Probiotic Effects on Multispecies Biofilm Composition, Architecture, and Caries Activity In Vitro
Abstract
1. Introduction
2. Materials and Methods
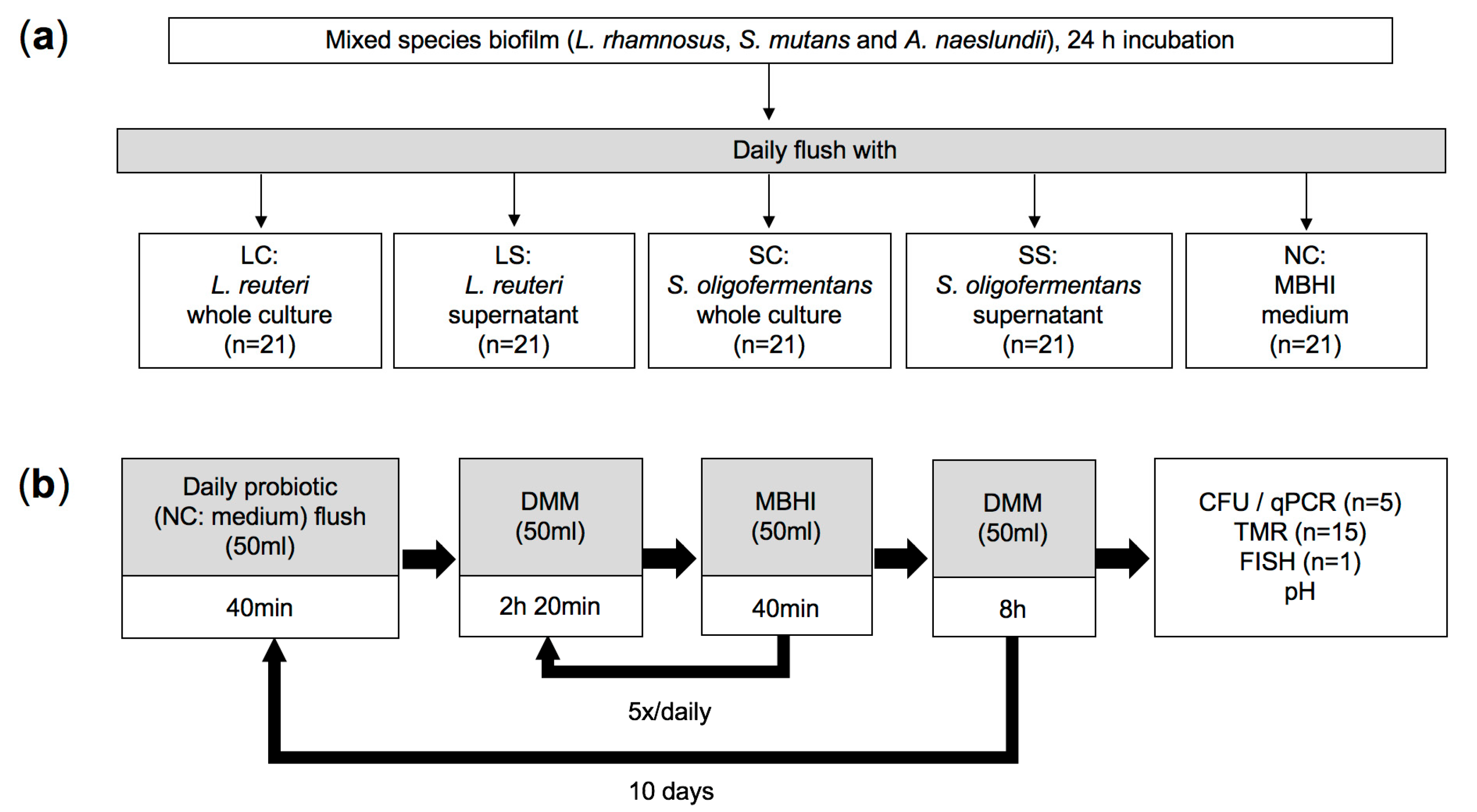
2.1. Sample Preparation
2.2. Bacterial Strains and Culture Conditions
2.3. Multispecies Biofilm Model
2.4. Quantification of Bacterial Numbers via CFU
2.5. DNA Isolation and Quantitative Polymerase Chain Reaction (qPCR)
2.6. Caries Activity Determination (pH Measurements and Transverse Microradiography)
2.7. Fluorescence In Situ Hybridization (FISH)
2.8. Statistical Analysis
3. Results
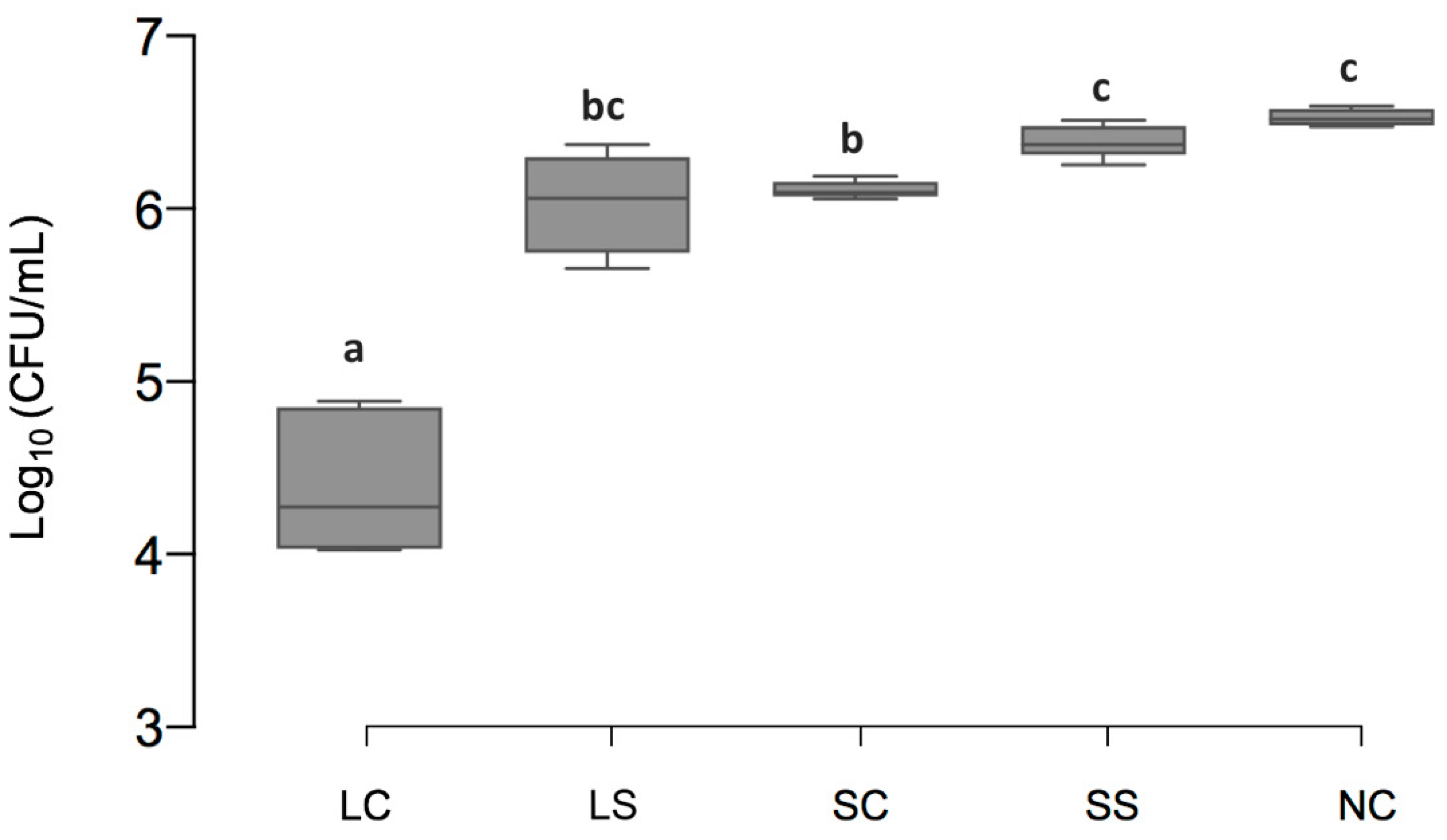
3.1. Viable Cell Counts of Biofilms
3.2. Cariogenic Bacterial Amounts
3.3. pH Value
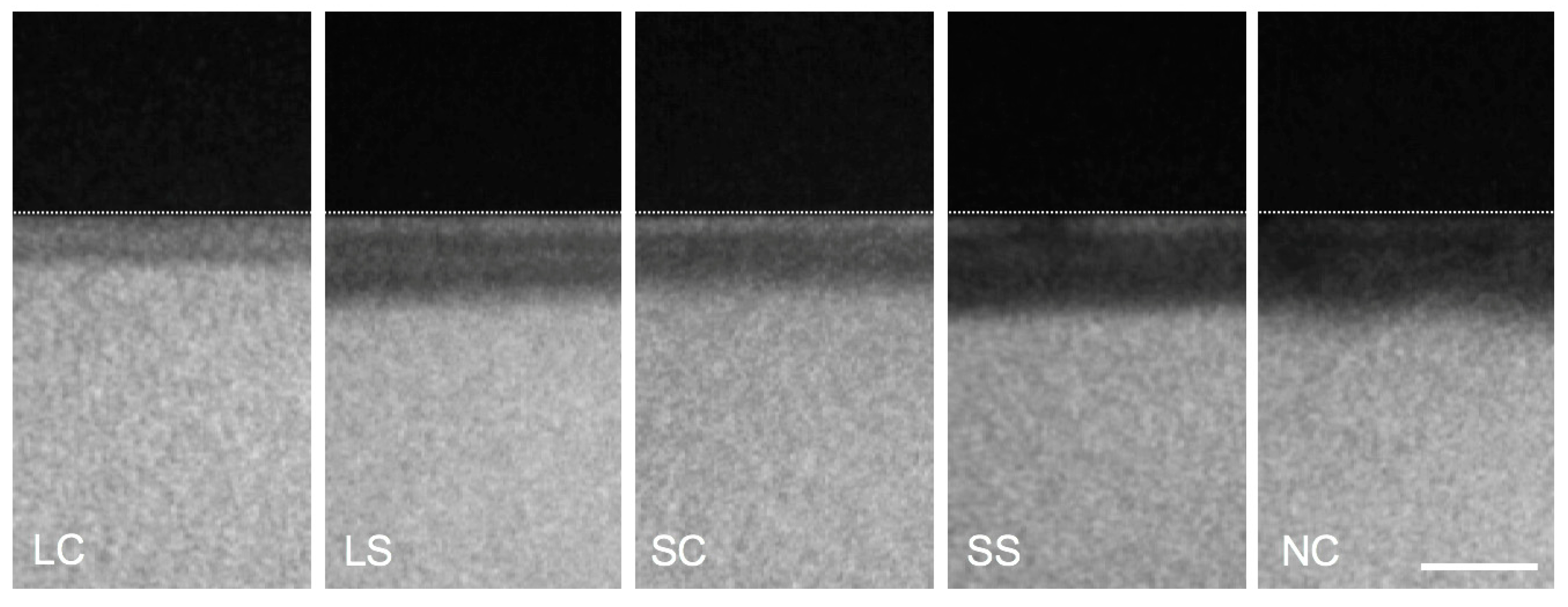
3.4. Mineral Loss of Enamel Lesions
3.5. Biofilm Structure and Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takahashi, N.; Nyvad, B. Caries ecology revisited: microbial dynamics and the caries process. Caries Res. 2008, 42, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Gareau, M.G.; Sherman, P.M.; Walker, W.A. Probiotics and the gut microbiota in intestinal health and disease. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 503. [Google Scholar] [CrossRef] [PubMed]
- Olle, B. Medicines from microbiota. Nat. Biotechnol. 2013, 31, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Gruner, D.; Paris, S.; Schwendicke, F. Probiotics for managing caries and periodontitis: systematic review and meta-analysis. J. Dent. 2016, 48, 16–25. [Google Scholar] [CrossRef]
- Bustamante, M.; Oomah, B.D.; Mosi-Roa, Y.; Rubilar, M.; Burgos-Díaz, C. Probiotics as an adjunct therapy for the treatment of halitosis, dental caries and periodontitis. Probiotics Antimicrob. Proteins 2020, 12, 325–334. [Google Scholar] [CrossRef]
- Zaura, E.; Twetman, S. Critical appraisal of oral pre- and probiotics for caries prevention and care. Caries Res. 2019, 53, 1–13. [Google Scholar] [CrossRef]
- Kang, M.S.; Oh, J.S.; Lee, H.C.; Lim, H.S.; Lee, S.W.; Yang, K.H.; Choi, N.K.; Kim, S.M. Inhibitory effect of Lactobacillus reuteri on periodontopathic and cariogenic bacteria. J. Microbiol. 2011, 49, 193–199. [Google Scholar] [CrossRef]
- Baca-Castanón, M.L.; De la Garza-Ramos, M.A.; Alcázar-Pizana, A.G.; Grondin, Y.; Coronado-Mendoza, A.; Sánchez-Najera, R.I.; Cárdenas-Estrada, E.; Medina-De la Garza, C.E.; Escamilla-García, E. Antimicrobial effect of Lactobacillus reuteri on cariogenic bacteria Streptococcus gordonii, Streptococcus mutans, and periodontal diseases Actinomyces naeslundii and Tannerella forsythia. Probiotics Antimicrob. Proteins 2015, 7, 1–8. [Google Scholar] [CrossRef]
- Krzyściak, W.; Kościelniak, D.; Papież, M.; Vyhouskaya, P.; Zagórska-Świeży, K.; Kołodziej, I.; Bystrowska, B.; Jurczak, A. Effect of a Lactobacillus salivarius probiotic on a double-species Streptococcus mutans and Candida albicans caries biofilm. Nutrients 2017, 9, 1242. [Google Scholar] [CrossRef]
- Bijle, M.N.A.; Yiu, C.K.Y.; Ekambaram, M. Can oral ADS activity or arginine levels be a caries risk indicator? A systematic review and meta-analysis. Clin. Oral Investig. 2018, 22, 583–596. [Google Scholar] [CrossRef]
- Reyes, E.; Martin, J.; Moncada, G.; Neira, M.; Palma, P.; Gordan, V.; Oyarzo, J.F.; Yevenes, I. Caries-free subjects have high levels of urease and arginine deiminase activity. J. Appl. Oral Sci. 2014, 22, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Meurman, J.H. Probiotics: do they have a role in oral medicine and dentistry? Eur. J. Oral Sci. 2005, 113, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Gutierrez, E.; Mayer, M.J.; Cotter, P.D.; Narbad, A. Gut microbiota as a source of novel antimicrobials. Gut Microbes 2019, 10, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Indira, M.; Venkateswarulu, T.C.; Peele, K.A.; Bobby, M.N.; Krupanidhi, S. Bioactive molecules of probiotic bacteria and their mechanism of action: a review. 3 Biotech. 2019, 9, 306. [Google Scholar] [CrossRef]
- La Fata, G.; Weber, P.; Mohajeri, M.H. Probiotics and the gut immune system: indirect regulation. Probiotics Antimicrob. Proteins 2018, 10, 11–21. [Google Scholar] [CrossRef]
- Talarico, T.L.; Casas, I.A.; Chung, T.C.; Dobrogosz, W.J. Production and isolation of reuterin, a growth inhibitor produced by Lactobacillus reuteri. Antimicrob. Agents Chemother. 1988, 32, 1854–1858. [Google Scholar] [CrossRef]
- Axelsson, L.T.; Chung, T.C.; Dobrogosz, W.J.; Lindgren, S.E. Production of a broad spectrum antimicrobial substance by Lactobacillus reuteri. Microb. Ecol. Health Dis. 1989, 2, 131–136. [Google Scholar] [CrossRef]
- Wasfi, R.; Abd El-Rahman, O.A.; Zafer, M.M.; Ashour, H.M. Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J. Cell Mol. Med. 2018, 22, 1972–1983. [Google Scholar] [CrossRef]
- Tong, H.; Chen, W.; Merritt, J.; Qi, F.; Shi, W.; Dong, X. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 2007, 63, 872–880. [Google Scholar] [CrossRef]
- Wong, L.; Sissons, C.H. Human dental plaque microcosm biofilms: effect of nutrient variation on calcium phosphate deposition and growth. Arch. Oral Biol. 2007, 52, 280–289. [Google Scholar] [CrossRef]
- Schwendicke, F.; Dörfer, C.; Kneist, S.; Meyer-Lueckel, H.; Paris, S. Cariogenic effects of probiotic Lactobacillus rhamnosus GG in a dental biofilm model. Caries Res. 2014, 48, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Millhouse, E.; Shaw, T.; Lappin, D.; Rajendran, R.; Bagg, J.; Lin, H.; Ramage, G. Evaluating Streptococcus mutans strain dependent characteristics in a polymicrobial biofilm community. Front. Microbiol. 2018, 9, 1498. [Google Scholar] [CrossRef] [PubMed]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J. Primer3 - new capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Paster, B.J.; Bartoszyk, I.M.; Dewhirst, F.E. Identification of oral streptococci using PCR-based, reverse-capture, checkerboard hybridization. Methods Cell Sci. 1998, 20, 223–231. [Google Scholar] [CrossRef]
- Gmür, R.; Lüthi-Schaller, H. A combined immunofluorescence and fluorescent in situ hybridization assay for single cell analyses of dental plaque microorganisms. J. Microbiol. Methods 2007, 69, 402–405. [Google Scholar] [CrossRef]
- Amann, R.I.; Binder, B.J.; Olson, R.J.; Chisholm, S.W.; Devereux, R.; Stahl, D.A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 1990, 56, 1919–1925. [Google Scholar] [CrossRef]
- Dige, I.; Raarup, M.K.; Nyengaard, J.R.; Kilian, M.; Nyvad, B. Actinomyces naeslundii in initial dental biofilm formation. Microbiology 2009, 155, 2116–2126. [Google Scholar] [CrossRef]
- Nascimento, M.M.; Alvarez, A.J.; Huang, X.; Browngardt, C.; Jenkins, R.; Sinhoreti, M.C.; Ribeiro, A.P.D.; Dilbone, D.A.; Richards, V.P.; Burne, R.A.; et al. Metabolic profile of supragingival plaque exposed to arginine and fluoride. J. Dent. Res. 2019, 98, 1245–1252. [Google Scholar] [CrossRef]
- Nascimento, M.M.; Gordan, V.V.; Garvan, C.W.; Browngardt, C.M.; Burne, R.A. Correlations of oral bacterial arginine and urea catabolism with caries experience. Oral Microbiol. Immunol. 2009, 24, 89–95. [Google Scholar] [CrossRef]
- Huang, X.; Schulte, R.M.; Burne, R.A.; Nascimento, M.M. Characterization of the arginolytic microflora provides insights into pH homeostasis in human oral biofilms. Caries Res. 2015, 49, 165–176. [Google Scholar] [CrossRef]
- Takahashi, N.; Nyvad, B. The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 2011, 90, 294–303. [Google Scholar] [CrossRef] [PubMed]
- Holling, C.S. Resilience and stability of ecological systems. Annu. Rev. Ecol. Syst. 1973, 4, 1–23. [Google Scholar] [CrossRef]
- Folke, C.; Carpenter, S.; Walker, B.; Scheffer, M.; Elmqvist, T.; Gunderson, L.; Holling, C.S. Regime shifts, resilience, and biodiversity in ecosystem management. Annu. Rev. Ecol. Evol. Syst. 2004, 35, 557–581. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef]
- Rosier, B.T.; Marsh, P.D.; Mira, A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res. 2018, 97, 371–380. [Google Scholar] [CrossRef]
- Yu, H.; Ganas, P.; Schwendicke, F. Environment-specific probiotic supernatants modify the metabolic activity and survival of Streptococcus mutans in vitro. Front. Microbiol. 2020, 11, 1447. [Google Scholar] [CrossRef]
- Shu, M.; Wong, L.; Miller, J.H.; Sissons, C.H. Development of multi-species consortia biofilms of oral bacteria as an enamel and root caries model system. Arch. Oral Biol. 2000, 45, 27–40. [Google Scholar] [CrossRef]
- Jiang, Q.; Stamatova, I.; Kainulainen, V.; Korpela, R.; Meurman, J.H. Interactions between Lactobacillus rhamnosus GG and oral micro-organisms in an in vitro biofilm model. BMC Microbiol. 2016, 16, 149. [Google Scholar] [CrossRef]
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563. [Google Scholar] [CrossRef]
- Lamont, R.J.; Koo, H.; Hajishengallis, G. The oral microbiota: dynamic communities and host interactions. Nat. Rev. Microbiol. 2018, 16, 745–759. [Google Scholar] [CrossRef]
- Takahashi, N. Oral microbiome metabolism: from “who are they?” to “what are they doing?”. J. Dent. Res. 2015, 94, 1628–1637. [Google Scholar] [CrossRef] [PubMed]
- Jones, S.E.; Versalovic, J. Probiotic Lactobacillus reuteri biofilms produce antimicrobial and anti-inflammatory factors. BMC Microbiol. 2009, 9, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Hou, Y.; Wu, G. Nutritionally nonessential amino acids: a misnomer in nutritional sciences. Adv. Nutr. 2017, 8, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.S.; Biswas, I. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl. Environ. Microbiol. 2011, 77, 2428–2434. [Google Scholar] [CrossRef]
- Kreth, J.; Zhang, Y.; Herzberg, M.C. Streptococcal antagonism in oral biofilms: Streptococcus sanguinis and Streptococcus gordonii interference with Streptococcus mutans. J. Bacteriol. 2008, 190, 4632–4640. [Google Scholar] [CrossRef]
- Klopper, K.B.; Deane, S.M.; Dicks, L.M. Aciduric strains of Lactobacillus reuteri and Lactobacillus rhamnosus, isolated from human feces, have strong adhesion and aggregation properties. Probiotics Antimicrob. Proteins 2018, 10, 89–97. [Google Scholar] [CrossRef]
- Russel, J.; Røder, H.L.; Madsen, J.S.; Burmølle, M.; Sørensen, S.J. Antagonism correlates with metabolic similarity in diverse bacteria. Proc. Natl. Acad. Sci. USA 2017, 114, 10684–10688. [Google Scholar] [CrossRef]
- Xie, Q.; Li, J.; Zhou, X. Anticaries effect of compounds extracted from Galla Chinensis in a multispecies biofilm model. Oral Microbiol. Immunol. 2008, 23, 459–465. [Google Scholar] [CrossRef]
- Yip, H.; Guo, J.; Wong, W. Protection offered by root-surface restorative materials against biofilm challenge. J. Dent. Res. 2007, 86, 431–435. [Google Scholar] [CrossRef]
- Bosch, J.J.T.; Angmar-Mansson, B. A review of quantitative methods for studies of mineral content of intra-oral incipient carious lesions. J. Dent. Res. 1991, 70, 2–14. [Google Scholar] [CrossRef]


| Group | L. rhamnosus | S. mutans | A. naeslundii |
|---|---|---|---|
| LC | 0.55 ± 0.10 a | 0.37 ± 0.11 a | 0.19 ± 0.04 a |
| LS | 4.03 ± 1.87 ab | 3.65 ± 1.86 abc | 0.36 ± 0.20 ab |
| SC | 2.23 ± 0.86 ab | 0.22 ± 0.14 ab | 0.09 ± 0.04 b |
| SS | 3.84 ± 1.61 ab | 2.76 ± 0.57 c | 0.43 ± 0.07 a |
| NC | 3.13 ± 0.87 b | 3.69 ± 1.38 c | 0.45 ± 0.10 a |
| Group | 16 h | 24 h |
|---|---|---|
| LC | 5.00 ± 0.02 a | 3.90 ± 0.03 a |
| LS | 3.90 ± 0.02 b | 3.53 ± 0.01 b |
| SC | 3.83 ± 0.01 cd | 3.75 ± 0.00 a |
| SS | 3.73 ± 0.03 d | 3.44 ± 0.01 c |
| NC | 3.94 ± 0.03 bc | 3.70 ± 0.01 d |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Z.; Schlafer, S.; Göstemeyer, G.; Schwendicke, F. Probiotic Effects on Multispecies Biofilm Composition, Architecture, and Caries Activity In Vitro. Microorganisms 2020, 8, 1272. https://doi.org/10.3390/microorganisms8091272
Chen Z, Schlafer S, Göstemeyer G, Schwendicke F. Probiotic Effects on Multispecies Biofilm Composition, Architecture, and Caries Activity In Vitro. Microorganisms. 2020; 8(9):1272. https://doi.org/10.3390/microorganisms8091272
Chicago/Turabian StyleChen, Zhihui, Sebastian Schlafer, Gerd Göstemeyer, and Falk Schwendicke. 2020. "Probiotic Effects on Multispecies Biofilm Composition, Architecture, and Caries Activity In Vitro" Microorganisms 8, no. 9: 1272. https://doi.org/10.3390/microorganisms8091272
APA StyleChen, Z., Schlafer, S., Göstemeyer, G., & Schwendicke, F. (2020). Probiotic Effects on Multispecies Biofilm Composition, Architecture, and Caries Activity In Vitro. Microorganisms, 8(9), 1272. https://doi.org/10.3390/microorganisms8091272






