Development of Real-Time and Colorimetric Loop Mediated Isothermal Amplification Assay for Detection of Xanthomonas gardneri
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culturing
2.2. DNA Isolation
2.3. Primer Design
2.4. Real-Time LAMP
2.5. Electrophoresis of LAMP
2.6. Colorimetric LAMP
2.7. Sensitivity and Specificity (Real-Time and Colorimetric LAMP)
2.8. LAMP Assay on Plant Tissues
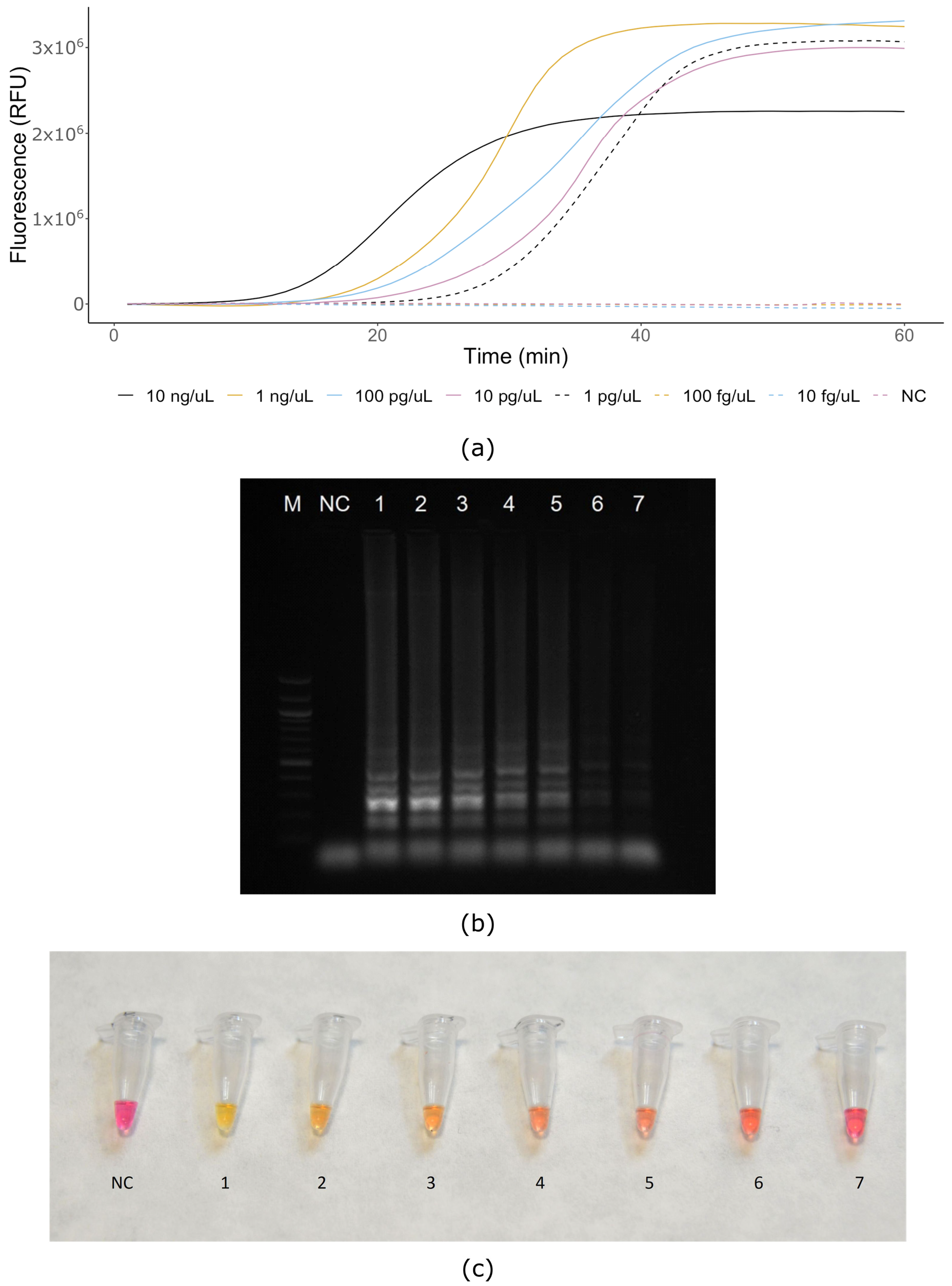
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Šutic, D.D. Bakterioze crvenog patlidžana = Bacteriosis of tomatoes; Institut za zaštitu bilja: Belgrade, Serbia, 1957; pp. 1–65. [Google Scholar]
- Dye, D.W. Cultural and biochemical reactions of additional Xanthomonas spp. N. Z. J. Sci. 1966, 9, 913. [Google Scholar]
- De Ley, J.; Segers, P.; Gillis, M. Intra- and Intergeneric Similarities of Chromobacterium and Janthinobacterium Ribosomal Ribonucleic Acid Cistrons. Int. J. Syst. Bacteriol. 1987, 28, 154–168. [Google Scholar] [CrossRef]
- Jones, J.B.; Bouzar, H.; Stall, R.E.; Almira, E.C.; Roberts, P.D.; Bowen, B.W.; Sudberry, J.; Strickler, P.M.; Chun, J. Systematic analysis of xanthomonads (Xanthomonas spp.) associated with pepper and tomato lesions. Int. J. Syst. Evol. Microbiol. 2000, 50, 1211–1219. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.B.; Lacy, G.H.; Bouzar, H.; Stall, R.E.; Schaad, N.W. Reclassification of the Xanthomonads Associated with Bacterial Spot Disease of Tomato and Pepper. Syst. Appl. Microbiol. 2004, 27, 755–762. [Google Scholar] [CrossRef]
- Hamza, A.A.; Robène-Soustrade, I.; Jouen, E.; Gagnevin, L.; Lefeuvre, P.; Chiroleu, F.; Pruvost, O. Genetic and Pathological Diversity Among Xanthomonas Strains Responsible for Bacterial Spot on Tomato and Pepper in the Southwest Indian Ocean Region. Plant Dis. 2010, 94, 993–999. [Google Scholar] [CrossRef]
- Barak, J.D.; Vancheva, T.; Lefeuvre, P.; Jones, J.B.; Timilsina, S.; Minsavage, G.V.; Koebnik, R. Whole-genome sequences of Xanthomonas euvesicatoria strains clarify taxonomy and reveal a stepwise erosion of type 3 effectors. Front. Plant Sci. 2016, 7, 1805. [Google Scholar] [CrossRef]
- Constantin, E.C.; Cleenwerck, I.; Maes, M.; Baeyen, S.; Van Malderghem, C.; De Vos, P.; Cottyn, B. Genetic characterization of strains named as Xanthomonas axonopodis pv. dieffenbachiae leads to a taxonomic revision of the X. axonopodis species complex. Plant Pathol. 2016, 65, 792–806. [Google Scholar] [CrossRef]
- Parkinson, N.; Aritua, V.; Heeney, J.; Cowie, C.; Bew, J.; Stead, D. Phylogenetic analysis of Xanthomonas species by comparison of partial gyrase B gene sequences. Int. J. Syst. Evol. Microbiol. 2007, 57, 2881–2887. [Google Scholar] [CrossRef]
- Young, J.M.; Park, D.C.; Shearman, H.M.; Fargier, E. A multilocus sequence analysis of the genus Xanthomonas. Syst. Appl. Microbiol. 2008, 31, 366–377. [Google Scholar] [CrossRef]
- Araújo, E.R.; Costa, J.R.; Ferreira, M.A.S.V.; Quezado-Duval, A.M. Widespread distribution of Xanthomonas perforans and limited presence of X. gardneri in Brazil. Plant Pathol. 2017, 66, 159–168. [Google Scholar] [CrossRef]
- Koenraadt, H.; Van Betteray, B.; Germain, R.; Hiddink, G.; Jones, J.B.; Oosterhof, J. Development of specific primers for the molecular detection of bacterial spot of pepper and tomato. Acta Hortic. 2009, 808, 99–102. [Google Scholar] [CrossRef]
- Strayer, A.L.; Jeyaprakash, A.; Minsavage, G.V.; Timilsina, S.; Vallad, G.E.; Jones, J.B.; Paret, M.L. A Multiplex Real-Time PCR Assay Differentiates Four Xanthomonas Species Associated with Bacterial Spot of Tomato. Plant Dis. 2016, 100, 1660–1668. [Google Scholar] [CrossRef] [PubMed]
- Cuppels, D.A.; Louws, F.J.; Ainsworth, T. Development and Evaluation of PCR-Based Diagnostic Assays for the Bacterial Speck and Bacterial Spot Pathogens of Tomato. Plant Dis. 2006, 90, 451–458. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Li, J.; Chen, Q. One new method of nucleic acid amplification—Loop-mediated isothermal amplification of DNA. Virol. Sin. 2008, 23, 167–172. [Google Scholar] [CrossRef]
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao, P.V.L.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef] [PubMed]
- Zanoli, L.M.; Spoto, G. Isothermal Amplification Methods for the Detection of Nucleic Acids in Microfluidic Devices. Biosensors 2013, 3, 18–43. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated reaction by loop-mediated isothermal amplification using loop primers. Mol. Cell. Probes 2002, 16, 223–229. [Google Scholar] [CrossRef]
- Aliotta, J.M.; Pelletier, J.J.; Ware, J.L.; Moran, L.S.; Benner, J.S.; Kong, H. Thermostable Bst DNA polymerase I lacks a 3′ → 5′ proofreading exonuclease activity. Genet. Anal. Biomol. Eng. 1996, 12, 185–195. [Google Scholar] [CrossRef]
- Hawwa, R.; Aikens, J.; Turner, R.J.; Santarsiero, B.D.; Mesecar, A.D. Structural basis for thermostability revealed through the identification and characterization of a highly thermostable phosphotriesterase-like lactonase from Geobacillus stearothermophilus. Arch. Biochem. Biophys. 2009, 488, 109–120. [Google Scholar] [CrossRef]
- Schaad, N.W.; Jones, J.B.; Chun, W. Laboratory Guide for the Identification of Plant. Pathogenic Bacteria, 3rd ed.; American Phytopathological Society, APS Press: St. Paul, MN, USA, 2001. [Google Scholar]
- King, E.O.; Ward, M.K.; Raney, D.E. Two simple media for the demonstration of pyocyanin and fluorescin. J. Lab. Clin. Med. 1954, 44, 301–307. [Google Scholar] [CrossRef] [PubMed]
- Roberts, S.J.; Koenraadt, H. 7-019a: Detection of Xanthomonas Campestris pv. Campestris on Brassica spp., 4th ed.; International Rules for Seed Testing, Chapter 7: Validated Seed Health Testing Methods; International Seed Testing Association: Bassersdorf, Switzerland, 2015. [Google Scholar]
- Bühlmann, A.; Pothier, J.F.; Tomlinson, J.A.; Frey, J.E.; Boonham, N.; Smits, T.H.M.; Duffy, B. Genomics-informed design of loop-mediated isothermal amplification for detection of phytopathogenic Xanthomonas arboricola pv. pruni at the intraspecific level. Plant Pathol. 2013, 62, 475–484. [Google Scholar] [CrossRef]
- Hodgetts, J.; Hall, J.; Karamura, G.; Grant, M.; Studholme, D.; Boonham, N.; Karamura, E.; Smith, J.J. Rapid, specific, simple, in-field detection of Xanthomonas campestris pathovar musacearum by loop-mediated isothermal amplification. J. Appl. Microbiol. 2015, 119, 1651–1658. [Google Scholar] [CrossRef] [PubMed]
- Lang, J.M.; Langlois, P.; Nguyen, M.H.R.; Triplett, L.R.; Purdie, L.; Holton, T.A.; Djikeng, A.; Vera Cruz, C.M.; Verdier, V.; Leach, J.E. Sensitive Detection of Xanthomonas oryzae Pathovars oryzae and oryzicola by Loop-Mediated Isothermal Amplification. Appl. Environ. Microbiol. 2014, 80, 4519–4530. [Google Scholar] [CrossRef] [PubMed]
- Langlois, P.A.; Snelling, J.; Hamilton, J.P.; Bragard, J.P.; Koebnik, R.; Verdier, V.; Triplett, L.R.; Blom, J.; Tisserat, N.A.; Leach, J.E. Characterization of the Xanthomonas translucens Complex Using Draft Genomes, Comparative Genomics, Phylogenetic Analysis, and Diagnostic LAMP Assays. Phytopathology 2017, 107, 519–527. [Google Scholar] [CrossRef]
- Rigano, L.A.; Marano, M.R.; Castagnaro, A.P.; Do Amaral, A.M.; Vojnov, A.A. Rapid and sensitive detection of Citrus Bacterial Canker by loop-mediated isothermal amplification combined with simple visual evaluation methods. BMC Microbiol. 2010, 10, 176. [Google Scholar] [CrossRef]
- Larrea-Sarmiento, A.; Dhakal, U.; Boluk, G.; Fatdal, L.; Alvarez, A.; Strayer-Scherer, A.; Paret, M.; Jones, J.; Jenkins, D.; Arif, M. Development of a genome-informed loop-mediated isothermal amplification assay for rapid and specific detection of Xanthomonas euvesicatoria. Sci. Rep. 2018, 8, 1–11. [Google Scholar] [CrossRef]
- Araújo, E.R.; Costa, J.R.; Ferreira, M.A.S.V.; Quezado-Duval, A.M. Simultaneous detection and identification of the Xanthomonas species complex associated with tomato bacterial spot using species-specific primers and multiplex PCR. J. Appl. Microbiol. 2012, 113, 1479–1490. [Google Scholar] [CrossRef]
- Obradovic, A.; Mavridis, A.; Rudolph, K.; Janse, J.D.; Arsenijevic, M.; Jones, J.B.; Minsavage, G.V.; Wang, J.F. Characterization and PCR-based typing of Xanthomonas campestris pv. vesicatoria from peppers and tomatoes in Serbia. Eur. J. Plant Pathol. 2004, 110, 285–292. [Google Scholar] [CrossRef]
- Wang, H.; Turechek, W.W. A loop-mediated isothermal amplification assay and sample preparation procedure for sensitive detection of Xanthomonas fragariae in strawberry. PLoS ONE 2016, 11, e0147122. [Google Scholar] [CrossRef]
- Kebede, M.; Timilsina, S.; Ayalew, A.; Admassu, B.; Potnis, N.; Minsavage, G.V.; Jones, J.B. Molecular characterization of Xanthomonas strains responsible for bacterial spot of tomato in Ethiopia. Eur. J. Plant Pathol. 2014, 140, 677–688. [Google Scholar] [CrossRef]
- Leite, R.P.; Minsavage, G.V.; Bonas, U.; Stall, R.E. Detection and identification of phytopathogenic Xanthomonas strains by amplification of DNA sequences related to the hrp genes of Xanthomonas campestris pv. vesicatoria. Appl. Environ. Microbiol. 1994, 60, 1068–1077. [Google Scholar] [CrossRef] [PubMed]
- Keremane, M.L.; Ramadugu, C.; Rodriguez, E.; Kubota, R.; Shibata, S.; Hall, R.F. A rapid field detection system for citrus huanglongbing associated‘Candidatus Liberibacter asiaticus’ from the psyllid vector, Diaphorina citri Kuwayama and its implications in disease management. Crop. Prot. 2015, 68, 41–48. [Google Scholar] [CrossRef]
- Golmohammadi, M.; Llop, P.; Scuderi, G.; Gell, I.; Graham, J.H.; Cubero, J. mRNA from selected genes is useful for specific detection and quantification of viable Xanthomonas citri subsp. citri. Plant Pathol. 2012, 61, 479–488. [Google Scholar] [CrossRef]
- Simões, T.H.; Gonçalves, E.R.; Rosato, Y.B.; Mehta, A. Differentiation of Xanthomonas species by PCR-RFLP of rpfB and atpD genes. FEMS Microbiol. Lett. 2007, 271, 33–39. [Google Scholar] [CrossRef][Green Version]


| Name 1 | Collection | Number in Collection | Geographic Origin | LAMP Result 5 |
|---|---|---|---|---|
| Xanthomonas gardneri | ||||
| X. gardneri2 | DSMZ | 19127 | Yugoslavia | + |
| X. gardneri | CFBP | 8588 | France (Réunion) | + |
| X. gardneri | CFBP | 7992 | France (Réunion) | + |
| X. gardneri | CFBP | 8120 | Costa Rica | + |
| Other (non-gardneri) Xanthomonas | ||||
| X. alfalfae subsp. alfalfae | CFBP | 3836 | Sudan | − |
| X. arboricola pv. pruni | BCCM/LMG | 854 | New Zealand | − |
| X. axonopodis pv. allii | CFBP | 6369 | Not specified (N.S.) 4 | − |
| X. axonopodis pv. carotoae | NCPPB | 3440 | Brazil | − |
| X. axonopodis pv. vesicatoria | CRI | 1013 | Czech Republic | − |
| X. campestris pv. incanae | HRI-W | 6377 | UK | − |
| X. campestris pv. phaseoli | NCAIM | B.01695 | Hungary | − |
| X. campestris pv. pisi | NCAIM | B.01393 | N.S. 4 | − |
| X. campestris pv. raphani | HRI-W | 8305 | UK | − |
| X. campestris pv. vesicatoria | BCCM/LMG | 934 | Brazil | − |
| X. campestris pv. vesicatoria | BCCM/LMG | 921 | USA (Long Island) | − |
| X. euvesicatoria | BCCM/LMG | 918 | India | − |
| X. euvesicatoria | BCCM/LMG | 922 | USA (Florida) | − |
| X. euvesicatoria | BCCM/LMG | 921 | USA (Long Island) | − |
| X. fragariae2 | CFBP | 6766 | USA | − |
| X. oryzae pv. Oryzicola 3 | CFBP | 2286 | N.S. 4 | − |
| X. perforans 9.22 | CFBP | 7293 | USA (Florida) | − |
| X. perforans 9.2 | CFBP | 8122 | Thailand | − |
| X. perforans2 | DSMZ | 18975 | USA | − |
| X. vesicatoria | BCCM/LMG | 925 | Hungary | − |
| X. vesicatoria2 | CFBP | 2537 | New Zealand | − |
| X. vesicatoria | BCCM/LMG | 920 | Italy | − |
| Other species | ||||
| Agrobacterium tumefaciens | CCM | 2835 | Czech Republic | − |
| Burkholderia glumae | BCCM/LMG | 20138 | Philippines (province Jalajala Riza) | − |
| B. glumae2 | BCCM/LMG | 2196 | Japan (Ehime) | − |
| Clavibacter michiganensis subsp. michiganensis | CFBP | 1460 | France | − |
| C. michiganensis subsp. sepedonicus | NCPPB | 3467 | Poland | − |
| Erwinia amylovora | CRI | Ea10/96 | Czech Republic | − |
| Pantoea agglomerans2 | CFBP | 3845 | N.S. 4 | − |
| Pseudomonas syringae pv. phaseolicola | CRI | 186/2 | Czech Republic | − |
| P. syringae pv. pisi | NCPPB | 3496 | USA | − |
| P. syringae pv. syringae | NCPPB | 2306 | Italy | − |
| P. syringae pv. tomato | CRI | 8119 | Czech Republic | − |
| Ralstonia pseudosolanacearum | CFBP | 3936 | China (Guangdong) | − |
| R. solanacearum | NCPPB | 2505 | Sweden | − |
| Stenotrophomonas sp. | NCPPB | 2859 | Turkey | − |
| Primer Name | Primer Length (nt) | Tm (°C) | Sequence (5′–3′) |
|---|---|---|---|
| F3 | 16 | 61.60 | CGGGGTGCAGGTCAGC |
| B3 | 15 | 61.13 | ACCGGCACCGCCAAG |
| FIP | 37 | - | CCACCTCGGCACGTTGCAGGCGAGGTATGCGAGTTGC |
| BIP | 35 | - | GCCGCCATCTCGCCTTGCGCCCCGATCCGATCACG |
| LB | 17 | 61.26 | CGAGCTGGTGGGCTTGT |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stehlíková, D.; Beran, P.; Cohen, S.P.; Čurn, V. Development of Real-Time and Colorimetric Loop Mediated Isothermal Amplification Assay for Detection of Xanthomonas gardneri. Microorganisms 2020, 8, 1301. https://doi.org/10.3390/microorganisms8091301
Stehlíková D, Beran P, Cohen SP, Čurn V. Development of Real-Time and Colorimetric Loop Mediated Isothermal Amplification Assay for Detection of Xanthomonas gardneri. Microorganisms. 2020; 8(9):1301. https://doi.org/10.3390/microorganisms8091301
Chicago/Turabian StyleStehlíková, Dagmar, Pavel Beran, Stephen P. Cohen, and Vladislav Čurn. 2020. "Development of Real-Time and Colorimetric Loop Mediated Isothermal Amplification Assay for Detection of Xanthomonas gardneri" Microorganisms 8, no. 9: 1301. https://doi.org/10.3390/microorganisms8091301
APA StyleStehlíková, D., Beran, P., Cohen, S. P., & Čurn, V. (2020). Development of Real-Time and Colorimetric Loop Mediated Isothermal Amplification Assay for Detection of Xanthomonas gardneri. Microorganisms, 8(9), 1301. https://doi.org/10.3390/microorganisms8091301





