Diversity, Antibiotic Resistance, and Biofilm-Forming Ability of Enterobacteria Isolated from Red Meat and Poultry Preparations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Microbiological Analysis
2.3. Species Identification
2.4. Antimicrobial Susceptibility Testing
2.5. Biofilm Determination
2.6. Statistical Analysis
3. Results and Discussion
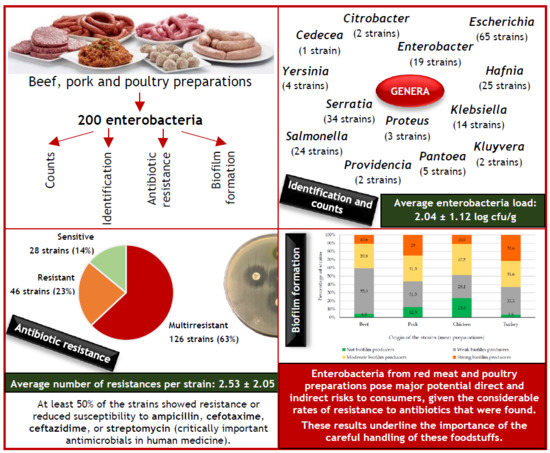
3.1. Enterobacteria Load and Identification
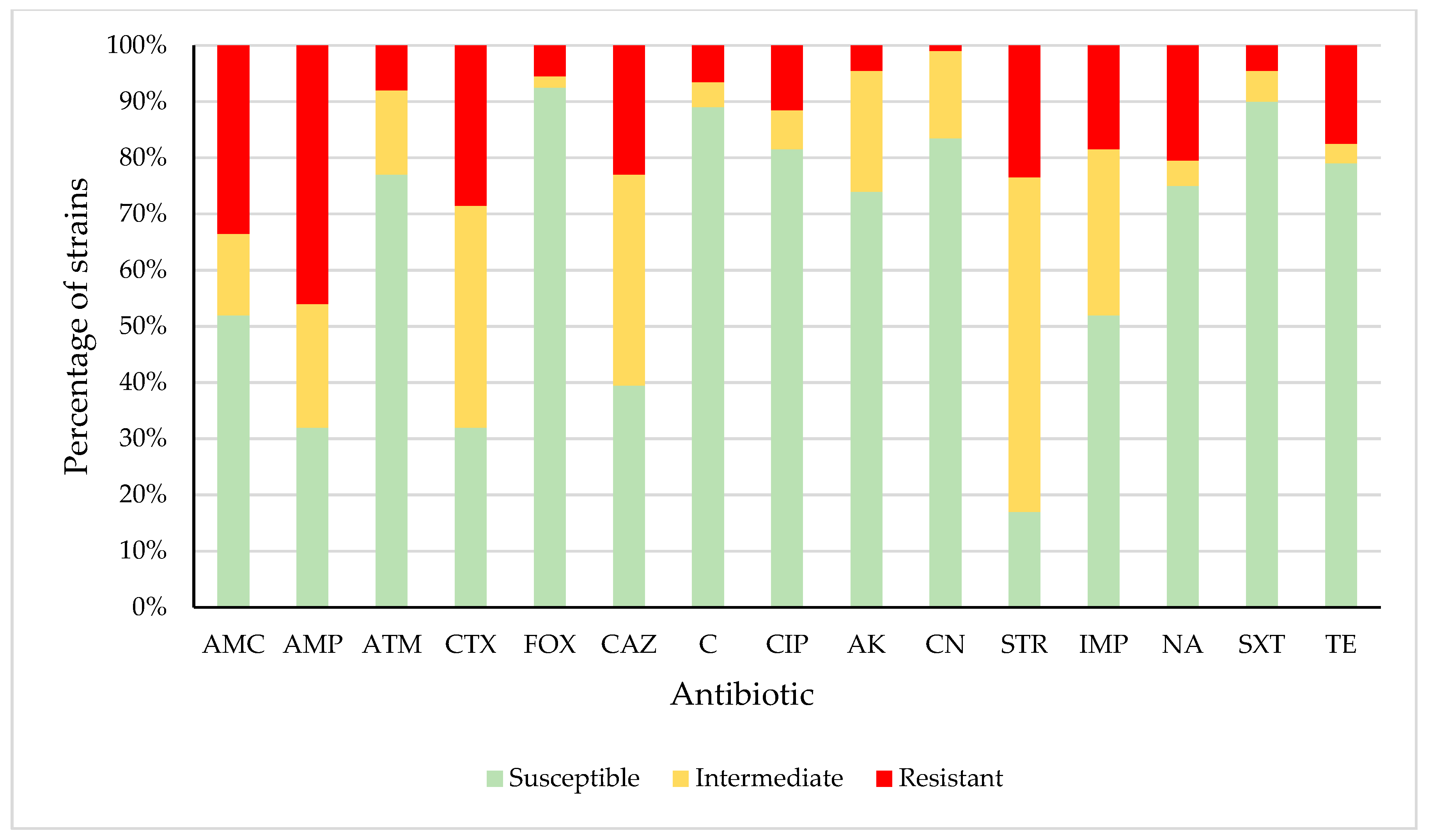
3.2. Antimicrobial Susceptibility
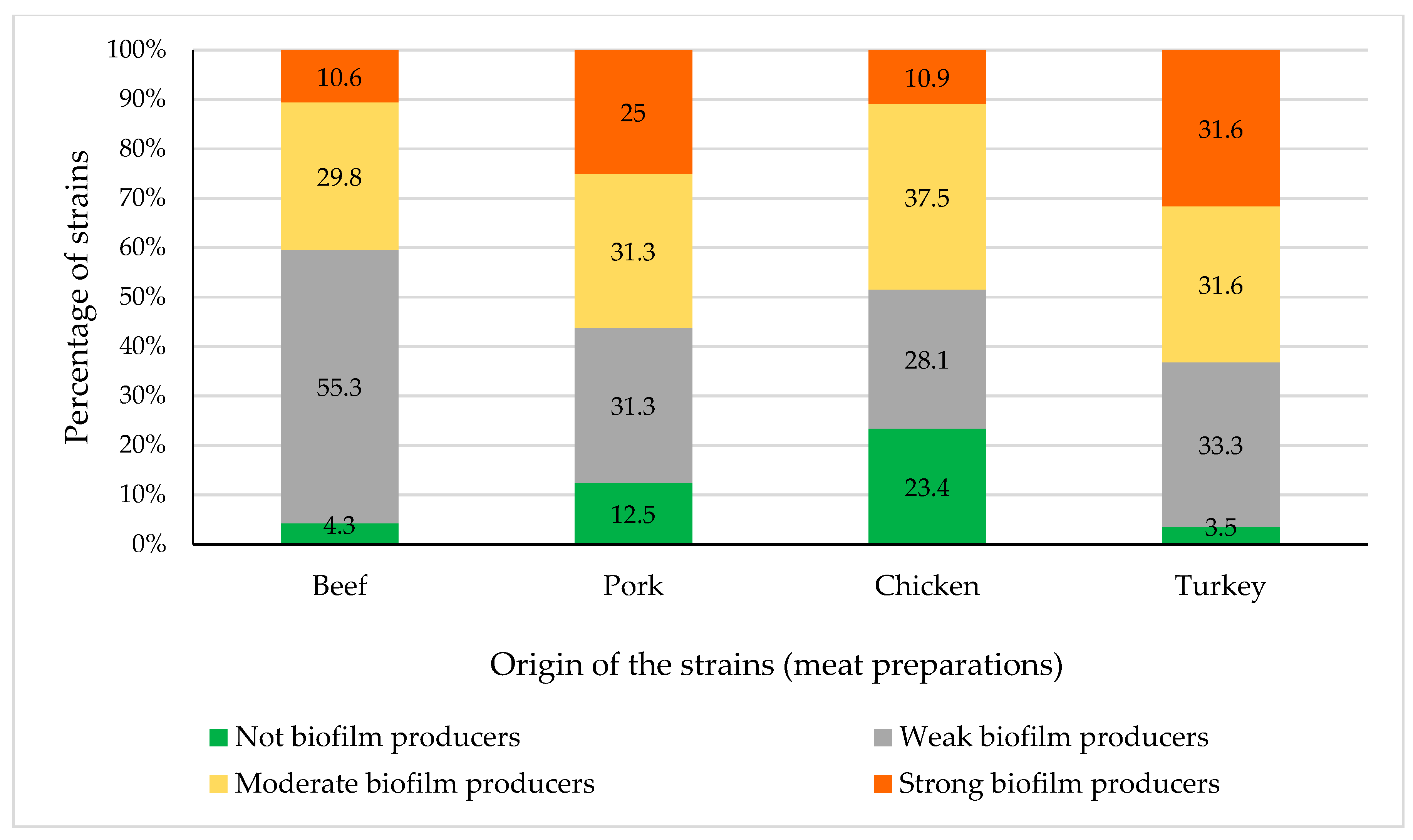
3.3. Biofilm Production
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- FAOSTAT. Available online: http://www.fao.org/faostat/es/#data (accessed on 30 June 2020).
- European Parliament. Regulation (EC) No 853/2004 of the European Parliament and of the Council of 29 April 2004 laying down specific hygiene rules for food of animal origin. Off. J. Eur. Union 2004, L139, 55–205. [Google Scholar]
- Del Río, E.; Panizo-Morán, M.; Prieto, M.; Alonso-Calleja, C.; Capita, R. Effect of various chemical decontamination treatments on natural microflora and sensory characteristics of poultry. Int. J. Food Microbiol. 2007, 115, 268–280. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Astorga, M.; Capita, R.; Alonso-Calleja, C.; Moreno, B.; García-Fernández, M.C. Microbiological quality of retail chicken by-products in Spain. Meat Sci. 2002, 62, 45–50. [Google Scholar] [CrossRef]
- Alonso-Calleja, C.; Martínez-Fernández, B.; Prieto, M.; Capita, R. Microbiological quality of vacuum-packed retail ostrich meat in Spain. Food Microbiol. 2004, 21, 241–246. [Google Scholar] [CrossRef]
- Rouger, A.; Tresse, O.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef]
- Capita, R.; Alonso-Calleja, C. Antibiotic-resistant bacteria: A challenge for the food industry. Crit. Rev. Food Sci. Nutr. 2013, 53, 11–48. [Google Scholar] [CrossRef]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Available online: https://amr-review.org/sites/default/files/160518_Final%20paper_with%20cover.pdf (accessed on 2 July 2020).
- OECD. Antimicrobial Resistance. Tackling the Burden in the European Union. Organization for Economic Coordination and Development. Available online: https://www.oecd.org/health/health-systems/AMR-Tackling-the-Burden-in-the-EU-OECD-ECDC-Briefing-Note-2019.pdf (accessed on 30 June 2020).
- SCENIHR. Assessment of the Antibiotic Resistance Effects of Biocides. Scientific Committee on Emerging and Newly Identified Health Risks. 19 January 2009. Available online: http://ec.europa.eu/health/ph_risk/committees/04_scenihr/docs/scenihr_o_021.pdf (accessed on 1 July 2020).
- Díez-García, M.; Capita, R.; Alonso-Calleja, C. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiol. 2012, 31, 173–180. [Google Scholar] [CrossRef]
- González-Machado, C.; Capita, R.; Riesco-Peláez, F.; Alonso-Calleja, C. Visualization and quantification of the cellular and extracellular components of Salmonella Agona biofilms at different stages of development. PLoS ONE 2018, 13, e0200011. [Google Scholar] [CrossRef]
- Rodríguez-Campos, D.; Rodríguez-Melcón, C.; Alonso-Calleja, C.; Capita, R. Persistent Listeria monocytogenes isolates from a poultry-processing facility form more biofilm but do not have a greater resistance to disinfectants than sporadic strains. Pathogens 2019, 8, 250. [Google Scholar] [CrossRef] [Green Version]
- Capita, R.; Riesco-Peláez, F.; Alonso-Hernando, A.; Alonso-Calleja, C. Exposure of Escherichia coli ATCC 12806 to sublethal concentrations of food-grade biocides influences its ability to form biofilm, resistance to antimicrobials, and ultrastructure. Appl. Environ. Microbiol. 2014, 80, 1268–1280. [Google Scholar] [CrossRef] [Green Version]
- Piercey, M.J.; Hingston, P.A.; Hansen, L.T. Genes involved in Listeria monocytogenes biofilm formation at a simulated food processing plant temperature of 15 °C. Int. J. Food Microbiol. 2016, 223, 63–74. [Google Scholar] [CrossRef]
- CLSI. M100 Performance Standars for Antimicrobial Susceptibility Testing, 29th ed.; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2019. [Google Scholar]
- Andritsos, N.D.; Mataragas, M.; Mavrou, E.; Stamatiou, A.; Drosinos, E.H. The microbiological condition of minced pork prepared at retail stores in Athens, Greece. Meat Sci. 2012, 91, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Scheutz, F.; Teel, L.D.; Beutin, L.; Piérard, D.; Buvens, G.; Karch, H.; Mellmann, A.; Caprioli, A.; Tozzoli, R.; Morabito, S.; et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J. Clin. Microbiol. 2012, 50, 2951–2963. [Google Scholar] [CrossRef] [Green Version]
- Wong, T.; Whyte, R.J.; Cornelius, A.J.; Hudson, J.A. Enumeration of Campylobacter and Salmonella on chicken packs. Br. Food J. 2004, 106, 651–662. [Google Scholar] [CrossRef]
- IFST. Development and use of microbiological criteria for foods. Food Sci. Technol. Today 1997, 11, 137–176. [Google Scholar]
- ICMSF. Microorganisms in Foods 8. Use of Data for Assessing Process Control and Product Acceptance; Springer: New York, NY, USA, 2011. [Google Scholar]
- Uyttendaele, M.; Jacxsens, L.; De Loy-Hendrickx, A.; Devlieghere, F.; Debevere, J. Microbiologische Richtwaarden & Wettelijke Microbiologische Criteria. Department of Food Safety and Food Quality, Ghent University. Available online: https://biblio.ugent.be/publication/1169787 (accessed on 1 July 2020).
- Cordero, J.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Microbial loads and antibiotic resistance patterns of Escherichia coli and Enterococcus faecalis isolates from the meat of wild and domestic pigeons. Foods 2019, 8, 536. [Google Scholar] [CrossRef] [Green Version]
- González-Gutiérrez, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Microbial load and antibiotic resistance in raw beef preparations from northwest Spain. Food Sci. Nutr. 2020, 8, 777–785. [Google Scholar] [CrossRef]
- Luning, P.A.; Jacxsens, L.; Rovira, J.; Osés, S.M.; Uyttendaele, M.; Marcelis, W.J. A concurrent diagnosis of microbiological food safety output and food safety management system performance: Cases from meat processing industries. Food Control 2011, 22, 555–565. [Google Scholar] [CrossRef]
- Doménech, E.; Jimenez-Belenguer, A.; Pérez, R.; Ferrús, M.A.; Escriche, I. Risk characterization of antimicrobial resistance of Salmonella in meat products. Food Control 2015, 57, 18–23. [Google Scholar] [CrossRef] [Green Version]
- Doménech, E.; Jimenez-Belenguer, A.; Amoros, J.A.; Ferrus, M.A.; Escriche, I. Prevalence and antimicrobial resistance of Listeria monocytogenes and Salmonella strains isolated in ready-to-eat foods in Eastern Spain. Food Control 2015, 47, 120–125. [Google Scholar] [CrossRef]
- Álvarez-Fernández, E.; Alonso-Calleja, C.; García-Fernández, C.; Capita, R. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from poultry in Spain: Comparison between 1993 and 2006. Int. J. Food Microbiol. 2012, 153, 281–287. [Google Scholar] [CrossRef] [PubMed]
- Zwe, Y.H.; Tang, V.C.Y.; Aung, K.T.; Gutiérrez, R.A.; Ng, L.C.; Yuk, H.-G. Prevalence, sequence types, antibiotic resistance and, gyrA mutations of Salmonella isolated from retail fresh chicken meat in Singapore. Food Control 2018, 90, 233–240. [Google Scholar] [CrossRef]
- Yildirim, Y.; Gonulalan, Z.; Pamuk, S.; Ertas, N. Incidence and antibiotic resistance of Salmonella spp. on raw chicken carcasses. Food Res. Int. 2011, 44, 725–728. [Google Scholar] [CrossRef]
- Capita, R.; Álvarez-Fernández, E.; Fernández-Buelta, E.; Manteca, J.; Alonso-Calleja, C. Decontamination treatments can increase the prevalence of resistance to antibiotics of Escherichia coli naturally present on poultry. Food Microbiol. 2013, 34, 112–117. [Google Scholar] [CrossRef]
- Gwida, M.; Hotzel, H.; Geue, L.; Tomaso, H. Occurrence of Enterobacteriaceae in raw meat and in human samples from Egyptian retail sellers. Int. Sch. Res. Not. 2014, 565671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stiles, M.E.; Ng, L.K. Enterobacteriaceae associated with meats and meat handling. Appl. Environ. Microbiol. 1981, 41, 867–872. [Google Scholar] [CrossRef] [Green Version]
- Farmer, J.J., III; Boatwright, K.D.; Janda, J.M. Enterobacteriaceae: Introduction and identification. In Manual of Clinical Microbiology, 9th ed.; Murray, P.R., Baron, E.J., Jorgensen, J., Pfaller, M.A., Landry, M.L., Eds.; ASM Press Inc.: Washington, DC, USA, 2007; pp. 649–669. [Google Scholar]
- Osman, E.A.; El-Amin, N.; Adrees, E.A.A.; Al-Hassan, L.; Mukhtar, M. Comparing conventional, biochemical and genotypic methods for accurate identification of Klebsiella pneumoniae in Sudan. Access Microbiol. 2020, 2. [Google Scholar] [CrossRef]
- Cosgrove, S.E. The relationship between antimicrobial resistance and patient outcomes: Mortality, length of hospital stay, and health care costs. Clin. Infect. Dis. 2006, 42, S82–S89. [Google Scholar] [CrossRef] [Green Version]
- Capita, R.; Cordero, J.; González-Molina, D.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C. Phylogenetic diversity, antimicrobial susceptibility and virulence characteristics of Escherichia coli isolates from pigeon meat. Antibiotics 2019, 8, 259. [Google Scholar] [CrossRef] [Green Version]
- Logue, C.M.; Sherwood, J.S.; Olah, P.A.; Elijah, L.M.; Dockter, M.R. The incidence of antimicrobial-resistant Salmonella spp. on freshly processed poultry from US Midwestern processing plants. J. Appl. Microbiol. 2003, 94, 16–24. [Google Scholar] [CrossRef]
- Raphael, E.; Riley, L.W. Infections caused by antimicrobial drug-resistant saprophytic Gram-negative bacteria in the environment. Front. Med. 2017, 4, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alonso-Hernando, A.; Prieto, M.; García-Fernández, C.; Alonso-Calleja, C.; Capita, R. Increase over time in the prevalence of multiple antibiotic resistance among isolates of Listeria monocytogenes from poultry in Spain. Food Control 2012, 23, 37–41. [Google Scholar] [CrossRef]
- Álvarez-Fernández, E.; Cancelo, A.; Díaz-Vega, C.; Capita, R.; Alonso-Calleja, C. Antimicrobial resistance in E. coli isolates from conventionally and organically reared poultry: A comparison of agar disc diffusion and Sensi Test Gram-negative methods. Food Control 2013, 30, 227–234. [Google Scholar] [CrossRef]
- Buzón-Durán, L.; Capita, R.; Alonso-Calleja, C. Microbial loads and antibiotic resistance patterns of Staphylococcus aureus in different types of raw poultry-based meat preparations. Poult. Sci. 2017, 96, 4046–4052. [Google Scholar] [CrossRef] [PubMed]
- Guerra, B.; Junker, E.; Schroeter, A.; Malorny, B.; Lehmann, S.; Helmuth, R. Phenotypic and genotypic characterization of antimicrobial resistance in German Escherichia coli isolates from cattle, swine and poultry. J. Antimicrob. Chemother. 2003, 52, 489–492. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Cordero, J.; Molina-González, D.; Poeta, P.; Igrejas, G.; Alonso-Calleja, C.; Capita, R. Antimicrobial resistance and virulence genes in enterococci from wild game meat in Spain. Food Microbiol. 2016, 53, 156–164. [Google Scholar] [CrossRef]
- Guerrero-Ramos, E.; Molina-González, D.; Blanco-Morán, S.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C.; Capita, R. Prevalence, antimicrobial resistance, and genotypic characterization of vancomycin-resistant enterococci in meat preparations. J. Food Prot. 2016, 79, 748–756. [Google Scholar] [CrossRef]
- Cameron, A.; McAllister, A.A. Antimicrobial usage and resistance in beef production. J. Anim. Sci. Biotechnol. 2016, 7, 68. [Google Scholar] [CrossRef] [Green Version]
- De Briyne, N.; Atkinson, J.; Pokludová, L.; Borriello, S.P. Antibiotics used most commonly to treat animals in Europe. Vet. Rec. 2014, 175, 325. [Google Scholar] [CrossRef] [Green Version]
- Roth, N.; Käsbohrer, A.; Mayrhofer, S.; Zitz, U.; Hofacre, C.; Doming, K.J. The application of antibiotics in broiler production and the resulting antibiotic resistance in Escherichia coli: A global overview. Poult. Sci. 2019, 98, 1791–1804. [Google Scholar] [CrossRef]
- WHO. Critically Important Antimicrobials for Human Medicine, 6th ed.; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- OIE. OIE List of Antimicrobial Agents of Veterinary Importance; World Organization for Animal Health: Paris, France, 2018. [Google Scholar]
- Nirwati, H.; Sinanjung, K.; Fahrunissa, F.; Wijaya, F.; Napitupulu, S.; Hati, V.P.; Hakim, M.S.; Meliala, A.; Aman, A.T.; Nuryastuti, T. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC Proc. 2019, 13, 20. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.; Devi, K.M.; Damrolien, S.; Devi, K.S.; Krossnunpuii, K.T. Biofilm production and its correlation with antibiotic resistance pattern among clinical isolates of Pseudomonas aeruginosa in a tertiary care hospital in north-East India. Int. J. Adv. Med. 2018, 5, 964–968. [Google Scholar] [CrossRef] [Green Version]
- Cepas, V.; López, Y.; Muñoz, E.; Rolo, D.; Ardanuy, C.; Martí, S.; Xercavins, M.; Horcajada, J.P.; Bosch, J.; Soto, S.M. Relationship between biofilm formation and antimicrobial resistance in Gram-Negative bacteria. Microb. Drug Resist. 2019, 25, 72–79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kragh, K.N.; Alhede, M.; Rybtke, M.; Stavnsberg, C.; Jensen, P.Ø.; Tolker-Nielsen, T.; Whiteley, M.; Bjarnsholta, T. The inoculation method could impact the outcome of microbiological experiments. Appl. Environ. Microbiol. 2018, 84, e02264-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kragh, K.N.; Alhede, M.; Kvich, L.; Bjarnsholt, T. Into the well—A close look at the complex structures of a microtiter biofilm and the crystal violet assay. Biofilm 2019, 1, 100006. [Google Scholar] [CrossRef]
| Enterobacterial Species | Origin (Number of Strains) | ||||
|---|---|---|---|---|---|
| Beef (47) | Pork (32) | Chicken (64) | Turkey (57) | Average Number of Antibiotic Resistances | |
| Cedecea lapagei | 1 | 2.00 ± 0.00 | |||
| Citrobacter braakii | 1 | 2.00 ± 0.00 | |||
| Citrobacter freundii | 1 | 1.00 ± 0.00 | |||
| Enterobacter aerogenes | 1 | 2.00 ± 0.00 | |||
| Enterobacter amnigenus | 1 | 3 | 4.75 ± 0.50 | ||
| Enterobacter asburiae | 4 | 1 | 4 | 1.11 ± 1.36 | |
| Enterobacter cloacae | 1 | 4 | 1.60 ± 0.55 | ||
| Escherichia coli | 1 | 25 | 29 | 3.95 ± 2.46 | |
| Escherichia vulneris | 5 | 5 | 1.10 ± 0.32 | ||
| Hafnia alvei | 10 | 8 | 3 | 4 | 2.84 ± 1.07 |
| Klebsiella oxytoca | 4 | 5 | 2.11 ± 1.05 | ||
| Klebsiella pneumoniae | 1 | 2 | 2 | 2.00 ± 1.87 | |
| Kluyvera spp. | 1 | 1 | 3.00 ± 1.41 | ||
| Pantoea spp. | 1 | 2 | 2 | 1.20 ± 1.64 | |
| Proteus vulgaris | 3 | 1.67 ± 0.58 | |||
| Providencia alcalifaciens | 1 | 1 | 1.00 ± 1.41 | ||
| Salmonella enterica | 4 | 10 | 5 | 5 | 2.17 ± 1.66 |
| Serratia liquefaciens | 13 | 3 | 6 | 10 | 1.59 ± 1.64 |
| Serratia marcescens | 1 | 2.00 ± 0.00 | |||
| Serratia plymuthica | 1 | 0.00 ± 0.00 | |||
| Yersinia spp. | 3 | 1 | 2.25 ± 3.86 | ||
| M | R | S | ||||
|---|---|---|---|---|---|---|
| Animal Species | No | % | No | % | No | % |
| Beef (n = 47) | 33 | 70% | 10 | 21% | 4 | 9% |
| Pork (n = 32) | 21 | 66% | 6 | 19% | 5 | 16% |
| Chicken (n = 64) | 35 | 55% | 18 | 28% | 11 | 17% |
| Turkey (n = 57) | 37 | 65% | 12 | 21% | 8 | 14% |
| Total (n = 200) | 126 | 63% | 46 | 23% | 28 | 14% |
| Number of Strains (%) | ||||
|---|---|---|---|---|
| Bacterial Species | Not Biofilm Producer | WeakBiofilm Producer | ModerateBiofilm Producer | StrongBiofilm Producer |
| Cedecea lapagei | 1 (100%) | |||
| Citrobacter braakii | 1 (100%) | |||
| Citrobacter freundii | 1 (100%) | |||
| Enterobacter aerogenes | 1 (100%) | |||
| Enterobacter amnigenus | 1 (25%) | 3 (75%) | ||
| Enterobacter asburiae | 5 (56%) | 3 (33%) | 1 (11%) | |
| Enterobacter cloacae | 2 (40%) | 3 (60%) | ||
| Escherichia coli | 5 (9%) | 23 (42%) | 14 (25%) | 13 (24%) |
| Escherichia vulneris | 1 (10%) | 5 (50%) | 2 (20%) | 2 (20%) |
| Hafnia alvei | 9 (36%) | 12 (48%) | 4 (16%) | |
| Klebsiella oxytoca | 3 (33%) | 4 (44%) | 2 (22%) | |
| Klebsiella pneumoniae | 1 (20%) | 4 (80%) | ||
| Kluyvera spp. | 2 (100%) | |||
| Pantoea spp. | 4 (80%) | 1 (20%) | ||
| Proteus vulgaris | 3 (100%) | |||
| Providencia alcalifaciens | 1 (50%) | 1 (50%) | ||
| Salmonella enterica | 5 (21%) | 5 (21%) | 12 (50%) | 2 (8%) |
| Serratia liquefaciens | 2 (6%) | 16 (50%) | 5 (16%) | 9 (28%) |
| Serratia marcescens | 1 (100%) | |||
| Serratia plymuthica | 1 (100%) | |||
| Yersinia spp. | 1 (25%) | 3 (75%) | ||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Capita, R.; Castaño-Arriba, A.; Rodríguez-Melcón, C.; Igrejas, G.; Poeta, P.; Alonso-Calleja, C. Diversity, Antibiotic Resistance, and Biofilm-Forming Ability of Enterobacteria Isolated from Red Meat and Poultry Preparations. Microorganisms 2020, 8, 1226. https://doi.org/10.3390/microorganisms8081226
Capita R, Castaño-Arriba A, Rodríguez-Melcón C, Igrejas G, Poeta P, Alonso-Calleja C. Diversity, Antibiotic Resistance, and Biofilm-Forming Ability of Enterobacteria Isolated from Red Meat and Poultry Preparations. Microorganisms. 2020; 8(8):1226. https://doi.org/10.3390/microorganisms8081226
Chicago/Turabian StyleCapita, Rosa, Ana Castaño-Arriba, Cristina Rodríguez-Melcón, Gilberto Igrejas, Patricia Poeta, and Carlos Alonso-Calleja. 2020. "Diversity, Antibiotic Resistance, and Biofilm-Forming Ability of Enterobacteria Isolated from Red Meat and Poultry Preparations" Microorganisms 8, no. 8: 1226. https://doi.org/10.3390/microorganisms8081226
APA StyleCapita, R., Castaño-Arriba, A., Rodríguez-Melcón, C., Igrejas, G., Poeta, P., & Alonso-Calleja, C. (2020). Diversity, Antibiotic Resistance, and Biofilm-Forming Ability of Enterobacteria Isolated from Red Meat and Poultry Preparations. Microorganisms, 8(8), 1226. https://doi.org/10.3390/microorganisms8081226