A Proof of Principle for the Detection of Viable Brucella spp. in Raw Milk by qPCR Targeting Bacteriophages
Abstract
1. Introduction
2. Materials and Methods
2.1. Phage Propagation
2.2. Phage DNA Isolation and Quantification
2.3. Brucella Phage Helicase (BPHeli) qPCR Assay Design and Running Conditions
2.4. Performance Indicators of the BPHeli qPCR Assay
2.5. Stability of the BPHeli qPCR Assay in Cow’s Milk
2.6. Phage Propagation and Growth of B. Microti in Raw Cow’s Milk
3. Results
3.1. Phage Propagation and DNA Isolation
3.2. Brucella Phage Helicase (BPHeli) qPCR Assay Performance
3.3. BPHeli qPCR Assay Stability in Cow’s Milk
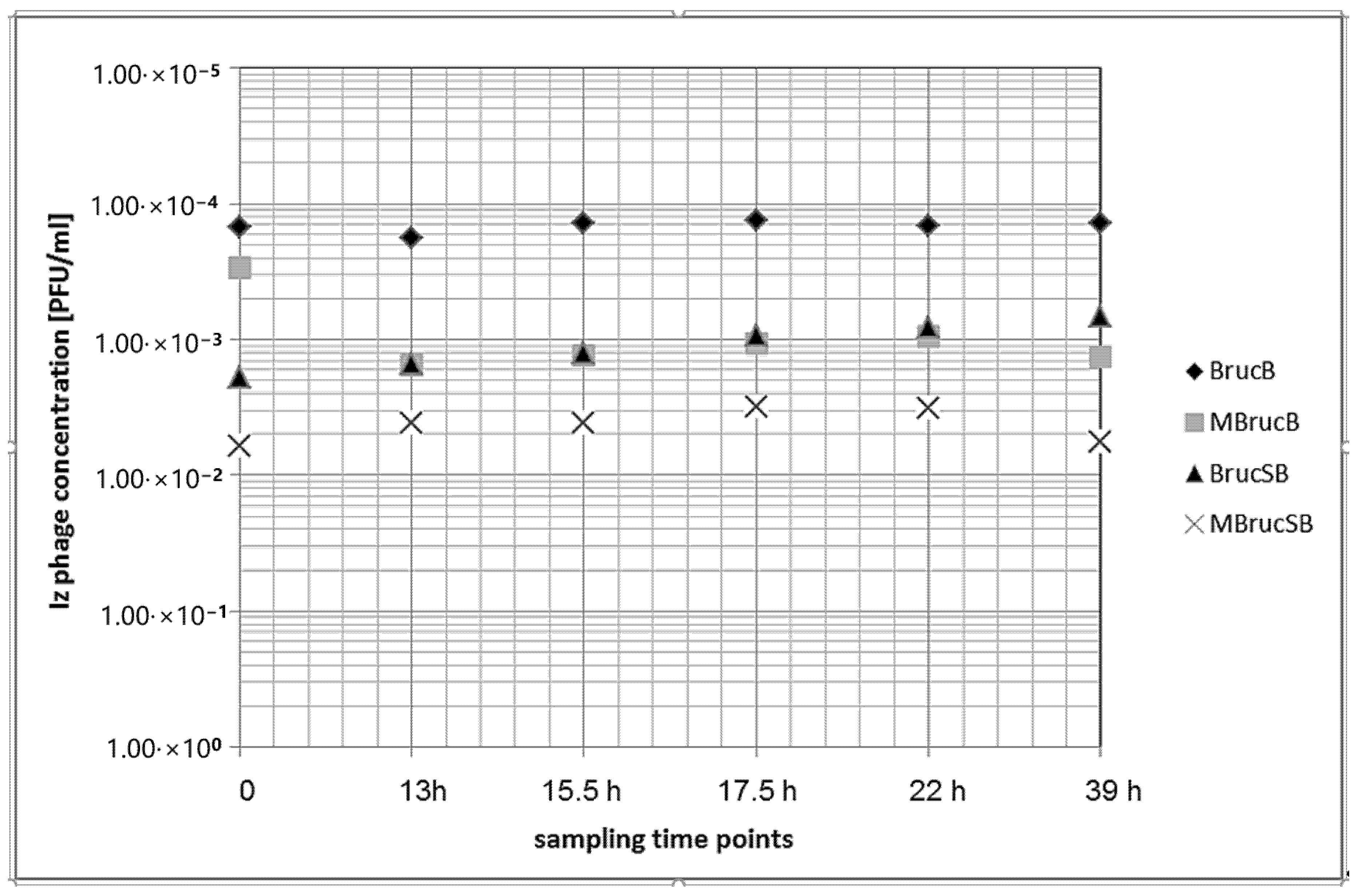
3.4. Growth of B. microti and Izv Phage Propagation in Raw Cow’s Milk
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Olsen, S.C.; Palmer, M.V. Advancement of knowledge of Brucella over the past 50 years. Vet. Pathol. 2014, 51, 1076–1089. [Google Scholar] [CrossRef]
- Pappas, G.; Papadimitriou, P.; Akritidis, N.; Christou, L.; Tsianos, E.V. The new global map of human brucellosis. Lancet Infect. Dis. 2006, 6, 91–99. [Google Scholar] [CrossRef]
- Jansen, W.; Linard, C.; Noll, M.; Noeckler, K.; Al Dahouk, S. Brucella-positive raw milk cheese sold on the inner European market: A public health threat due to illegal import? Food Control 2019, 100, 130–137. [Google Scholar] [CrossRef]
- Franc, K.A.; Krecek, R.C.; Hasler, B.N.; Arenas-Gamboa, A.M. Brucellosis remains a neglected disease in the developing world: A call for interdisciplinary action. BMC Public Health 2018, 18, 125. [Google Scholar] [CrossRef]
- Dadar, M.; Shahali, Y.; Whatmore, A.M. Human brucellosis caused by raw dairy products: A review on the occurrence, major risk factors and prevention. Int. J. Food Microbiol. 2019, 292, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Cross, A.R.; Baldwin, V.M.; Roy, S.; Essex-Lopresti, A.E.; Prior, J.L.; Harmer, N.J. Zoonoses under our noses. Microb. Infect. 2019, 21, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Godfroid, J.; DeBolle, X.; Roop, R.M.; O’Callaghan, D.; Tsolis, R.M.; Baldwin, C.; Santos, R.L.; McGiven, J.; Olsen, S.; Nymo, I.H.; et al. The quest for a true one health perspective of brucellosis. Rev. Sci. Technol. 2014, 33, 521–538. [Google Scholar] [CrossRef]
- Galińska, E.M.; Zagórski, J. Brucellosis in humans-etiology, diagnostics, clinical forms. Ann. Agric. Environ. Med. 2013, 20, 233–238. [Google Scholar]
- Alton, G.G.; Jones, L.M.; Angus, R.D.; Verger, J.M. (Eds.) Techniques for the Brucellosis Laboratory; Institute National de la Recherche Agronomique: Paris, France, 1988. [Google Scholar]
- Al Dahouk, S.; Tomaso, H.; Noeckler, K.; Neubauer, H.; Frangoulidis, D. Laboratory-based diagnosis of brucellosis-a review of the literature. Part I: Techniques for direct detection and identification of Brucella spp. Clin. Lab. 2003, 49, 487–505. [Google Scholar]
- Al Dahouk, S.; Tomaso, H.; Noeckler, K.; Neubauer, H. The detection of Brucella spp. using PCR-ELISA and real-time PCR assays. Clin. Lab. 2004, 50, 387–394. [Google Scholar]
- Kaden, R.; Ferrari, S.; Jinnerot, T.; Lindberg, M.; Wahab, T.; Lavander, M. Brucella abortus: Determination of survival times and evaluation of methods for detection in several matrices. BMC Infect. Dis. 2018, 18, 259. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z.; Zhang, Y.; Bai, L.; Zhao, Y.; Liu, C.; Ma, A.; Yu, H. Polymerase chain reaction-based assays for the diagnosis of human brucellosis. Ann. Clin. Microbiol. Antimicrob. 2014, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, E.A.; Vasin, A.V.; Sandybaev, N.T.; Klotchenko, S.A.; Plotnikova, M.A.; Chervyakova, O.V.; Sansyzbay, A.R.; Kiselev, O.I. Current methods of human and animal brucellosis diagnostics. Adv. Infect. Dis. 2013, 03, 177–184. [Google Scholar] [CrossRef]
- Vrioni, G.; Pappas, G.; Priavali, E.; Gartzonika, C.; Levidiotou, S. An eternal microbe: Brucella DNA load persists for years after clinical cure. Clin. Infect. Dis. 2008, 46, e131–e136. [Google Scholar] [CrossRef] [PubMed]
- Castano, M.J.; Solera, J. Chronic brucellosis and persistence of Brucella melitensis DNA. J. Clin. Microbiol. 2009, 47, 2084–2089. [Google Scholar] [CrossRef]
- Gwida, M.; El-Ashker, M.; Melzer, F.; El-Diasty, M.; El-Beskawy, M.; Neubauer, H. Use of serology and real time PCR to control an outbreak of bovine brucellosis at a dairy cattle farm in the Nile Delta region, Egypt. Ir. Vet. J. 2015, 69, 3. [Google Scholar] [CrossRef]
- Hinić, V.; Brodard, I.; Thomann, A.; Holub, M.; Miserez, R.; Abril, C. IS711-based real-time PCR assay as a tool for detection of Brucella spp. in wild boars and comparison with bacterial isolation and serology. BMC Vet. Res. 2009, 5, 22. [Google Scholar] [CrossRef]
- Rigby, C.E.; Cerqueira-Campos, M.L.; Kelly, H.A.; Surujballi, O.P. Properties and partial genetic characterization of Nepean phage and other lytic phages of Brucella species. Can. J. Vet. Res. 1989, 53, 319–325. [Google Scholar]
- Jones, L.M.; Merz, G.S.; Wilson, J.B. Phage typing reactions on Brucella species. Appl. Microbiol. 1968, 16, 1179–1190. [Google Scholar] [CrossRef]
- Jones, L.M. Comparison of phage typing with standard methods of species differentiation in brucellae. Bull. World Health Organ. 1960, 23, 130–133. [Google Scholar]
- Morris, J.A.; Corbel, M.J.; Phillip, J.I. Characterization of three phages lytic for Brucella species. J. Gen. Virol. 1973, 20, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Farlow, J.; Filippov, A.A.; Sergueev, K.V.; Hang, J.; Kotorashvili, A.; Nikolich, M.P. Comparative whole genome analysis of six diagnostic brucellaphages. Gene 2014, 541, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Goellner, C.; Jaeckel, C.; Scholz, H.C.; Noeckler, K.; Reetz, J.; Al Dahouk, S.; Hertwig, S. Genetic diversity of Brucella reference and non-reference phages and its impact on Brucella-typing. Front. Microbiol. 2017, 8, 408. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Poshtiban, S.; Evoy, S. Recent advances in bacteriophage based biosensors for food-borne pathogen detection. Sensors 2013, 13, 1763–1786. [Google Scholar] [CrossRef] [PubMed]
- Pujato, S.A.; Quiberoni, A.; Mercanti, D.J. Bacteriophages on dairy foods. J. Appl. Microbiol. 2019, 126, 14–30. [Google Scholar] [CrossRef]
- Bai, J.; Kim, Y.T.; Ryu, S.; Lee, J.H. Biocontrol and rapid detection of food-borne pathogens using bacteriophages and endolysins. Front. Microbiol. 2016, 7, 474. [Google Scholar] [CrossRef]
- Fernandez, L.; Escobedo, S.; Gutierrez, D.; Portilla, S.; Martinez, B.; Garcia, P.; Rodriguez, A. Bacteriophages in the dairy environment: From enemies to allies. Antibiotics 2017, 6, 27. [Google Scholar] [CrossRef]
- Schmelcher, M.; Loessner, M.J. Application of bacteriophages for detection of foodborne pathogens. Bacteriophage 2014, 4, e28137. [Google Scholar] [CrossRef]
- Wei, S.; Chelliah, R.; Rubab, M.; Oh, D.H.; Uddin, M.J.; Ahn, J. Bacteriophages as potential tools for detection and control of Salmonella spp. in food systems. Microorganisms 2019, 7, 570. [Google Scholar] [CrossRef]
- Koressaar, T.; Remm, M. Enhancements and modifications of primer design program primer3. Bioinformatics 2007, 23, 1289–1291. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3-new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Hammerl, J.A.; Al Dahouk, S.; Noeckler, K.; Goellner, C.; Appel, B.; Hertwig, S. F1 and Tbilisi are closely related brucellaphages exhibiting some distinct nucleotide variations which determine the host specificity. Genome Announc. 2014, 2. [Google Scholar] [CrossRef] [PubMed]
- Tevdoradze, E.; Farlow, J.; Kotorashvili, A.; Skhirtladze, N.; Antadze, I.; Gunia, S.; Balarjishvili, N.; Kvachadze, L.; Kutateladze, M. Whole genome sequence comparison of ten diagnostic brucellaphages propagated on two Brucella abortus hosts. Virol. J. 2015, 12, 66. [Google Scholar] [CrossRef] [PubMed]
- Jansen, W.; Mueller, A.; Grabowski, N.T.; Kehrenberg, C.; Muylkens, B.; Al Dahouk, S. Foodborne diseases do not respect borders: Zoonotic pathogens and antimicrobial resistant bacteria in food products of animal origin illegally imported into the European Union. Vet. J. 2019, 244, 75–82. [Google Scholar] [CrossRef]
- Matero, P.; Hemmila, H.; Tomaso, H.; Piiparinen, H.; Rantakokko-Jalava, K.; Nuotio, L.; Nikkari, S. Rapid field detection assays for Bacillus anthracis, Brucella spp. Francisella tularensis and Yersinia pestis. Clin. Microbiol. Infect. 2011, 17, 34–43. [Google Scholar] [CrossRef]
- Sergueev, K.V.; Filippov, A.A.; Nikolich, M.P. Highly sensitive bacteriophage-based detection of Brucella abortus in mixed culture and spiked blood. Viruses 2017, 9, 144. [Google Scholar] [CrossRef]
- Peng, X.; Nguyen, A.; Ghosh, D. Quantification of M13 and T7 bacteriophages by TaqMan and SYBR green qPCR. J. Virol. Methods 2018, 252, 100–107. [Google Scholar] [CrossRef]
- Cormier, J.; Janes, M. A double layer plaque assay using spread plate technique for enumeration of bacteriophage MS2. J. Virol. Methods 2014, 196, 86–92. [Google Scholar] [CrossRef]
- Tawil, N.; Sacher, E.; Mandeville, R.; Meunier, M. Bacteriophages: Biosensing tools for multi-drug resistant pathogens. Analyst 2014, 139, 1224–1236. [Google Scholar] [CrossRef]
- Luo, J.; Jiang, M.; Xiong, J.; Li, J.; Zhang, X.; Wei, H.; Yu, J. Exploring a phage-based real-time PCR assay for diagnosing Acinetobacter baumannii bloodstream infections with high sensitivity. Anal. Chim. Acta 2018, 1044, 147–153. [Google Scholar] [CrossRef]
- Scholz, H.C.; Hubalek, Z.; Sedlacek, I.; Vergnaud, G.; Tomaso, H.; Al Dahouk, S.; Melzer, F.; Kaempfer, P.; Neubauer, H.; Cloeckaert, A.; et al. Brucella microti sp. nov., isolated from the common vole Microtus arvalis. Int. J. Syst. Evol. Microbiol. 2008, 58, 375–382. [Google Scholar] [CrossRef] [PubMed]
- Al Dahouk, S.; Hofer, E.; Tomaso, H.; Vergnaud, G.; Le Fleche, P.; Cloeckaert, A.; Koylass, M.S.; Whatmore, A.M.; Noeckler, K.; Scholz, H.C. Intraspecies biodiversity of the genetically homologous species Brucella microti. Appl. Environ. Microbiol. 2012, 78, 1534–1543. [Google Scholar] [CrossRef] [PubMed]
- Seleem, M.N.; Boyle, S.M.; Sriranganathan, N. Brucellosis: A re-emerging zoonosis. Vet. Microbiol. 2010, 140, 392–398. [Google Scholar] [CrossRef] [PubMed]
- De Massis, F.; Di Girolamo, A.; Petrini, A.; Pizzigallo, E.; Giovannini, A. Correlation between animal and human brucellosis in Italy during the period 1997–2002. Clin. Microbiol. Infect. 2005, 11, 632–636. [Google Scholar] [CrossRef] [PubMed]
- Hesse, S.; Rajaure, M.; Wall, E.; Johnson, J.; Bliskovsky, V.; Gottesman, S.; Adhya, S. Phage resistance in multidrug-resistant Klebsiella pneumoniae ST258 evolves via diverse mutations that culminate in impaired adsorption. mBio 2020, 11. [Google Scholar] [CrossRef]
- Chen, L.; Fan, J.; Yan, T.; Liu, Q.; Yuan, S.; Zhang, H.; Yang, J.; Deng, D.; Huang, S.; Ma, Y. Isolation and characterization of specific phages to prepare a cocktail preventing Vibrio sp. Va-F3 infections in shrimp (Litopenaeus vannamei). Front. Microbiol. 2019, 10, 2337. [Google Scholar] [CrossRef]
- Sharma, S.C.; Memic, A.; Rupasinghe, C.N.; Duc, A.C.; Spaller, M.R. T7 phage display as a method of peptide ligand discovery for PDZ domain proteins. Biopolymers 2009, 92, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wang, D.; Chen, J.; Sela, D.A.; Nugen, S.R. Development of a novel bacteriophage based biomagnetic separation method as an aid for sensitive detection of viable Escherichia coli. Analyst 2016, 141, 1009–1016. [Google Scholar] [CrossRef]

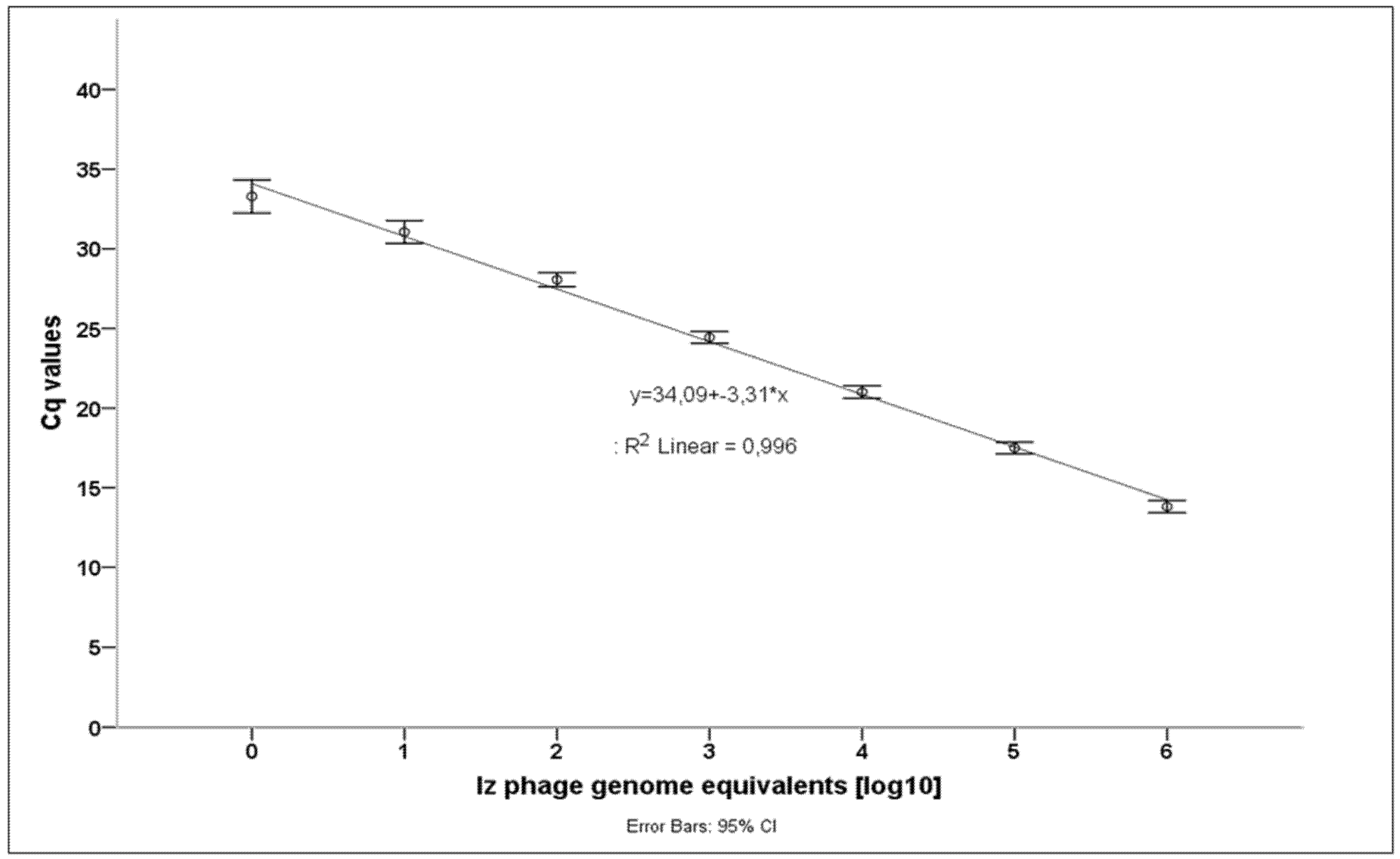
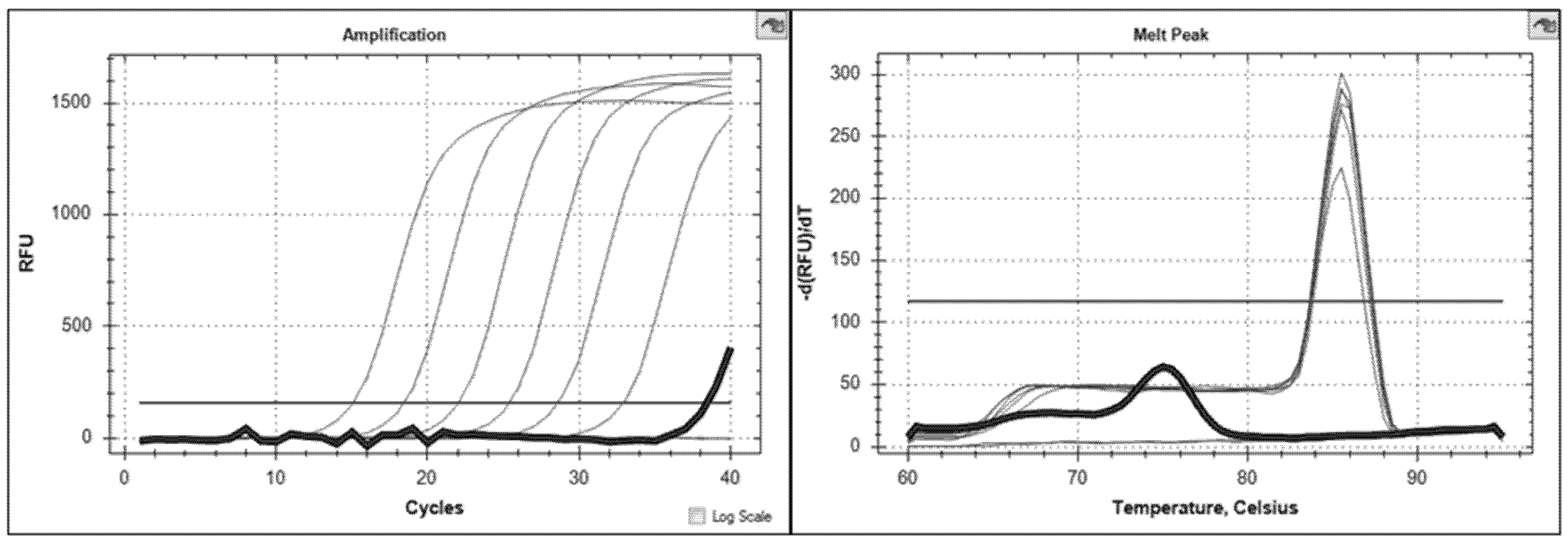
| Izv Genome Equivalents [log10] | 0 | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|---|
| Mean Cq-value | 33.29 | 31.06 | 28.06 | 24.44 | 21.01 | 17.49 | 13.81 |
| Standard deviation | 0.45 | 0.32 | 0.21 | 0.17 | 0.19 | 0.17 | 0.18 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Projahn, M.; Hammerl, J.A.; Dieckmann, R.; Dahouk, S.A. A Proof of Principle for the Detection of Viable Brucella spp. in Raw Milk by qPCR Targeting Bacteriophages. Microorganisms 2020, 8, 1326. https://doi.org/10.3390/microorganisms8091326
Projahn M, Hammerl JA, Dieckmann R, Dahouk SA. A Proof of Principle for the Detection of Viable Brucella spp. in Raw Milk by qPCR Targeting Bacteriophages. Microorganisms. 2020; 8(9):1326. https://doi.org/10.3390/microorganisms8091326
Chicago/Turabian StyleProjahn, Michaela, Jens A. Hammerl, Ralf Dieckmann, and Sascha Al Dahouk. 2020. "A Proof of Principle for the Detection of Viable Brucella spp. in Raw Milk by qPCR Targeting Bacteriophages" Microorganisms 8, no. 9: 1326. https://doi.org/10.3390/microorganisms8091326
APA StyleProjahn, M., Hammerl, J. A., Dieckmann, R., & Dahouk, S. A. (2020). A Proof of Principle for the Detection of Viable Brucella spp. in Raw Milk by qPCR Targeting Bacteriophages. Microorganisms, 8(9), 1326. https://doi.org/10.3390/microorganisms8091326





