Generation of a Non-Transgenic Genetically Improved Yeast Strain for Wine Production from Nitrogen-Deficient Musts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Yeast Populations and Mating Design
2.2. Microscale Fermentations with Synthetic Must
2.3. Heritability Analysis
2.4. Pilot Scale Fermentations with Natural Grape Must
2.5. Sensory Analysis
3. Results
3.1. Genetic Improvement Program to Obtain Strains that Consumes Less Nitrogen
3.2. Generation of a F2 Population of Improved Strains
3.3. Chemical and Sensory Analysis of Wine Produced by the Improved Strain “686”
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pretorius, I.S. Tailoring wine yeast for the new millennium: Novel approaches to the ancient art of winemaking. Yeast 2000, 16, 675–729. [Google Scholar] [CrossRef]
- Marsit, S.; Dequin, S. Diversity and adaptive evolution of Saccharomyces wine yeast: A review. FEMS Yeast Res. 2015, 15, fov067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dequin, S.; Casaregola, S. The genomes of fermentative Saccharomyces. Comptes Rendus Biol. 2011, 334, 687–693. [Google Scholar] [CrossRef] [PubMed]
- Querol, A. Adaptive evolution of wine yeast. Int. J. Food Microbiol. 2003, 86, 3–10. [Google Scholar] [CrossRef]
- Gobert, A.; Tourdot-Marechal, R.; Sparrow, C.; Morge, C.; Alexandre, H. Influence of nitrogen status in wine alcoholic fermentation. Food Microbiol. 2019, 83, 71–85. [Google Scholar] [CrossRef]
- Barbosa, C.; Falco, V.; Mendes-Faia, A.; Mendes-Ferreira, A. Nitrogen addition influences formation of aroma compounds, volatile acidity and ethanol in nitrogen deficient media fermented by Saccharomyces cerevisiae wine strains. J. Biosci. Bioeng. 2009, 108, 99–104. [Google Scholar] [CrossRef]
- Vilanova, M.; Ugliano, M.; Varela, C.; Siebert, T.; Pretorius, I.S.; Henschke, P.A. Assimilable nitrogen utilisation and production of volatile and non-volatile compounds in chemically defined medium by Saccharomyces cerevisiae wine yeasts. Appl. Microbiol. Biotechnol. 2007, 77, 145–157. [Google Scholar] [CrossRef] [Green Version]
- Broach, J.R. Nutritional control of growth and development in yeast. Genetics 2012, 192, 73–105. [Google Scholar] [CrossRef] [Green Version]
- Zhang, W.; Du, G.; Zhou, J.; Chen, J. Regulation of sensing, transportation, and catabolism of nitrogen sources in saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 2018, 82. [Google Scholar] [CrossRef] [Green Version]
- Taillandier, P.; Ramon Portugal, F.; Fuster, A.; Strehaiano, P. Effect of ammonium concentration on alcoholic fermentation kinetics by wine yeasts for high sugar content. Food Microbiol. 2007, 24, 95–100. [Google Scholar] [CrossRef] [Green Version]
- Conrad, M.; Schothorst, J.; Kankipati, H.N.; Van Zeebroeck, G.; Rubio-Texeira, M.; Thevelein, J.M. Nutrient sensing and signaling in the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 2014, 38, 254–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef] [Green Version]
- Martínez, C.; García, V.; González, D.; Jara, M.; Aguilera, M.; Ganga, M.A. Gene expression of specific enological traits in wine fermentation. Electron. J. Biotechnol. 2013, 16, 13. [Google Scholar] [CrossRef]
- Varela, C.; Pizarro, F.; Agosin, E. Biomass content governs fermentation rate in nitrogen-deficient wine musts. Appl. Environ. Microbiol. 2004, 70, 3392–3400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arias-Gil, M.; Garde-Cerdán, T.; Ancín-Azpilicueta, C. Influence of addition of ammonium and different amino acid concentrations on nitrogen metabolism in spontaneous must fermentation. Food Chem. 2007, 103, 1312–1318. [Google Scholar] [CrossRef]
- Garde-Cerdán, T.; Ancín-Azpilicueta, C. Effect of the addition of different quantities of amino acids to nitrogen-deficient must on the formation of esters, alcohols, and acids during wine alcoholic fermentation. LWT 2008, 41, 501–510. [Google Scholar] [CrossRef]
- Mendes-Ferreira, A.; Mendes-Faia, A.; Leao, C. Growth and fermentation patterns of Saccharomyces cerevisiae under different ammonium concentrations and its implications in winemaking industry. J. Appl. Microbiol. 2004, 97, 540–545. [Google Scholar] [CrossRef]
- Hernandez-Orte, P.; Ibarz, M.J.; Cacho, J.; Ferreira, V. Effect of the addition of ammonium and amino acids to musts of Airen variety on aromatic composition and sensory properties of the obtained wine. Food Chem. 2005, 89, 163–174. [Google Scholar] [CrossRef]
- Gutierrez, A.; Beltran, G.; Warringer, J.; Guillamon, J.M. Genetic basis of variations in nitrogen source utilization in four wine commercial yeast strains. PLoS ONE 2013, 8, e67166. [Google Scholar] [CrossRef] [Green Version]
- Jara, M.; Cubillos, F.A.; Garcia, V.; Salinas, F.; Aguilera, O.; Liti, G.; Martinez, C. Mapping genetic variants underlying differences in the central nitrogen metabolism in fermenter yeasts. PLoS ONE 2014, 9, e86533. [Google Scholar] [CrossRef] [Green Version]
- Kessi-Perez, E.I.; Araos, S.; Garcia, V.; Salinas, F.; Abarca, V.; Larrondo, L.F.; Martinez, C.; Cubillos, F.A. RIM15 antagonistic pleiotropy is responsible for differences in fermentation and stress response kinetics in budding yeast. FEMS Yeast Res. 2016, 16, fow021. [Google Scholar] [CrossRef] [Green Version]
- Brice, C.; Sanchez, I.; Tesniere, C.; Blondin, B. Assessing the mechanisms responsible for differences between nitrogen requirements of saccharomyces cerevisiae wine yeasts in alcoholic fermentation. Appl. Environ. Microbiol. 2014, 80, 1330–1339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzano, M.; Medrala, D.; Giusto, C.; Bartolomeoli, I.; Urso, R.; Comi, G. Classical and molecular analyses to characterize commercial dry yeasts used in wine fermentations. J. Appl. Microbiol. 2006, 100, 599–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Querol, A.; Barrio, E.; Huerta, T.; Ramon, D. Molecular monitoring of wine fermentations conducted by active dry yeast strains. Appl. Environ. Microbiol. 1992, 58, 2948–2953. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guimarães, T.M.; Moriel, D.G.; Machado, I.P.; Fadel Picheth, C.M.T.; Bonfim, T.M.B. Isolation and characterization of Saccharomyces cerevisiae strains of winery interest. Braz. J. Pharm. Sci. 2006, 42, 119–126. [Google Scholar] [CrossRef]
- Husnik, J.I.; Delaquis, P.J.; Cliff, M.A.; van Vuuren, H.J.J. Functional analyses of the malolactic wine yeast ML01. Am. J. Enol. Vitic. 2007, 58, 42–52. [Google Scholar]
- Coulon, J.; Husnik, J.I.; Inglis, D.L.; van der Merwe, G.K.; Lonvaud, A.; Erasmus, D.J.; van Vuuren, H.J.J. Metabolic engineering of Saccharomyces cerevisiae to minimize the production of ethyl carbamate in wine. Am. J. Enol. Vitic. 2006, 57, 113–124. [Google Scholar]
- Marullo, P.; Bely, M.; Masneuf-Pomarede, I.; Pons, M.; Aigle, M.; Dubourdieu, D. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 2006, 6, 268–279. [Google Scholar] [CrossRef] [Green Version]
- Marullo, P.; Mansour, C.; Dufour, M.; Albertin, W.; Sicard, D.; Bely, M.; Dubourdieu, D. Genetic improvement of thermo-tolerance in wine Saccharomyces cerevisiae strains by a backcross approach. FEMS Yeast Res. 2009, 9, 1148–1160. [Google Scholar] [CrossRef] [Green Version]
- Hart, R.S.; Jolly, N.P.; Ndimba, B.K. Characterisation of hybrid yeasts for the production of varietal Sauvignon blanc wine—A review. J. Microbiol. Methods 2019, 165, 105699. [Google Scholar] [CrossRef]
- Perez-Torrado, R.; Barrio, E.; Querol, A. Alternative yeasts for winemaking: Saccharomyces non-cerevisiae and its hybrids. Crit. Rev. Food Sci. Nutr. 2018, 58, 1780–1790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peris, D.; Perez-Torrado, R.; Hittinger, C.T.; Barrio, E.; Querol, A. On the origins and industrial applications of Saccharomyces cerevisiae x Saccharomyces kudriavzevii hybrids. Yeast 2018, 35, 51–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cubillos, F.A.; Brice, C.; Molinet, J.; Tisne, S.; Abarca, V.; Tapia, S.M.; Oporto, C.; Garcia, V.; Liti, G.; Martinez, C. Identification of nitrogen consumption genetic variants in yeast through QTL mapping and bulk segregant RNA-Seq analyses. G3 Genes Genomes Genet. 2017, 7, 1693–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kessi-Perez, E.I.; Salinas, F.; Gonzalez, A.; Su, Y.; Guillamon, J.M.; Hall, M.N.; Larrondo, L.F.; Martinez, C. KAE1 allelic variants affect TORC1 activation and fermentation kinetics in saccharomyces cerevisiae. Front. Microbiol. 2019, 10, 1686. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Molinet, J.; Cubillos, F.A.; Salinas, F.; Liti, G.; Martinez, C. Genetic variants of TORC1 signaling pathway affect nitrogen consumption in Saccharomyces cerevisiae during alcoholic fermentation. PLoS ONE 2019, 14, e0220515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salinas, F.; Cubillos, F.A.; Soto, D.; Garcia, V.; Bergstrom, A.; Warringer, J.; Ganga, M.A.; Louis, E.J.; Liti, G.; Martinez, C. The genetic basis of natural variation in oenological traits in Saccharomyces cerevisiae. PLoS ONE 2012, 7, e49640. [Google Scholar] [CrossRef] [Green Version]
- Kessi-Perez, E.I.; Salinas, F.; Molinet, J.; Gonzalez, A.; Muniz, S.; Guillamon, J.M.; Hall, M.N.; Larrondo, L.F.; Martinez, C. Indirect monitoring of TORC1 signalling pathway reveals molecular diversity among different yeast strains. Yeast 2019, 36, 65–74. [Google Scholar] [CrossRef]
- Garcia, V.; Rivera, J.; Contreras, A.; Ganga, M.A.; Martinez, C. Development and characterization of hybrids from native wine yeasts. Braz. J. Microbiol. 2012, 43, 482–489. [Google Scholar] [CrossRef] [Green Version]
- Contreras, A.; Garcia, V.; Salinas, F.; Urzua, U.; Ganga, M.A.; Martinez, C. Identification of genes related to nitrogen uptake in wine strains of Saccharomyces cerevisiae. World J. Microbiol. Biotechnol. 2012, 28, 1107–1113. [Google Scholar] [CrossRef]
- Legras, J.L.; Karst, F. Optimisation of interdelta analysis for Saccharomyces cerevisiae strain characterisation. FEMS Microbiol. Lett. 2003, 221, 249–255. [Google Scholar] [CrossRef] [Green Version]
- Xufre, A.; Albergaria, H.; Girio, F.; Spencer-Martins, I. Use of interdelta polymorphisms of Saccharomyces cerevisiae strains to monitor population evolution during wine fermentation. J. Ind. Microbiol. Biotechnol. 2011, 38, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Bradbury, J.E.; Richards, K.D.; Niederer, H.A.; Lee, S.A.; Rod Dunbar, P.; Gardner, R.C. A homozygous diploid subset of commercial wine yeast strains. Antonie Leeuwenhoek 2006, 89, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Field, D.; Wills, C. Abundant microsatellite polymorphism in Saccharomyces cerevisiae, and the different distributions of microsatellites in eight prokaryotes and S. cerevisiae, result from strong mutation pressures and a variety of selective forces. Proc. Natl. Acad. Sci. USA 1998, 95, 1647–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perez, M.A.; Gallego, F.J.; Martinez, I.; Hidalgo, P. Detection, distribution and selection of microsatellites (SSRs) in the genome of the yeast Saccharomyces cerevisiae as molecular markers. Lett. Appl. Microbiol. 2001, 33, 461–466. [Google Scholar] [CrossRef]
- Rossignol, T.; Dulau, L.; Julien, A.; Blondin, B. Genome-wide monitoring of wine yeast gene expression during alcoholic fermentation. Yeast 2003, 20, 1369–1385. [Google Scholar] [CrossRef]
- Nissen, T.L.; Schulze, U.; Nielsen, J.; Villadsen, J. Flux distributions in anaerobic, glucose-limited continuous cultures of Saccharomyces cerevisiae. Microbiology 1997, 143, 203–218. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Alonso, S.; Hermosin-Gutierrez, I.; Garcia-Romero, E. Simultaneous HPLC analysis of biogenic amines, amino acids, and ammonium ion as aminoenone derivatives in wine and beer samples. J. Agric. Food Chem. 2007, 55, 608–613. [Google Scholar] [CrossRef]
- R-Core-Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org/ (accessed on 8 May 2020).
- Henderson, C.R. A simple method for computing the inverse of a numerator relationship matrix used in prediction of breeding values. Biometrics 1976, 32, 69–83. [Google Scholar] [CrossRef]
- Meyer, K. Dfreml, 3rd ed. In Proceedings of the Sixth Wrold Congress on Livestock Production, Armidale, Australia, 11–16 January 1998 [CD-ROM]; University of New England: Armidale, Australia, 1998. [Google Scholar]
- Boldman, K.G.; Kriese, L.A.; Van Vleck, L.D.; Van Tassell, C.P.; Kachman, S.D. A Manual for Use of MTDFREML. A Set of Programs to Obtain eEstimates of Variances and Covariances; USDA, ARS: Lincoln, NE, USA, 1995.
- Ihaka, R.; Gentleman, R. R: A language for data analysis and graphics. J. Comput. Graph. Stat. 1996, 5, 299–314. [Google Scholar]
- Lawless, H.T.; Heymann, H. Sensory Evaluation of Food: Principles and Practices; Springer Science & Business Media: New York, NY, USA, 2010. [Google Scholar]
- Noble, A.C.; Arnold, R.A.; Buechsenstein, J.; Leach, E.J.; Schmidt, J.O.; Stern, P.M. Modification of a standarized system of wine aroma terminology. Am. J. Enol. Vitic. 1987, 38, 143–146. [Google Scholar]
- ISO. ISO 3591. Sensory Analysis-Apparatus: Wine Tasting Glass; International Organization for Standarization: Geneva, Switzerland, 1977. [Google Scholar]
- Jackson, R.S. Wine Tasting: A Professional Handbook; Academic Press: San Diego, CA, USA, 2009. [Google Scholar]
- Colonna, A.E.; Adams, D.O.; Noble, A.C. Comparison of procedures for reducing astringency carry-over effects in evaluation of red wines. Aust. J. Grape Wine Res. 2004, 10, 26–31. [Google Scholar] [CrossRef]
- Falconer, D.S.; Mackay, T.F.C. Introduction to Quantitative Genetics, 4th ed.; Longmans Green: Harlow, UK, 1996. [Google Scholar]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar] [PubMed]
- Hearne, C.M.; Ghosh, S.; Todd, J.A. Microsatellites for linkage analysis of genetic traits. Trends Genet. 1992, 8, 288–294. [Google Scholar] [CrossRef]
- Crepin, L.; Nidelet, T.; Sanchez, I.; Dequin, S.; Camarasa, C. Sequential use of nitrogen compounds by Saccharomyces cerevisiae during wine fermentation: A model based on kinetic and regulation characteristics of nitrogen permeases. Appl. Environ. Microbiol. 2012, 78, 8102–8111. [Google Scholar] [CrossRef] [Green Version]
- Swiegers, J.H.; Bartowsky, E.J.; Henschke, P.A.; Pretorius, I.S. Yeast and bacterial modulation of wine aroma and flavour. Aust. J. Grape Wine Res. 2005, 11, 139–173. [Google Scholar] [CrossRef]
- Carrau, F.M.; Medina, K.; Farina, L.; Boido, E.; Henschke, P.A.; Dellacassa, E. Production of fermentation aroma compounds by Saccharomyces cerevisiae wine yeasts: Effects of yeast assimilable nitrogen on two model strains. FEMS Yeast Res. 2008, 8, 1196–1207. [Google Scholar] [CrossRef] [Green Version]
- Fairbairn, S.; McKinnon, A.; Musarurwa, H.T.; Ferreira, A.C.; Bauer, F.F. The impact of single amino acids on growth and volatile aroma production by saccharomyces cerevisiae Strains. Front. Microbiol. 2017, 8, 2554. [Google Scholar] [CrossRef] [Green Version]
- Hernandez-Orte, P.; Ibarz, M.J.; Cacho, J.; Ferreira, V. Addition of amino acids to grape juice of the Merlot variety: Effect on amino acid uptake and aroma generation during alcoholic fermentation. Food Chem. 2006, 98, 300–310. [Google Scholar] [CrossRef]
- Hernandez-Orte, P.; Bely, M.; Cacho, J.; Ferreira, V. Impact of ammonium additions on volatile acidity, ethanol, and aromatic compound production by different Saccharomyces cerevisiae strains during fermentation in controlled synthetic media. Aust. J. Grape Wine Res. 2006, 12, 150–160. [Google Scholar] [CrossRef]
- Romagnoli, G.; Luttik, M.A.; Kotter, P.; Pronk, J.T.; Daran, J.M. Substrate specificity of thiamine pyrophosphate-dependent 2-oxo-acid decarboxylases in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2012, 78, 7538–7548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crepin, L.; Truong, N.M.; Bloem, A.; Sanchez, I.; Dequin, S.; Camarasa, C. Management of multiple nitrogen sources during wine fermentation by saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carrau, F.M.; Medina, K.; Boido, E.; Farina, L.; Gaggero, C.; Dellacassa, E.; Versini, G.; Henschke, P.A. De novo synthesis of monoterpenes by Saccharomyces cerevisiae wine yeasts. FEMS Microbiol. Lett. 2005, 243, 107–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]


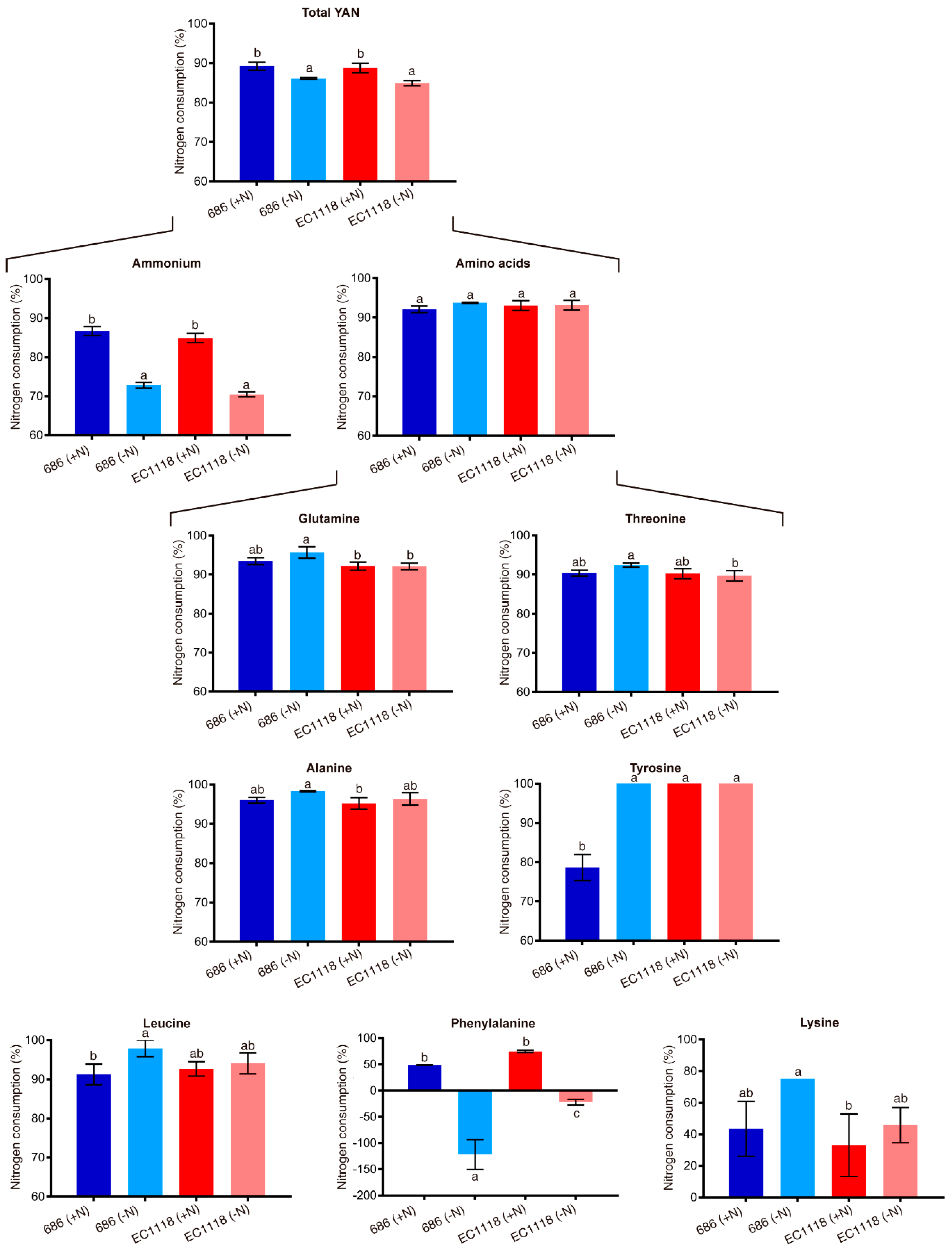
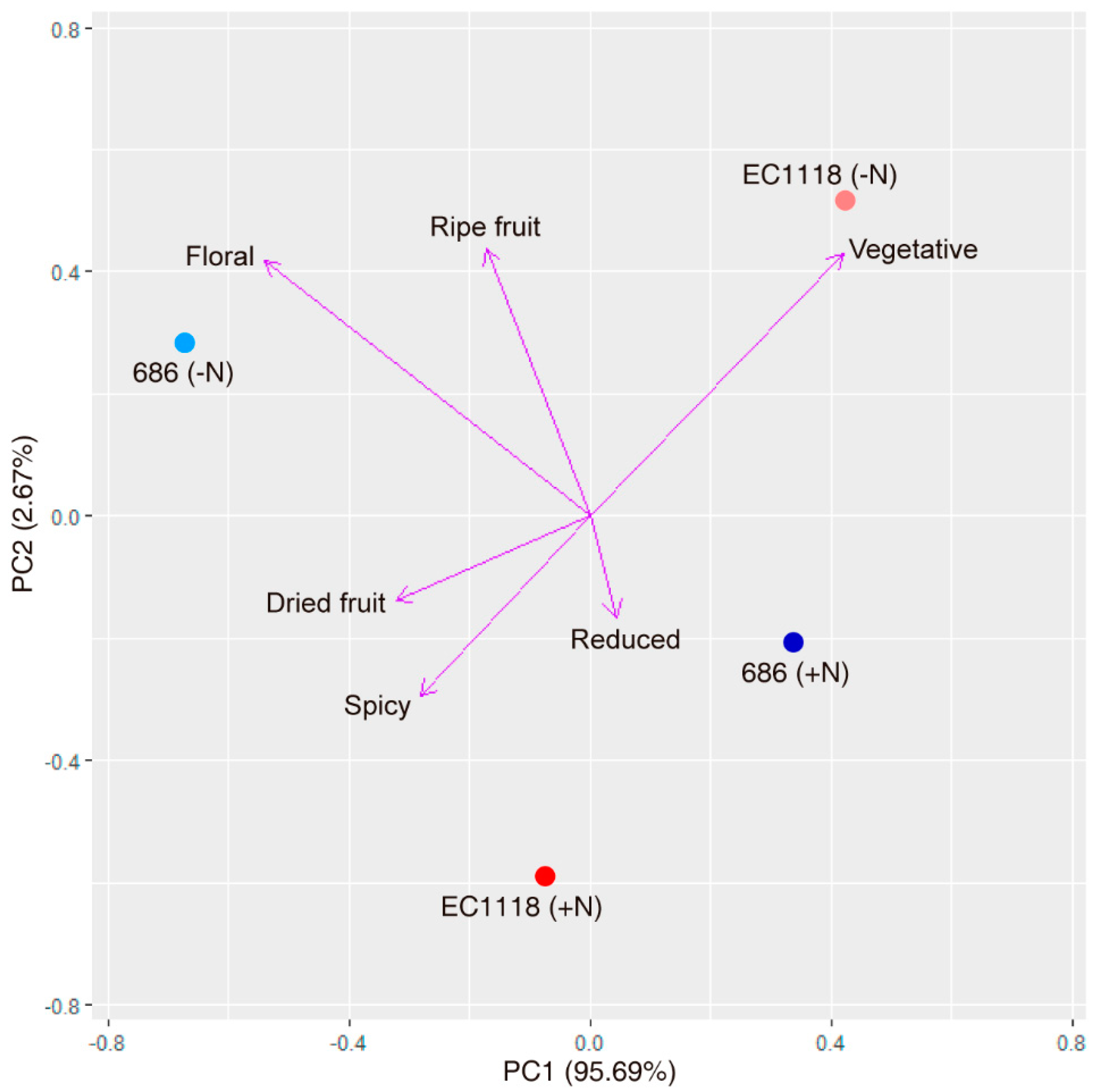
| Trait | YAN | CO2 | ACE | GLU | FRU | GLY | ETH |
|---|---|---|---|---|---|---|---|
| YAN | 0.9707 | 0.9057 | 0.8007 | 0.9597 | 0.2909 | N.D. | 0.8886 |
| CO2 | 0.9926 | 0.9247 | 0.8134 | 0.8023 | 0.9075 | 0.9998 | 0.8638 |
| ACE | 0.9995 | 0.9575 | 0.9297 | 0.9936 | 0.9999 | 0.9610 | 1.0000 |
| GLU | 0.9965 | 0.9996 | 0.9825 | 0.7278 | 0.0794 | 0.9969 | 0.7947 |
| FRU | 0.9888 | 0.7535 | 0.9998 | 0.0249 | 0.7225 | 1.0000 | 0.9375 |
| GLY | 0.9999 | N.D. | 0.9959 | 0.9288 | 0.9996 | 0.8627 | 0.0268 |
| ETH | 0.9999 | 0.9693 | 0.9661 | 0.6314 | 0.9858 | 0.3866 | 0.7540 |
| Average | 0.9926 | 0.9184 | 0.9269 | 0.7241 | 0.7122 | 0.8678 | 0.7522 |
| Trait | YAN | CO2 | ACE | GLU | FRU | GLY | ETH |
|---|---|---|---|---|---|---|---|
| YAN | −0.0456 | 0.0291 | 0.2510 | −0.0016 | −0.0174 | −0.2551 | |
| CO2 | 0.0013 | 0.4172 | −0.1330 | −0.5841 | N.D. | 0.2406 | |
| ACE | 0.0421 | 0.3714 | −0.6822 | −0.5950 | 0.8395 | 0.5915 | |
| GLU | 0.3099 | −0.1394 | −0.6798 | 0.9028 | −0.7310 | −0.9229 | |
| FRU | 0.2089 | −0.3546 | −0.5951 | −0.9999 | −0.0235 | −0.9283 | |
| GLY | −0.0174 | N.D. | 0.8708 | −0.7748 | −0.3546 | 0.6530 | |
| ETH | −0.2753 | 0.1963 | 0.6018 | −0.9232 | −0.9347 | −1.0000 |
| Strain | Acetic Acid Produced ± SD (g/L) | Residual Glucose ± SD (g/L) | Residual Fructose ± SD (g/L) | Glycerol Produced ± SD (g/L) | Ethanol Produced ± SD (% v/v) |
|---|---|---|---|---|---|
| A17 α | 2.00 ± 0.20 a | 6.04 ± 8.64 a | 28.33 ± 20.42 a | 10.07 ± 1.42 a | 10.15 ± 2.05 a |
| A4 α | 2.29 ± 0.06 a | 0.05 ± 0.08 a | 9.09 ± 10.74 a | 11.46 ± 0.66 a | 10.34 ± 0.81 a |
| 476 β | 2.30 ± 0.10 a | 0.00 ± 0.00 a | 3.52 ± 4.84 a | 10.15 ± 0.67 a | 10.92 ± 0.72 a |
| Strain | Acetic Acid Produced ± SD (g/L) | Residual Glucose ± SD (g/L) | Residual Fructose ± SD (g/L) | Glycerol Produced ± SD (g/L) | Ethanol Produced ± SD (% v/v) |
|---|---|---|---|---|---|
| A3 α | 2.34 ± 0.07 a | 0.00 ± 0.00 a | 1.10 ± 0.47 a | 11.06 ± 0.66 a | 11.94 ± 0.58 a |
| A91 α | 2.15 ± 0.01 b | 1.71 ± 1.87 a | 23.19 ± 11.54 b | 10.05 ± 0.87 a | 10.95 ± 0.99 a |
| 686 β | 2.41 ± 0.13 a | 0.00 ± 0.00 a | 0.87 ± 0.57 a | 9.45 ± 0.45 a | 12.01 ± 0.41 a |
| Strain | Nitrogen Supplementation | Overall Aroma ± SD | Bitterness ± SD | Acidity ± SD | Concentration ± SD |
|---|---|---|---|---|---|
| 686 | Yes | 3.82 ± 2.38 a | 3.97 ± 0.57 a | 6.07 ± 0.83 a | 4.48 ± 1.66 a |
| No | 6.47 ± 2.37 b | 3.79 ± 0.83 a | 4.43 ± 1.75 ab | 6.57 ± 0.84 b | |
| EC1118 | Yes | 5.46 ± 1.06 ab | 1.58 ± 1.25 b | 3.99 ± 1.42 b | 4.26 ± 0.93 a |
| No | 3.55 ± 0.99 a | 3.52 ± 0.66 a | 5.74 ± 1.78 ab | 4.31 ± 1.89 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kessi-Pérez, E.I.; Molinet, J.; García, V.; Aguilera, O.; Cepeda, F.; López, M.E.; Sari, S.; Cuello, R.; Ciklic, I.; Rojo, M.C.; et al. Generation of a Non-Transgenic Genetically Improved Yeast Strain for Wine Production from Nitrogen-Deficient Musts. Microorganisms 2020, 8, 1194. https://doi.org/10.3390/microorganisms8081194
Kessi-Pérez EI, Molinet J, García V, Aguilera O, Cepeda F, López ME, Sari S, Cuello R, Ciklic I, Rojo MC, et al. Generation of a Non-Transgenic Genetically Improved Yeast Strain for Wine Production from Nitrogen-Deficient Musts. Microorganisms. 2020; 8(8):1194. https://doi.org/10.3390/microorganisms8081194
Chicago/Turabian StyleKessi-Pérez, Eduardo I., Jennifer Molinet, Verónica García, Omayra Aguilera, Fernanda Cepeda, María Eugenia López, Santiago Sari, Raúl Cuello, Iván Ciklic, María Cecilia Rojo, and et al. 2020. "Generation of a Non-Transgenic Genetically Improved Yeast Strain for Wine Production from Nitrogen-Deficient Musts" Microorganisms 8, no. 8: 1194. https://doi.org/10.3390/microorganisms8081194
APA StyleKessi-Pérez, E. I., Molinet, J., García, V., Aguilera, O., Cepeda, F., López, M. E., Sari, S., Cuello, R., Ciklic, I., Rojo, M. C., Combina, M., Araneda, C., & Martínez, C. (2020). Generation of a Non-Transgenic Genetically Improved Yeast Strain for Wine Production from Nitrogen-Deficient Musts. Microorganisms, 8(8), 1194. https://doi.org/10.3390/microorganisms8081194






