malloci, M.; Perdomo, L.; Veerasamy, M.; Andriantsitohaina, R.; Simard, G.; Martínez, M.C. Extracellular Vesicles: Mechanisms in Human Health and Disease. Antioxid. Redox Signal. 2019, 30, 813–856. [Google Scholar] [CrossRef]
- Bitto, N.J.; Kaparakis-Liaskos, M. The Therapeutic Benefit of Bacterial Membrane Vesicles. Int. J. Mol. Sci. 2017, 18, 1287. [Google Scholar] [CrossRef]
- Schwechheimer, C.; Kuehn, M.J. Outer-membrane vesicles from Gram-negative bacteria: Biogenesis and functions. Nat. Rev. Genet. 2015, 13, 605–619. [Google Scholar] [CrossRef]
- Fiocca, R.; Necchi, V.; Sommi, P.; Ricci, V.; Telford, J.; Cover, T.L.; Solcia, E. Release of Helicobacter pylori vacuolating cytotoxin by both a specific secretion pathway and budding of outer membrane vesicles. Uptake of released toxin and vesicles by gastric epithelium. J. Pathol. 1999, 188, 220–226. [Google Scholar] [CrossRef]
- Kaparakis, M.; Turnbull, L.; Carneiro, L.; Firth, S.; Coleman, H.A.; Parkington, H.C.; Le Bourhis, L.; Karrar, A.; Viala, J.; Mak, J.; et al. Bacterial membrane vesicles deliver peptidoglycan to NOD1 in epithelial cells. Cell. Microbiol. 2010, 12, 372–385. [Google Scholar] [CrossRef]
- Shen, Y.; Torchia, M.L.G.; Lawson, G.W.; Karp, C.L.; Ashwell, J.D.; Mazmanian, S.K. Outer membrane vesicles of a human commensal mediate immune regulation and disease protection. Cell Host Microbe 2012, 12, 509–520. [Google Scholar] [CrossRef]
- Ismail, S.; Hampton, M.B.; Keenan, J. Helicobacter pylori Outer Membrane Vesicles Modulate Proliferation and Interleukin-8 Production by Gastric Epithelial Cells. Infect. Immun. 2003, 71, 5670–5675. [Google Scholar] [CrossRef]
- Parker, H.; Keenan, J.I. Composition and function of Helicobacter pylori outer membrane vesicles. Microbes Infect. 2012, 14, 9–16. [Google Scholar] [CrossRef]
- Sgouras, D.; Tegtmeyer, N.; Wessler, S. Activity and Functional Importance of Helicobacter pylori Virulence Factors. Adv. Exp. Med. Biol. 2019, 35–56. [Google Scholar] [CrossRef]
- Wessler, S.; Backert, S. Molecular mechanisms of epithelial-barrier disruption by Helicobacter pylori. Trends Microbiol. 2008, 16, 397–405. [Google Scholar] [CrossRef]
- Keenan, J.I.; Allardyce, R.A.; Bagshaw, P.F. Dual silver staining to characterise Helicobacter spp. outer membrane components. J. Immunol. Methods 1997, 209, 17–24. [Google Scholar] [CrossRef]
- Keenan, J.; Day, T.; Neal, S.; Cook, B.; Perez-Perez, G.; Allardyce, R.; Bagshaw, P. A role for the bacterial outer membrane in the pathogenesis of Helicobacter pylori infection. FEMS Microbiol. Lett. 2000, 182, 259–264. [Google Scholar] [CrossRef]
- Hynes, S.O.; Keenan, J.I.; Ferris, J.A.; Annuk, H.; Moran, A.P. Lewis Epitopes on Outer Membrane Vesicles of Relevance to Helicobacter pylori Pathogenesis. Helicobacter 2005, 10, 146–156. [Google Scholar] [CrossRef]
- Choi, H.-I.; Choi, J.-P.; Seo, J.; Kim, B.J.; Rho, M.; Han, J.K.; Kim, J.G. Helicobacter pylori-derived extracellular vesicles increased in the gastric juices of gastric adenocarcinoma patients and induced inflammation mainly via specific targeting of gastric epithelial cells. Exp. Mol. Med. 2017, 49, e330. [Google Scholar] [CrossRef]
- Grande, R.; Di Marcantonio, M.C.; Robuffo, I.; Pompilio, A.; Celia, C.; Di Marzio, L.; Paolino, D.; Codagnone, M.; Muraro, R.; Stoodley, P.; et al. Helicobacter pylori ATCC 43629/NCTC 11639 Outer Membrane Vesicles (OMVs) from Biofilm and Planktonic Phase Associated with Extracellular DNA (eDNA). Front. Microbiol. 2015, 6. [Google Scholar] [CrossRef]
- Heczko, U.; Smith, V.C.; Meloche, R.M.; Buchan, A.M.J.; Finlay, B.B. Characteristics of Helicobacter pylori attachment to human primary antral epithelial cells. Microbes Infect. 2000, 2, 1669–1676. [Google Scholar] [CrossRef]
- Sommi, P.; Ricci, V.; Fiocca, R.; Necchi, V.; Romano, M.; Telford, J.L.; Solcia, E.; Ventura, U. Persistence of Helicobacter pylori VacA toxin and vacuolating potential in cultured gastric epithelial cells. Am. J. Physiol. Content 1998, 275, G681–G688. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Woo, T.; Kurata, S.; Zaman, C.; Hojo, F.; Hanawa, T.; Kato, S.; Kamiya, S. Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe 2011, 17, 388–390. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Fukutomi, T.; Hanawa, T.; Kurata, S.; Zaman, C.; Hojo, F.; Kamiya, S. Diversification of the AlpB Outer Membrane Protein of Helicobacter pylori Affects Biofilm Formation and Cellular Adhesion. J. Bacteriol. 2016, 199, e00729-16. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Kurata, S.; Fukuda, M.; Kawakami, H.; Ochiai, K.; Hanawa, T.; Kamiya, S. Outer Membrane Vesicles of Helicobacter pylori TK1402 are Involved in Biofilm Formation. BMC Microbiol. 2009, 9, 197. [Google Scholar] [CrossRef]
- Ronci, M.; Del Prete, S.; Puca, V.; Carradori, S.; Carginale, V.; Muraro, R.; Mincione, G.; Aceto, A.; Sisto, F.; Supuran, C.T.; et al. Identification and characterization of the α-CA in the outer membrane vesicles produced by Helicobacter pylori. J. Enzym. Inhib. Med. Chem. 2019, 34, 189–195. [Google Scholar] [CrossRef]
- Rozo, A.J.; Cox, M.H.; Devitt, A.; Rothnie, A.J.; Goddard, A.D. Biophysical analysis of lipidic nanoparticles. Methods 2020. [Google Scholar] [CrossRef]
- Mullaney, E.; Brown, P.; Smith, S.M.; Botting, C.H.; Yamaoka, Y.; Terres, A.M.; Kelleher, D.P.; Windle, H.J. Proteomic and functional characterization of the outer membrane vesicles from the gastric pathogen Helicobacter pylori. Proteom. Clin. Appl. 2009, 3, 785–796. [Google Scholar] [CrossRef]
- Zavan, L.; Bitto, N.J.; Johnston, E.L.; Greening, D.W.; Kaparakis-Liaskos, M. Helicobacter pylori Growth Stage Determines the Size, Protein Composition, and Preferential Cargo Packaging of Outer Membrane Vesicles. Proteomics 2018, 19, e1800209. [Google Scholar] [CrossRef]
- Turner, L.; Bitto, N.J.; Steer, D.L.; Lo, C.; D’Costa, K.; Ramm, G.; Shambrook, M.; Hill, A.F.; Ferrero, R.L.; Kaparakis-Liaskos, M. Helicobacter pylori Outer Membrane Vesicle Size Determines Their Mechanisms of Host Cell Entry and Protein Content. Front. Immunol. 2018, 9, 9. [Google Scholar] [CrossRef]
- Olofsson, A.; Vallström, A.; Petzold, K.; Tegtmeyer, N.; Schleucher, J.; Carlsson, S.; Haas, R.; Backert, S.; Wai, S.N.; Gröbner, G.; et al. Biochemical and functional characterization of Helicobacter pylori vesicles. Mol. Microbiol. 2010, 77, 1539–1555. [Google Scholar] [CrossRef]
- Turner, L.; Praszkier, J.; Hutton, M.L.; Steer, D.; Ramm, G.; Kaparakis-Liaskos, M.; Ferrero, R.L. Increased Outer Membrane Vesicle Formation in a Helicobacter pylori tolB Mutant. Helicobacter 2015, 20, 269–283. [Google Scholar] [CrossRef]
- Penfold, C.N.; Li, C.; Zhang, Y.; Vankemmelbeke, M.; James, R. Colicin A binds to a novel binding site of TolA in the Escherichia coli periplasm. Biochem. Soc. Trans. 2012, 40, 1469–1474. [Google Scholar] [CrossRef]
- Godlewska, R.; Wiśniewska, K.; Pietras, Z.; Jagusztyn-Krynicka, E.K. Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: Function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol. Lett. 2009, 298, 1–11. [Google Scholar] [CrossRef]
- Liu, Q.; Li, X.; Zhang, Y.; Song, Z.; Li, R.; Ruan, H.; Huang, X. Orally-administered outer-membrane vesicles from Helicobacter pylori reduce H. pylori infection via Th2-biased immune responses in mice. Pathog. Dis. 2019, 77. [Google Scholar] [CrossRef]
- Yonezawa, H.; Osaki, T.; Kamiya, S. Biofilm Formation by Helicobacter pylori and Its Involvement for Antibiotic Resistance. BioMed. Res. Int. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Carron, M.A.; Tran, V.R.; Sugawa, C.; Coticchia, J.M. Identification of Helicobacter pylori Biofilms in Human Gastric Mucosa. J. Gastrointest. Surg. 2006, 10, 712–717. [Google Scholar] [CrossRef]
- Coticchia, J.; Sugawa, C.; Tran, V.; Gurrola, J.; Kowalski, E.; Carron, M. Presence and Density of Helicobacter pylori Biofilms in Human Gastric Mucosa in Patients with Peptic Ulcer Disease. J. Gastrointest. Surg. 2006, 10, 883–889. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Keenan, J.I.; Davis, K.A.; Beaugie, C.R.; McGovern, J.J.; Moran, A.P. Alterations in Helicobacter pylori outer membrane and outer membrane vesicle-associated lipopolysaccharides under iron-limiting growth conditions. Innate Immun. 2008, 14, 279–290. [Google Scholar] [CrossRef]
- Koeppen, K.; Hampton, T.H.; Jarek, M.; Scharfe, M.; Gerber, S.A.; Mielcarz, D.W.; Demers, E.G.; Dolben, E.L.; Hammond, J.H.; Hogan, D.A.; et al. A Novel Mechanism of Host-Pathogen Interaction through sRNA in Bacterial Outer Membrane Vesicles. PLoS Pathog. 2016, 12, e1005672. [Google Scholar] [CrossRef]
- Polakovicova, I.; Jerez, S.; Wichmann, I.A.; Sandoval-Bórquez, A.; Carrasco-Véliz, N.; Corvalán, A.L. Role of microRNAs and Exosomes in Helicobacter pylori and Epstein-Barr Virus Associated Gastric Cancers. Front. Microbiol. 2018, 9, 636. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Song, Z.; Li, R.; Ruan, H.; Liu, Q.; Huang, X. sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL-8 secretion in human cells. Int. J. Med. Microbiol. 2020, 310, 151356. [Google Scholar] [CrossRef]
- Lee, H.-J. Microbe-Host Communication by Small RNAs in Extracellular Vesicles: Vehicles for Transkingdom RNA Transportation. Int. J. Mol. Sci. 2019, 20, 1487. [Google Scholar] [CrossRef]
- Patton, J.G.; Franklin, J.L.; Weaver, A.M.; Vickers, K.; Zhang, B.; Coffey, R.J.; Ansel, K.M.; Blelloch, R.; Goga, A.; Huang, B.; et al. Biogenesis, delivery, and function of extracellular RNA. J. Extracell. Vesicles 2015, 4, 27494. [Google Scholar] [CrossRef]
- Parker, H.; Chitcholtan, K.; Hampton, M.B.; Keenan, J.I. Uptake of Helicobacter pylori Outer Membrane Vesicles by Gastric Epithelial Cells. Infect. Immun. 2010, 78, 5054–5061. [Google Scholar] [CrossRef]
- Olofsson, A.; Skalman, L.N.; Obi, I.; Lundmark, R.; Arnqvist, A. Uptake of Helicobacter pylori Vesicles Is Facilitated by Clathrin-Dependent and Clathrin-Independent Endocytic Pathways. mBio 2014, 5, e00979-14. [Google Scholar] [CrossRef]
- Irving, A.T.; Mimuro, H.; Kufer, T.A.; Lo, C.Y.; Wheeler, R.; Turner, L.; Thomas, B.J.; Malosse, C.; Gantier, M.P.; Casillas, L.N.; et al. The Immune Receptor NOD1 and Kinase RIP2 Interact with Bacterial Peptidoglycan on Early Endosomes to Promote Autophagy and Inflammatory Signaling. Cell Host Microbe 2014, 15, 623–635. [Google Scholar] [CrossRef]
- Heusermann, W.; Hean, J.; Trojer, D.; Steib, E.; Von Bueren, S.; Graff-Meyer, A.; Genoud, C.; Martin, K.; Pizzato, N.; Voshol, J.; et al. Exosomes surf on filopodia to enter cells at endocytic hot spots, traffic within endosomes, and are targeted to the ER. J. Cell Biol. 2016, 213, 173–184. [Google Scholar] [CrossRef]
- Chitcholtan, K.; Hampton, M.B.; Keenan, J.I. Outer membrane vesicles enhance the carcinogenic potential of Helicobacter pylori. Carcinogenesis 2008, 29, 2400–2405. [Google Scholar] [CrossRef]
- Van Der Pol, L.; Stork, M.; Van Der Ley, P. Outer membrane vesicles as platform vaccine technology. Biotechnol. J. 2015, 10, 1689–1706. [Google Scholar] [CrossRef]
- Lekmeechai, S.; Su, Y.-C.; Brant, M.; Alvarado-Kristensson, M.; Vallström, A.; Obi, I.; Arnqvist, A.; Riesbeck, K. Helicobacter pylori Outer Membrane Vesicles Protect the Pathogen From Reactive Oxygen Species of the Respiratory Burst. Front. Microbiol. 2018, 9, 9. [Google Scholar] [CrossRef]
- Eaton, K.A.; Brooks, C.L.; Morgan, D.R.; Krakowka, S. Essential role of urease in pathogenesis of gastritis induced by Helicobacter pylori in gnotobiotic piglets. Infect. Immun. 1991, 59, 2470–2475. [Google Scholar] [CrossRef]
- Hathroubi, S.; Servetas, S.L.; Windham, I.; Merrell, D.S.; Ottemann, K.M. Helicobacter pylori Biofilm Formation and Its Potential Role in Pathogenesis. Microbiol. Mol. Biol. Rev. 2018, 82, e00001-18. [Google Scholar] [CrossRef]
- Winter, J.; Letley, D.; Rhead, J.; Atherton, J.; Robinson, K. Helicobacter pylori Membrane Vesicles Stimulate Innate Pro- and Anti-Inflammatory Responses and Induce Apoptosis in Jurkat T Cells. Infect. Immun. 2014, 82, 1372–1381. [Google Scholar] [CrossRef]
- Ayala, G.; Torres, L.; Espinosa, M.; Fierros-Zarate, G.; Maldonado, V.; Melendez-Zajgla, J. External membrane vesicles from Helicobacter pylori induce apoptosis in gastric epithelial cells. FEMS Microbiol. Lett. 2006, 260, 178–185. [Google Scholar] [CrossRef]
- Ilver, D.; Barone, S.; Mercati, D.; Lupetti, P.; Telford, J.L. Helicobacter pylori toxin VacA is transferred to host cells via a novel contact-dependent mechanism. Cell. Microbiol. 2004, 6, 167–174. [Google Scholar] [CrossRef]
- Molinari, M.; Galli, C.; Norais, N.; Telford, J.L.; Rappuoli, R.; Luzio, J.P.; Montecucco, C. Vacuoles Induced by Helicobacter pylori Toxin Contain Both Late Endosomal and Lysosomal Markers. J. Biol. Chem. 1997, 272, 25339–25344. [Google Scholar] [CrossRef]
- Wang, Y.-H.; Wu, J.-J.; Lei, H.-Y. The Autophagic Induction in Helicobacter pylori-Infected Macrophage. Exp. Biol. Med. 2009, 234, 171–180. [Google Scholar] [CrossRef]
- Galmiche, A.; Rassow, J.; Doye, A.; Cagnol, S.; Chambard, J.C.; Contamin, S.; De Thillot, V.; Just, I.; Ricci, V.; Solcia, E.; et al. The N-terminal 34 kDa fragment of Helicobacter pylori vacuolating cytotoxin targets mitochondria and induces cytochrome c release. EMBO J. 2000, 19, 6361–6370. [Google Scholar] [CrossRef]
- Papini, E.; Satin, B.; Norais, N.; De Bernard, M.; Telford, J.L.; Rappuoli, R.; Montecucco, C. Selective increase of the permeability of polarized epithelial cell monolayers by Helicobacter pylori vacuolating toxin. J. Clin. Investig. 1998, 102, 813–820. [Google Scholar] [CrossRef]
- Keenan, J.; Allardyce, R.A. Iron influences the expression of Helicobacter pylori outer membrane vesicle-associated virulence factors. Eur. J. Gastroenterol. Hepatol. 2000, 12, 1267–1273. [Google Scholar] [CrossRef]
- Ricci, V.; Chiozzi, V.; Necchi, V.; Oldani, A.; Romano, M.; Solcia, E.; Ventura, U. Free-soluble and outer membrane vesicle-associated VacA from Helicobacter pylori: Two forms of release, a different activity. Biochem. Biophys. Res. Commun. 2005, 337, 173–178. [Google Scholar] [CrossRef]
- Yamasaki, E.; Wada, A.; Kumatori, A.; Nakagawa, I.; Funao, J.; Nakayama, M.; Hisatsune, J.; Kimura, M.; Moss, J.; Hirayama, T. Helicobacter pylori Vacuolating Cytotoxin Induces Activation of the Proapoptotic Proteins Bax and Bak, Leading to Cytochrome c Release and Cell Death, Independent of Vacuolation. J. Biol. Chem. 2006, 281, 11250–11259. [Google Scholar] [CrossRef]
- Hoy, B.; Löwer, M.; Weydig, C.; Carra, G.; Tegtmeyer, N.; Geppert, T.; Schröder, P.; Sewald, N.; Backert, S.; Schneider, G.; et al. Helicobacter pylori HtrA is a new secreted virulence factor that cleaves E-cadherin to disrupt intercellular adhesion. EMBO Rep. 2010, 11, 798–804. [Google Scholar] [CrossRef]
- Tegtmeyer, N.; Wessler, S.; Necchi, V.; Rohde, M.; Harrer, A.; Rau, T.T.; Asche, C.I.; Boehm, M.; Loessner, H.; Figueiredo, C.; et al. Helicobacter pylori Employs a Unique Basolateral Type IV Secretion Mechanism for CagA Delivery. Cell Host Microbe 2017, 22, 552.e5–560.e5. [Google Scholar] [CrossRef]
- Zhang, G.; Ducatelle, R.; Pasmans, F.; D’Herde, K.; Huang, L.; Smet, A.; Haesebrouck, F.; Flahou, B. Effects of Helicobacter suis γ-Glutamyl Transpeptidase on Lymphocytes: Modulation by Glutamine and Glutathione Supplementation and Outer Membrane Vesicles as a Putative Delivery Route of the Enzyme. PLoS ONE 2013, 8, e77966. [Google Scholar] [CrossRef]
- Hatakeyama, M. Oncogenic mechanisms of the Helicobacter pylori CagA protein. Nat. Rev. Cancer 2004, 4, 688–694. [Google Scholar] [CrossRef]
- Poppe, M.; Feller, S.M.; Romer, G.; Wessler, S. Phosphorylation of Helicobacter pylori CagA by c-Abl leads to cell motility. Oncogene 2006, 26, 3462–3472. [Google Scholar] [CrossRef]
- Mueller, D.; Tegtmeyer, N.; Brandt, S.; Yamaoka, Y.; De Poire, E.; Sgouras, D.; Wessler, S.; Torres, J.; Smolka, A.; Backert, S. c-Src and c-Abl kinases control hierarchic phosphorylation and function of the CagA effector protein in Western and East Asian Helicobacter pylori strains. J. Clin. Investig. 2012, 122, 1553–1566. [Google Scholar] [CrossRef]
- Krisch, L.M.; Posselt, G.; Hammerl, P.; Wessler, S. CagA Phosphorylation in Helicobacter pylori-Infected B Cells Is Mediated by the Nonreceptor Tyrosine Kinases of the Src and Abl Families. Infect. Immun. 2016, 84, 2671–2680. [Google Scholar] [CrossRef]
- Selbach, M.; Moese, S.; Hauck, C.R.; Meyer, T.F.; Backert, S. Src Is the Kinase of the Helicobacter pylori CagA Protein in Vitro and in Vivo. J. Biol. Chem. 2002, 277, 6775–6778. [Google Scholar] [CrossRef]
- Stein, M.; Bagnoli, F.; Halenbeck, R.; Rappuoli, R.; Fantl, W.J.; Covacci, A. c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 2002, 43, 971–980. [Google Scholar] [CrossRef]
- Selbach, M.; Paul, F.E.; Brandt, S.; Guye, P.; Daumke, O.; Backert, S.; Dehio, C.; Mann, M. Host Cell Interactome of Tyrosine-Phosphorylated Bacterial Proteins. Cell Host Microbe 2009, 5, 397–403. [Google Scholar] [CrossRef]
- Shimoda, A.; Ueda, K.; Nishiumi, S.; Murata-Kamiya, N.; Mukai, S.-A.; Sawada, S.-I.; Azuma, T.; Hatakeyama, M.; Akiyoshi, K. Exosomes as nanocarriers for systemic delivery of the Helicobacter pylori virulence factor CagA. Sci. Rep. 2016, 6, 18346. [Google Scholar] [CrossRef]
- Ko, S.H.; Rho, D.J.; Jeon, J.I.; Kim, Y.-J.; Woo, H.A.; Kim, N.; Kim, J.M. Crude Preparations of Helicobacter pylori Outer Membrane Vesicles Induce Upregulation of Heme Oxygenase-1 via Activating Akt-Nrf2 and mTOR–IκB Kinase–NF-κB Pathways in Dendritic Cells. Infect. Immun. 2016, 84, 2162–2174. [Google Scholar] [CrossRef]
- Turkina, M.V.; Olofsson, A.; Magnusson, K.-E.; Arnqvist, A.; Vikström, E. Helicobacter pylori vesicles carrying CagA localize in the vicinity of cell–cell contacts and induce histone H1 binding to ATP in epithelial cells. FEMS Microbiol. Lett. 2015, 362, 362. [Google Scholar] [CrossRef]
- Fischer, W.; Püls, J.; Buhrdorf, R.; Gebert, B.; Odenbreit, S.; Haas, R. Systematic mutagenesis of the Helicobacter pylori cag pathogenicity island: Essential genes for CagA translocation in host cells and induction of interleukin-8. Mol. Microbiol. 2002, 42, 1337–1348. [Google Scholar] [CrossRef]
- Viala, J.; Chaput, C.; Boneca, I.G.; Cardona, A.; Girardin, S.E.; Moran, A.P.; Athman, R.; Mémet, S.; Huerre, M.R.; Coyle, A.J.; et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 2004, 5, 1166–1174. [Google Scholar] [CrossRef]
- Pfannkuch, L.; Hurwitz, R.; Traulsen, J.; Sigulla, J.; Poeschke, M.; Matzner, L.; Kosma, P.; Schmid, M.; Meyer, T.F. ADP heptose, a novel pathogen-associated molecular pattern identified in Helicobacter pylori. FASEB J. 2019, 33, 9087–9099. [Google Scholar] [CrossRef]
- Zimmermann, S.; Pfannkuch, L.; Al-Zeer, M.A.; Bartfeld, S.; Koch, M.; Liu, J.; Rechner, C.; Soerensen, M.; Sokolova, O.; Zamyatina, A.; et al. ALPK1- and TIFA-Dependent Innate Immune Response Triggered by the Helicobacter pylori Type IV Secretion System. Cell Rep. 2017, 20, 2384–2395. [Google Scholar] [CrossRef]
- Ko, S.H.; Jeon, J.I.; Kim, Y.-J.; Yoon, H.J.; Kim, H.; Kim, N.; Kim, J.S.; Kim, J.M. Helicobacter pylori Outer Membrane Vesicle Proteins Induce Human Eosinophil Degranulation via a β2 Integrin CD11/CD18- and ICAM-1-Dependent Mechanism. Mediat. Inflamm. 2015, 2015, 301716. [Google Scholar] [CrossRef]
- Hock, B.D.; McKenzie, J.L.; Keenan, J.I. Helicobacter pylori outer membrane vesicles inhibit human T cell responses via induction of monocyte COX-2 expression. Pathog. Dis. 2017, 75. [Google Scholar] [CrossRef]
- Keenan, J.I.; Allardyce, R.; Bagshaw, P.F. Lack of protection following immunisation with H. pylori outer membrane vesicles highlights antigenic differences between H. felis and H. pylori. FEMS Microbiol. Lett. 1998, 161, 21–27. [Google Scholar] [CrossRef][Green Version]
- Keenan, J.; Oliaro, J.; Domigan, N.; Potter, H.; Aitken, G.; Allardyce, R.; Roake, J. Immune Response to an 18-Kilodalton Outer Membrane Antigen Identifies Lipoprotein 20 as a Helicobacter pylori Vaccine Candidate. Infect. Immun. 2000, 68, 3337–3343. [Google Scholar] [CrossRef][Green Version]
- Keenan, J.; Rijpkema, S.; Durrani, Z.; Roake, J.A. Differences in immunogenicity and protection in mice and guinea pigs following intranasal immunization with Helicobacter pylori outer membrane antigens. FEMS Immunol. Med. Microbiol. 2003, 36, 199–205. [Google Scholar] [CrossRef]
- Jain, S.; Pillai, J. Bacterial membrane vesicles as novel nanosystems for drug delivery. Int. J. Nanomed. 2017, 12, 6329–6341. [Google Scholar] [CrossRef]
- Pathirana, R.D.; Kaparakis-Liaskos, M. Bacterial membrane vesicles: Biogenesis, immune regulation and pathogenesis. Cell. Microbiol. 2016, 18, 1518–1524. [Google Scholar] [CrossRef]
- Tan, K.; Li, R.; Huang, X.; Liu, Q. Outer Membrane Vesicles: Current Status and Future Direction of These Novel Vaccine Adjuvants. Front. Microbiol. 2018, 9, 783. [Google Scholar] [CrossRef]
- Irene, C.; Fantappiè, L.; Caproni, E.; Zerbini, F.; Anesi, A.; Tomasi, M.; Zanella, I.; Stupia, S.; Prete, S.; Valensin, S.; et al. Bacterial outer membrane vesicles engineered with lipidated antigens as a platform for Staphylococcus aureus vaccine. Proc. Natl. Acad. Sci. USA 2019, 116, 21780–21788. [Google Scholar] [CrossRef]
- Toledo, A.; Coleman, J.L.; Kuhlow, C.J.; Crowley, J.T.; Benach, J.L. The Enolase of Borrelia burgdorferi Is a Plasminogen Receptor Released in Outer Membrane Vesicles. Infect. Immun. 2011, 80, 359–368. [Google Scholar] [CrossRef]
- Gankema, H.; Wensink, J.; Guinée, P.A.M.; Jansen, W.H.; Witholt, B. Some characteristics of the outer membrane material released by growing enterotoxigenic Escherichia coli. Infect. Immun. 1980, 29, 704–713. [Google Scholar]
- Horstman, A.L.; Kuehn, M.J. Enterotoxigenic Escherichia coli Secretes Active Heat-labile Enterotoxin via Outer Membrane Vesicles. J. Biol. Chem. 2000, 275, 12489–12496. [Google Scholar] [CrossRef]
- Kato, S.; Kowashi, Y.; DeMuth, D.R. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 2002, 32, 1–13. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).