Phenotypic Characterization and Transformation Attempts Reveal Peculiar Traits of Xylella fastidiosa Subspecies pauca Strain De Donno
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Growth Curve, Biofilm, Settling Rate, and Twitching Motility Measurements of X. fastidiosa Strains
Statistical Analysis
2.3. Transformation Protocols
2.3.1. Plasmid Constructs
2.3.2. Electroporation
2.3.3. Observation of Fluorescence
2.3.4. Natural Competence
2.3.5. DNA Extraction
2.3.6. PCR Amplification and Primer Design
2.3.7. Analysis of Restriction–Modification Systems
2.3.8. Analysis of Sequence Variations in Genes Involved in Natural Competence and Twitching Motility
3. Results
3.1. Phenotypic Differences between X. fastidiosa Strains De Donno and Temecula1
3.1.1. Growth Rate
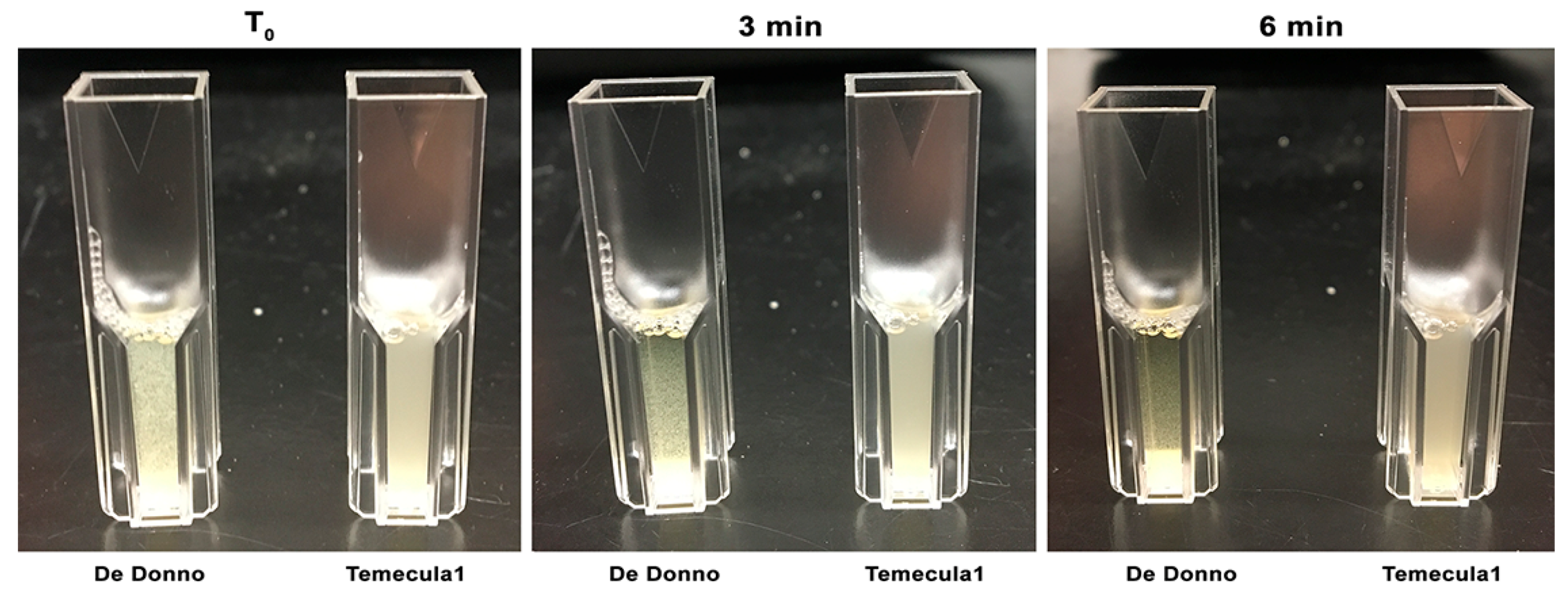
3.1.2. Settling Rate
3.1.3. Biofilm Formation
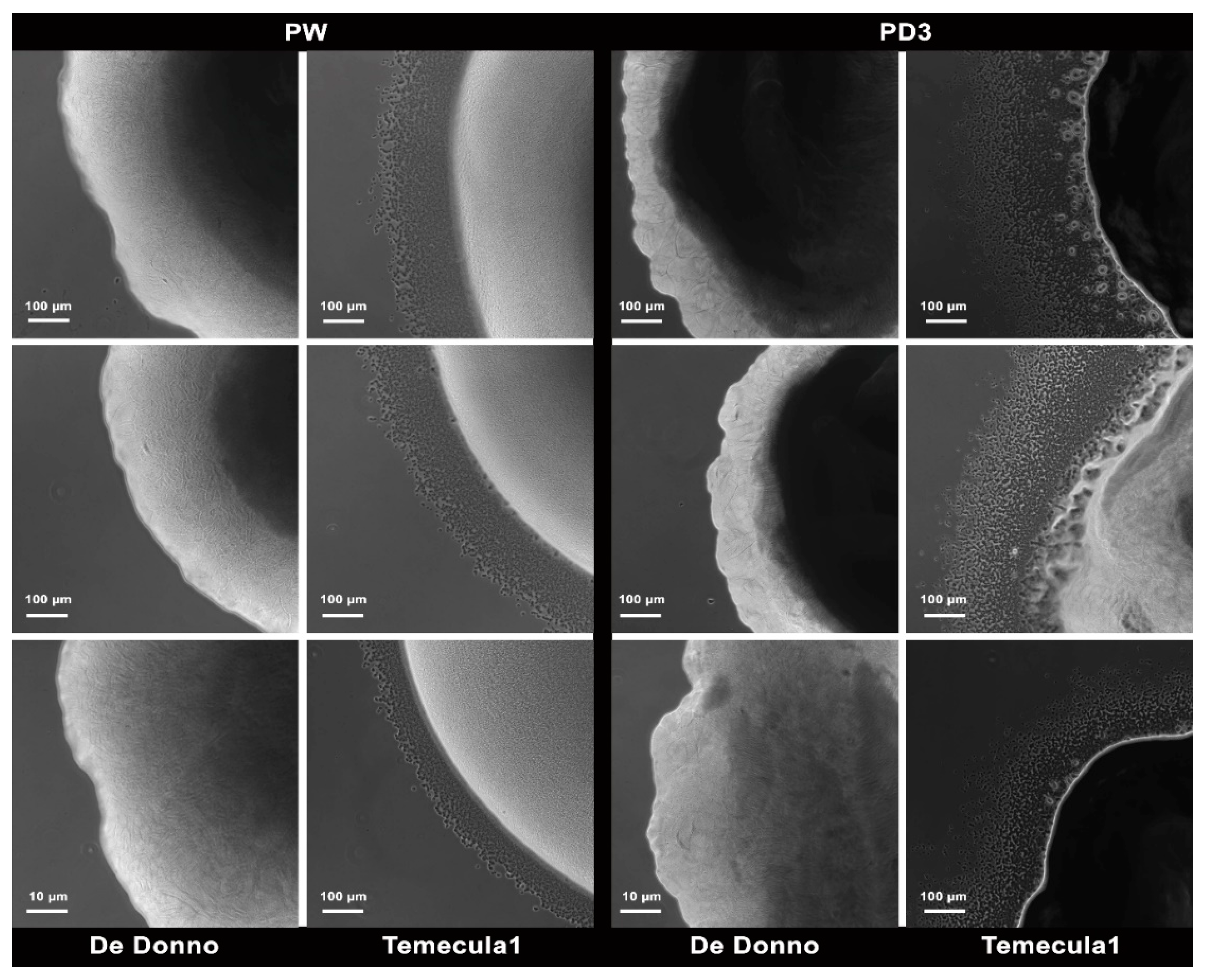
3.1.4. Twitching Motility
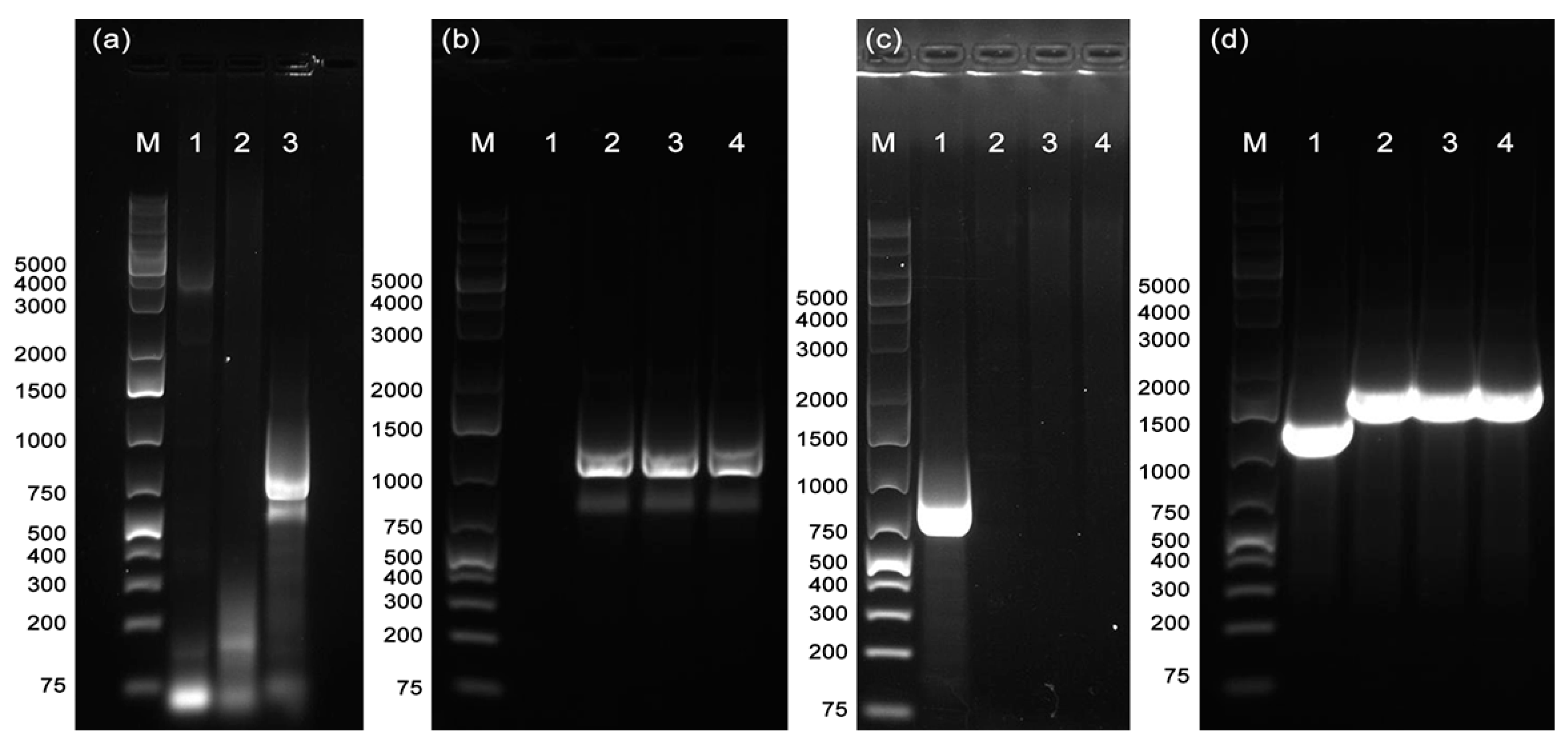
3.2. Attempts to Transform X. fastidiosa Strain De Donno
3.2.1. Transformation by Electroporation
3.2.2. Assessment of Stable Integration of the kan-gfp Cassette into the X. fastidiosa De Donno Chromosome
3.2.3. Assessment of Recombination in the rpfF Knockout Strain DDrpfF-
3.2.4. Natural Transformation
3.2.5. In Silico Prediction of the Effects of Sequence Variations in Genes Involved in Natural Competence and Twitching Motility
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority. Update of the Xylella spp. host plant database–systematic literature search up to 30 June 2019. EFSA J. 2020, 18, e06114. [Google Scholar]
- Almeida, R.P.P. Xylella fastidiosa vector transmission biology. In Vector-Mediated Transmission of Plant Pathogens; The American Phytopathological Association: St. Paul, MN, USA, 2016; pp. 165–173. [Google Scholar]
- Hopkins, D.L.; Purcell, A.H. Xylella fastidiosa: Cause of Pierce’s disease of grapevine and other emergent diseases. Plant Dis. 2002, 86, 1056–1066. [Google Scholar] [CrossRef]
- Saponari, M.; Boscia, D.; Altamura, G.; Loconsole, G.; Zicca, S.; D’Attoma, G.; Morelli, M.; Palmisano, F.; Saponari, A.; Tavano, D. Isolation and pathogenicity of Xylella fastidiosa associated to the olive quick decline syndrome in southern Italy. Sci. Rep. 2017, 7, 17723. [Google Scholar] [CrossRef]
- Saponari, M.; Giampetruzzi, A.; Loconsole, G.; Boscia, D.; Saldarelli, P. Xylella fastidiosa in olive in Apulia: Where we stand. Phytopathology 2019, 109, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Van der Werf, W.; Cendoya, M.; Mourits, M.; Navas-Cortés, J.A.; Vicent, A.; Lansink, A.O. Impact of Xylella fastidiosa subspecies pauca in European olives. Proc. Natl. Acad. Sci. USA 2020, 117, 9250–9259. [Google Scholar] [CrossRef]
- Hopkins, D.L. Xylella fastidiosa: Xylem-limited bacterial pathogen of plants. Annu. Rev. Phytopathol. 1989, 27, 271–290. [Google Scholar] [CrossRef]
- Tyson, G.E.; Stojanovic, B.J.; Kuklinski, R.F.; DiVittorio, T.J.; Sullivan, M.L. Scanning electron microscopy of Piercés disease bacterium in petiolar xylem of grape leaves. Phytopathology 1985, 75, 264–269. [Google Scholar] [CrossRef]
- Chatterjee, S.; Newman, K.L.; Lindow, S.E. Cell-to-cell signaling in Xylella fastidiosa suppresses movement and xylem vessel colonization in grape. Mol. Plant-Microbe Interact. 2008, 21, 1309–1315. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Sun, Y.; Walker, M.A.; Labavitch, J.M. Vascular occlusions in grapevines with Pierce’s disease make disease symptom development worse. Plant Physiol. 2013, 161, 1529–1541. [Google Scholar] [CrossRef]
- Newman, K.L.; Almeida, R.P.P.; Purcell, A.H.; Lindow, S.E. Cell-cell signaling controls Xylella fastidiosa interactions with both insects and plants. Proc. Natl. Acad. Sci. USA 2004, 101, 1737–1742. [Google Scholar] [CrossRef]
- Beaulieu, E.D.; Ionescu, M.; Chatterjee, S.; Yokota, K.; Trauner, D.; Lindow, S. Characterization of a diffusible signaling factor from Xylella fastidiosa. mBio 2013, 4. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Li, J.-L.; Lindow, S.E. RpfF-dependent regulon of Xylella fastidiosa. Phytopathology 2012, 102, 1045–1053. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, J.F.; Matthews, M.A.; Greve, L.C.; Labavitch, J.M.; Rost, T.L. Grapevine susceptibility to Pierce’s disease II: Progression of anatomical symptoms. Am. J. Enol. Vitic. 2004, 55, 238–245. [Google Scholar]
- Tyree, M.T.; Zimmermann, M.H. Hydraulic architecture of woody shoots. In Xylem Structure and the Ascent of Sap; Springer: Berlin/Heidelberg, Germany, 2002; pp. 143–174. [Google Scholar]
- Meng, Y.; Li, Y.; Galvani, C.D.; Hao, G.; Turner, J.N.; Burr, T.J.; Hoch, H. Upstream migration of Xylella fastidiosa via pilus-driven twitching motility. J. Bacteriol. 2005, 187, 5560–5567. [Google Scholar] [CrossRef]
- Mattick, J.S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 2002, 56, 289–314. [Google Scholar] [CrossRef]
- Huang, B.; Whitchurch, C.B.; Mattick, J.S. FimX, a multidomain protein connecting environmental signals to twitching motility in Pseudomonas aeruginosa. J. Bacteriol. 2003, 185, 7068–7076. [Google Scholar] [CrossRef]
- Liu, H.; Kang, Y.; Genin, S.; Schell, M.A.; Denny, T.P. Twitching motility of Ralstonia solanacearum requires a type IV pilus system. Microbiology 2001, 147, 3215–3229. [Google Scholar] [CrossRef]
- Feil, H.; Feil, W.S.; Lindow, S.E. Contribution of fimbrial and afimbrial adhesins of Xylella fastidiosa to attachment to surfaces and virulence to grape. Phytopathology 2007, 97, 318–324. [Google Scholar] [CrossRef]
- Guilhabert, M.R.; Kirkpatrick, B.C. Identification of Xylella fastidiosa antivirulence genes: Hemagglutinin adhesins contribute to X. fastidiosa biofilm maturation and colonization and attenuate virulence. Mol. Plant-Microbe Interact. 2005, 18, 856–868. [Google Scholar] [CrossRef][Green Version]
- Caserta, R.; Takita, M.; Targon, M.; Rosselli-Murai, L.; De Souza, A.; Peroni, L.; Stach-Machado, D.; Andrade, A.; Labate, C.; Kitajima, E. Expression of Xylella fastidiosa fimbrial and afimbrial proteins during biofilm formation. Appl. Environ. Microbiol. 2010, 76, 4250–4259. [Google Scholar] [CrossRef]
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422. [Google Scholar] [CrossRef] [PubMed]
- Cruz, L.F.; Cobine, P.A.; De La Fuente, L. Calcium increases Xylella fastidiosa surface attachment, biofilm formation, and twitching motility. Appl. Environ. Microbiol. 2012, 78, 1321–1331. [Google Scholar] [CrossRef] [PubMed]
- Sicard, A.; Zeilinger, A.R.; Vanhove, M.; Schartel, T.E.; Beal, D.J.; Daugherty, M.P.; Almeida, R.P. Xylella fastidiosa: Insights into an emerging plant pathogen. Annu. Rev. Phytopathol. 2018, 56, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Kung, S.H.; Almeida, R.P. Natural competence and recombination in the plant pathogen Xylella fastidiosa. Appl. Environ. Microbiol. 2011, 77, 5278–5284. [Google Scholar] [CrossRef] [PubMed]
- Kung, S.H.; Almeida, R.P. Biological and genetic factors regulating natural competence in a bacterial plant pathogen. Microbiology 2014, 160, 37–46. [Google Scholar] [CrossRef]
- Thomas, C.M.; Nielsen, K.M. Mechanisms of, and barriers to, horizontal gene transfer between bacteria. Nat. Rev. Microbiol. 2005, 3, 711–721. [Google Scholar] [CrossRef] [PubMed]
- Potnis, N.; Kandel, P.P.; Merfa, M.V.; Retchless, A.C.; Parker, J.K.; Stenger, D.C.; Almeida, R.P.; Bergsma-Vlami, M.; Westenberg, M.; Cobine, P.A. Patterns of inter-and intrasubspecific homologous recombination inform eco-evolutionary dynamics of Xylella fastidiosa. ISME J. 2019, 13, 2319–2333. [Google Scholar] [CrossRef] [PubMed]
- Almeida, R.P.; Nascimento, F.E.; Chau, J.; Prado, S.S.; Tsai, C.-W.; Lopes, S.A.; Lopes, J.R. Genetic structure and biology of Xylella fastidiosa strains causing disease in citrus and coffee in Brazil. Appl. Environ. Microbiol. 2008, 74, 3690–3701. [Google Scholar] [CrossRef][Green Version]
- Scally, M.; Schuenzel, E.L.; Stouthamer, R.; Nunney, L. Multilocus sequence type system for the plant pathogen Xylella fastidiosa and relative contributions of recombination and point mutation to clonal diversity. Appl. Environ. Microbiol. 2005, 71, 8491–8499. [Google Scholar] [CrossRef] [PubMed]
- Baltrus, D.A.; Guillemin, K.; Phillips, P.C. Natural transformation increases the rate of adaptation in the human pathogen Helicobacter pylori. Evol. Int. J. Org. Evol. 2008, 62, 39–49. [Google Scholar] [CrossRef]
- Nunney, L.; Hopkins, D.L.; Morano, L.D.; Russell, S.E.; Stouthamer, R. Intersubspecific recombination in Xylella fastidiosa strains native to the United States: Infection of novel hosts associated with an unsuccessful invasion. Appl. Environ. Microbiol. 2014, 80, 1159–1169. [Google Scholar] [CrossRef] [PubMed]
- Nunney, L.; Schuenzel, E.L.; Scally, M.; Bromley, R.E.; Stouthamer, R. Large-scale intersubspecific recombination in the plant-pathogenic bacterium Xylella fastidiosa is associated with the host shift to mulberry. Appl. Environ. Microbiol. 2014, 80, 3025–3033. [Google Scholar] [CrossRef] [PubMed]
- Nunney, L.; Yuan, X.; Bromley, R.E.; Stouthamer, R. Detecting genetic introgression: High levels of intersubspecific recombination found in Xylella fastidiosa in Brazil. Appl. Environ. Microbiol. 2012, 78, 4702–4714. [Google Scholar] [CrossRef] [PubMed]
- Friesen, T.L.; Stukenbrock, E.H.; Liu, Z.; Meinhardt, S.; Ling, H.; Faris, J.D.; Rasmussen, J.B.; Solomon, P.S.; McDonald, B.A.; Oliver, R.P. Emergence of a new disease as a result of interspecific virulence gene transfer. Nat. Genet. 2006, 38, 953–956. [Google Scholar] [CrossRef]
- Kandel, P.P.; Almeida, R.P.P.; Cobine, P.A.; De La Fuente, L. Natural competence rates are variable among Xylella fastidiosa strains and homologous recombination occurs in vitro between subspecies fastidiosa and multiplex. Mol. Plant-Microbe Interact. 2017, 30, 589–600. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kandel, P.P.; Lopez, S.M.; Almeida, R.P.P.; De La Fuente, L. Natural competence of Xylella fastidiosa occurs at a high frequency inside microfluidic chambers mimicking the bacterium’s natural habitats. Appl. Environ. Microbiol. 2016, 82, 5269–5277. [Google Scholar] [CrossRef]
- Kung, S.H.; Retchless, A.C.; Kwan, J.Y.; Almeida, R.P. Effects of DNA size on transformation and recombination efficiencies in Xylella fastidiosa. Appl. Environ. Microbiol. 2013, 79, 1712–1717. [Google Scholar] [CrossRef]
- Roberts, R.J.; Vincze, T.; Posfai, J.; Macelis, D. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007, 35, D269–D270. [Google Scholar] [CrossRef]
- Moreira, L.M.; De Souza, R.F.; Digiampietri, L.A.; Da Silva, A.C.; Setubal, J.C. Comparative analyses of Xanthomonas and Xylella complete genomes. OMICS. 2005, 9, 43–76. [Google Scholar] [CrossRef]
- Niza, B.; Merfa, M.V.; Alencar, V.C.; Menegidio, F.B.; Nunes, L.R.; Machado, M.A.; Takita, M.A.; de Souza, A.A. Draft genome sequence of 11399, a transformable citrus-pathogenic strain of Xylella fastidiosa. Genome Announc. 2016, 4. [Google Scholar] [CrossRef]
- Kobayashi, I. Behavior of restriction–modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001, 29, 3742–3756. [Google Scholar] [CrossRef] [PubMed]
- Guilhabert, M.R.; Kirkpatrick, B.C. Transformation of Xylella fastidiosa with broad host range RSF1010 derivative plasmids. Mol. Plant Pathol. 2003, 4, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Román Ecija, M.; Landa, B.B.; Navas Cortés, J.A.; Gómez, L.; Fuente, L. Phenotypic characterization of two Spanish strains of Xylella fastidiosa subsp. multiplex ST6 differing in plasmid content. In Proceedings of the 2nd European Conference on Xylella fastidiosa (How Research Can Support Solutions), Ajaccio, France, 29–30 October 2019. [Google Scholar]
- Cattò, C.; De Vincenti, L.; Cappitelli, F.; D’Attoma, G.; Saponari, M.; Villa, F.; Forlani, F. Non-Lethal Effects of N-Acetylcysteine on Xylella fastidiosa Strain De Donno Biofilm Formation and Detachment. Microorganisms 2019, 7, 656. [Google Scholar] [CrossRef] [PubMed]
- Giampetruzzi, A.; D’Attoma, G.; Zicca, S.; Abou Kubaa, R.; Rizzo, D.; Boscia, D.; Saldarelli, P.; Saponari, M. Draft genome sequence resources of three strains (TOS4, TOS5, and TOS14) of Xylella fastidiosa infecting different host plants in the newly discovered outbreak in Tuscany, Italy. Phytopathology 2019, 109, 1516–1518. [Google Scholar] [CrossRef] [PubMed]
- Van Sluys, M.A.; De Oliveira, M.C.; Monteiro-Vitorello, C.B.; Miyaki, C.Y.; Furlan, L.R.; Camargo, L.E.A.; Da Silva, A.C.R.; Moon, D.H.; Takita, M.A.; Lemos, E.G.M. Comparative analyses of the complete genome sequences of Pierce’s disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol. 2003, 185, 1018–1026. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Saponari, M.; Almeida, R.P.P.; Essakhi, S.; Boscia, D.; Loconsole, G.; Saldarelli, P. Complete genome sequence of the olive-infecting strain Xylella fastidiosa subsp. pauca De Donno. Genome Announc. 2017, 5. [Google Scholar] [CrossRef]
- Davis, M.; Davis, M.J.; Thomson, S.V. Isolation media for the Pierce’s disease bacterium. Phytopathology 1980, 70, 425–429. [Google Scholar] [CrossRef]
- Davis, M.J.; French, W.J.; Schaad, N.W. Axenic culture of the bacteria associated with phony disease of peach and plum leaf scald. Curr. Microbiol. 1981, 6, 309–314. [Google Scholar] [CrossRef]
- Allaire, J. RStudio: Integrated development environment for R. Bostonma 2012, 770, 394. [Google Scholar]
- McDonald, J.H. Handbook of Biological Statistics; Sparky House Publishing: Baltimore, MD, USA, 2009; Volume 2. [Google Scholar]
- Wickham, H. ggplot2. Wiley Interdiscip. Rev. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Newman, K.L.; Almeida, R.P.P.; Purcell, A.H.; Lindow, S.E. Use of a green fluorescent strain for analysis of Xylella fastidiosa colonization of Vitis vinifera. Appl. Environ. Microbiol. 2003, 69, 7319–7327. [Google Scholar] [CrossRef] [PubMed]
- Kandel, P.P.; Chen, H.; De La Fuente, L. A short protocol for gene knockout and complementation in Xylella fastidiosa shows that one of the type IV pilin paralogs (PD1926) is needed for twitching while another (PD1924) affects pilus number and location. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.M.; Raju, B.C.; Nyland, G.; Lowe, S.K. Medium for isolation and growth of bacteria associated with plum leaf scald and phony peach diseases. Appl. Environ. Microbiol. 1981, 42, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Loconsole, G.; Boscia, D.; Palmisano, F.; Savino, V.; Potere, O.; Martelli, G.P.; Saponari, M. A Xylella fastidiosa strain with unique biology and phylogeny is associated with a severe disease of olive in Southern Apulia. J. Plant Pathol. 2014, 96, S4. [Google Scholar]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. Bmc Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- REBASE. The Restriction Enzyme Database. Available online: http://rebase.neb.com/rebase/rebase.html (accessed on 5 August 2020).
- Ensembl Bacteria. Available online: https://bacteria.ensembl.org/index.html (accessed on 25 October 2020).
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- PROVEAN (Protein Variation Effect Analyzer). Available online: provean.jcvi.org (accessed on 5 August 2020).
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef]
- Stoodley, P.; Cargo, R.; Rupp, C.J.; Wilson, S.; Klapper, I. Biofilm material properties as related to shear-induced deformation and detachment phenomena. J. Ind. Microbiol. Biotechnol. 2002, 29, 361–367. [Google Scholar] [CrossRef]
- Janissen, R.; Murillo, D.M.; Niza, B.; Sahoo, P.K.; Nobrega, M.M.; Cesar, C.L.; Temperini, M.L.; Carvalho, H.F.; De Souza, A.A.; Cotta, M.A. Spatio-temporal distribution of different extracellular polymeric substances and filamentation mediate Xylella fastidiosa adhesion and biofilm formation. Sci. Rep. 2015, 5, 9856. [Google Scholar] [CrossRef]
- Marques, L.; Ceri, H.; Manfio, G.; Reid, D.; Olson, M. Characterization of biofilm formation by Xylella fastidiosa in vitro. Plant Dis. 2002, 86, 633–638. [Google Scholar] [CrossRef]
- Matsumoto, A.; Igo, M.M. Species-specific type II restriction-modification system of Xylella fastidiosa Temecula1. Appl. Environ. Microbiol. 2010, 76, 4092–4095. [Google Scholar] [CrossRef]
- O’ Leary, M.; Burbank, L.; Stenger, D.C. Distinct genetic lineages of Xylella fastidiosa carry conserved Type I Restriction-Modification systems with diverse specificity subunits. In Proceedings of the APS Annual Meeting “Plant Health 2020”, Online, 10–14 August 2020; p. 16338. [Google Scholar]
- Goosens, V.J.; Busch, A.; Georgiadou, M.; Castagnini, M.; Forest, K.T.; Waksman, G.; Pelicic, V. Reconstitution of a minimal machinery capable of assembling periplasmic type IV pili. Proc. Natl. Acad. Sci. USA 2017, 114, E4978–E4986. [Google Scholar] [CrossRef] [PubMed]
- De La Fuente, L.; Montanes, E.; Meng, Y.; Li, Y.; Burr, T.J.; Hoch, H.; Wu, M. Assessing adhesion forces of type I and type IV pili of Xylella fastidiosa bacteria by use of a microfluidic flow chamber. Appl. Environ. Microbiol. 2007, 73, 2690–2696. [Google Scholar] [CrossRef] [PubMed]
- Saponari, M.; Boscia, D.; Del Castillo, B.B.L.; Jacques, M.A.; Marco, E.; Poliakoff, F. Emerge of Xylella fastidiosa in Europe. Phytopathology 2018, 108, 1. [Google Scholar]
- Giampetruzzi, A.; Velasco-Amo, M.P.; Marco-Noales, E.; Montes-Borrego, M.; Roman-Ecija, M.; Navarro, I.; Monterde, A.; Barbé, S.; Almeida, R.P.P.; Saldarelli, P. Draft genome resources of two strains (“ESVL” and “IVIA5901”) of Xylella fastidiosa associated with almond leaf scorch disease in Alicante, Spain. Phytopathology 2019, 109, 219–221. [Google Scholar] [CrossRef] [PubMed]
- Landa, B.B.; Velasco-Amo, M.P.; Marco-Noales, E.; Olmo, D.; López, M.M.; Navarro, I.; Monterde, A.; Barbé, S.; Montes-Borrego, M.; Román-Écija, M. Draft genome sequence of Xylella fastidiosa subsp. fastidiosa strain IVIA5235, isolated from Prunus avium in Mallorca Island, Spain. Microbiol. Resour. Announc. 2018, 7, e01222-18. [Google Scholar] [CrossRef]
- Gomila, M.; Moralejo, E.; Busquets, A.; Segui, G.; Olmo, D.; Nieto, A.; Juan, A.; Lalucat, J. Draft genome resources of two strains of Xylella fastidiosa XYL1732/17 and XYL2055/17 isolated from Mallorca vineyards. Phytopathology 2019, 109, 222–224. [Google Scholar] [CrossRef]
- Della Coletta-Filho, H.; Takita, M.A.; de Souza, A.A.; Aguilar-Vildoso, C.I.; Machado, M.A. Differentiation of strains of Xylella fastidiosa by a variable number of tandem repeat analysis. Appl. Environ. Microbiol. 2001, 67, 4091–4095. [Google Scholar] [CrossRef][Green Version]
- Loenen, W.A.; Dryden, D.T.; Raleigh, E.A.; Wilson, G.G. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014, 42, 20–44. [Google Scholar] [CrossRef]
- Waldron, D.E.; Lindsay, J.A. Sau1: A novel lineage-specific type I restriction-modification system that blocks horizontal gene transfer into Staphylococcus aureus and between S. aureus isolates of different lineages. J. Bacteriol. 2006, 188, 5578–5585. [Google Scholar] [CrossRef]
- Chen, I.; Dubnau, D. DNA uptake during bacterial transformation. Nat. Rev. Microbiol. 2004, 2, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Seitz, P.; Blokesch, M. Cues and regulatory pathways involved in natural competence and transformation in pathogenic and environmental Gram-negative bacteria. FEMS Microbiol. Rev. 2013, 37, 336–363. [Google Scholar] [CrossRef]
- Muranaka, L.S.; Giorgiano, T.E.; Takita, M.A.; Forim, M.R.; Silva, L.F.; Coletta-Filho, H.D.; Machado, M.A.; de Souza, A.A. N-Acetylcysteine in agriculture, a novel use for an old molecule: Focus on controlling the plant–pathogen Xylella fastidiosa. PLoS ONE 2013, 8, e72937. [Google Scholar] [CrossRef]
- Nunes, L.R.; Rosato, Y.B.; Muto, N.H.; Yanai, G.M.; da Silva, V.S.; Leite, D.B.; Gonçalves, E.R.; de Souza, A.A.; Coletta-Filho, H.D.; Machado, M.A. Microarray analyses of Xylella fastidiosa provide evidence of coordinated transcription control of laterally transferred elements. Genome Res. 2003, 13, 570–578. [Google Scholar] [CrossRef]
- Leite, B.; Andersen, P.C.; Ishida, M.L. Colony aggregation and biofilm formation in xylem chemistry-based media for Xylella fastidiosa. FEMS Microbiol. Lett. 2004, 230, 283–290. [Google Scholar] [CrossRef][Green Version]
- Bi, J.; Dumenyo, C.; Hernandez-Martinez, R.; Cooksey, D.; Toscano, N. Effect of host plant xylem fluid on growth, aggregation, and attachment of Xylella fastidiosa. J. Chem. Ecol. 2007, 33, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Andersen, P.C.; Brodbeck, B.V.; Oden, S.; Shriner, A.; Leite, B. Influence of xylem fluid chemistry on planktonic growth, biofilm formation and aggregation of Xylella fastidiosa. FEMS Microbiol. Lett. 2007, 274, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Zaini, P.A.; De La Fuente, L.; Hoch, H.C.; Burr, T.J. Grapevine xylem sap enhances biofilm development by Xylella fastidiosa. FEMS Microbiol. Lett. 2009, 295, 129–134. [Google Scholar] [CrossRef]
- Cogan, N.; Donahue, M.; Whidden, M.; De La Fuente, L. Pattern formation exhibited by biofilm formation within microfluidic chambers. Biophys. J. 2013, 104, 1867–1874. [Google Scholar] [CrossRef]
- Giampetruzzi, A.; Morelli, M.; Saponari, M.; Loconsole, G.; Chiumenti, M.; Boscia, D.; Savino, V.N.; Martelli, G.P.; Saldarelli, P. Transcriptome profiling of two olive cultivars in response to infection by the CoDiRO strain of Xylella fastidiosa subsp. pauca. BMC Genom. 2016, 17, 1–18. [Google Scholar] [CrossRef]
- De Pascali, M.; Vergine, M.; Sabella, E.; Aprile, A.; Nutricati, E.; Nicolì, F.; Buja, I.; Negro, C.; Miceli, A.; Rampino, P. Molecular effects of Xylella fastidiosa and drought combined stress in olive trees. Plants 2019, 8, 437. [Google Scholar] [CrossRef] [PubMed]
- Sabella, E.; Luvisi, A.; Aprile, A.; Negro, C.; Vergine, M.; Nicolì, F.; Miceli, A.; De Bellis, L. Xylella fastidiosa induces differential expression of lignification related-genes and lignin accumulation in tolerant olive trees cv. Leccino. J. Plant Physiol. 2018, 220, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Saponari, M.; Altamura, G.; Abou Kubaa, R.; Montilon, V.; Saldarelli, P.; Specchia, F.; Palmisano, F.; Silletti, M.R.; Pollastro, P.; Zicca, S.; et al. Further acquisition on the response of a large number of olive cultivars to infections caused by Xylella fastidiosa subsp. pauca, ST53. In Proceedings of the 2nd European Conference on Xylella fastidiosa (How Research Can Support Solutions), Ajaccio, France, 29–30 October 2019. [Google Scholar]
- Giampetruzzi, A.; Baptista, P.; Morelli, M.; Cameirão, C.; Lino Neto, T.; Costa, D.; D’Attoma, G.; Abou Kubaa, R.; Altamura, G.; Saponari, M. Differences in the Endophytic Microbiome of Olive Cultivars Infected by Xylella fastidiosa across Seasons. Pathogens 2020, 9, 723. [Google Scholar] [CrossRef] [PubMed]



| Name | Sequence 5′-3′ a | Target | Amplicon Size (bp) |
|---|---|---|---|
| Up_F | CATTGACAGGAGACAGAAAGA | Upstream region of rpfF | 827 |
| Up_R | GCAACACCTTCTTCACGAGGCAGACTGTTGTTCTCCGTAATAGTAGTC | ||
| Down_F | GAGATTTTGAGACACAACGTGGCTTACTCAAAGCTGTGCTGATG | Downstream region of rpfF | 819 |
| Down_R | GAGTGCTGGTTGCTGATG | ||
| Kan_F | GTCTGCCTCGTGAAG | Kanamycin resistance gene | 1203 |
| Kan_R | AAGCCACGTTGTGT | ||
| RpfF_F | ATGTCCGCTGTACAT CCCATTCCT | rpfF gene | 803 |
| RpfF_R | GCGCTCCATAGTTCGGAGTGATTT | ||
| RpfFMut_F | GAAGCGGACATTAGCGTTAC | Recombination region in DDrpfF- mutant | 1697 (1370 b) |
| RpfFMut_R | GCTCGGTCATCTTGGTTTAATG | ||
| Int_F | TAGGGGGTCATCGTGACTTGC | Integration site in DDgfp mutant | 750 |
| Int_R | CCTCGAGCAAGACGTTTCCC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Attoma, G.; Morelli, M.; De La Fuente, L.; Cobine, P.A.; Saponari, M.; de Souza, A.A.; De Stradis, A.; Saldarelli, P. Phenotypic Characterization and Transformation Attempts Reveal Peculiar Traits of Xylella fastidiosa Subspecies pauca Strain De Donno. Microorganisms 2020, 8, 1832. https://doi.org/10.3390/microorganisms8111832
D’Attoma G, Morelli M, De La Fuente L, Cobine PA, Saponari M, de Souza AA, De Stradis A, Saldarelli P. Phenotypic Characterization and Transformation Attempts Reveal Peculiar Traits of Xylella fastidiosa Subspecies pauca Strain De Donno. Microorganisms. 2020; 8(11):1832. https://doi.org/10.3390/microorganisms8111832
Chicago/Turabian StyleD’Attoma, Giusy, Massimiliano Morelli, Leonardo De La Fuente, Paul A. Cobine, Maria Saponari, Alessandra Alves de Souza, Angelo De Stradis, and Pasquale Saldarelli. 2020. "Phenotypic Characterization and Transformation Attempts Reveal Peculiar Traits of Xylella fastidiosa Subspecies pauca Strain De Donno" Microorganisms 8, no. 11: 1832. https://doi.org/10.3390/microorganisms8111832
APA StyleD’Attoma, G., Morelli, M., De La Fuente, L., Cobine, P. A., Saponari, M., de Souza, A. A., De Stradis, A., & Saldarelli, P. (2020). Phenotypic Characterization and Transformation Attempts Reveal Peculiar Traits of Xylella fastidiosa Subspecies pauca Strain De Donno. Microorganisms, 8(11), 1832. https://doi.org/10.3390/microorganisms8111832








