How Do Soil Bacterial Diversity and Community Composition Respond under Recommended and Conventional Nitrogen Fertilization Regimes?
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Sites
2.2. Experimental Design
2.3. Sampling, Determination of Soil Bio-Chemical Properties and Maize Yield
2.4. Bioinformatics Analyses
2.4.1. DNA Extraction and PCR Amplification
2.4.2. Illumina MiSeq Sequencing
2.5. Statistical Analyses
3. Results
3.1. Soil Chemical, Biological Properties, and Crop Yield
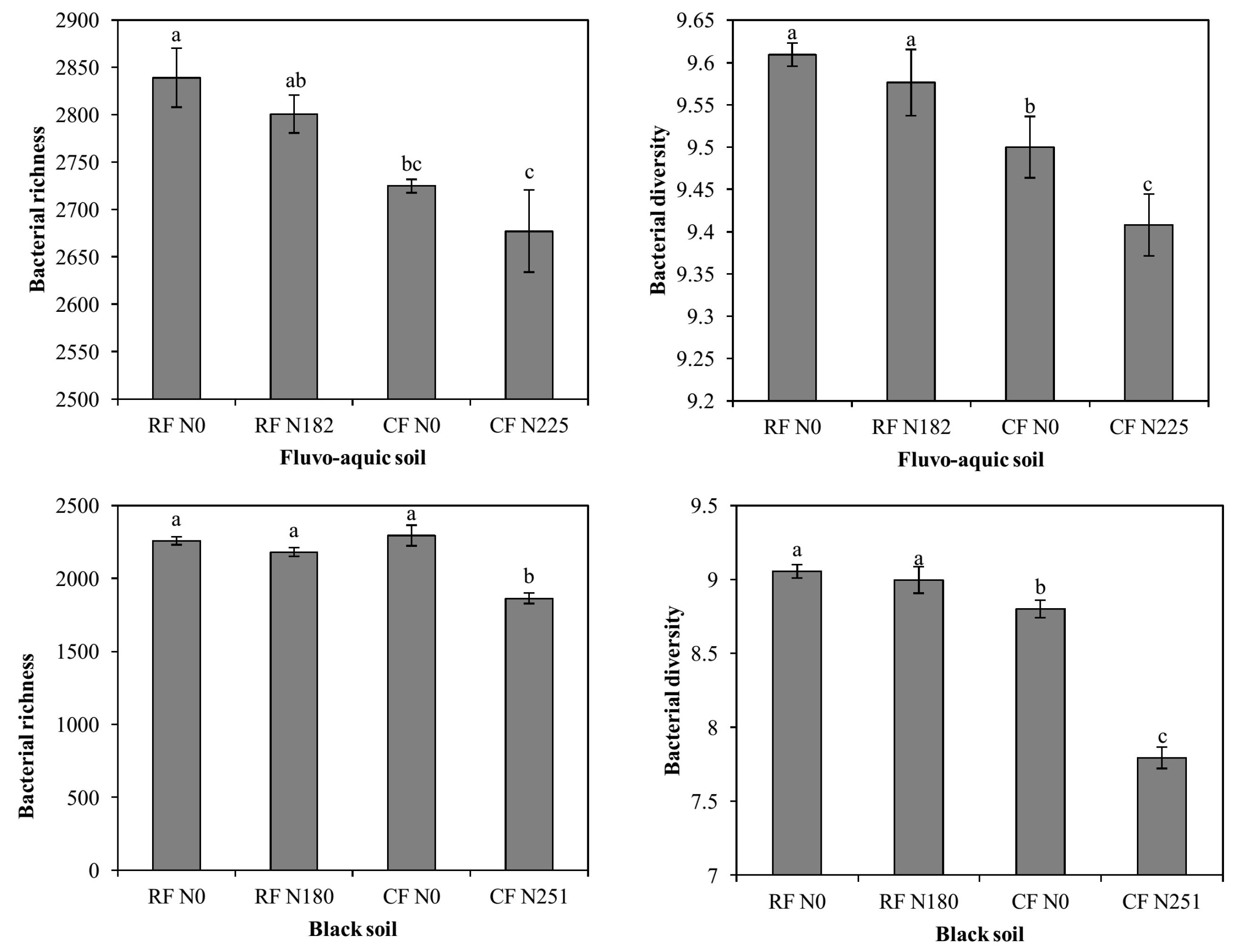
3.2. Bacterial Alpha Diversity under Long-Term Fertilization
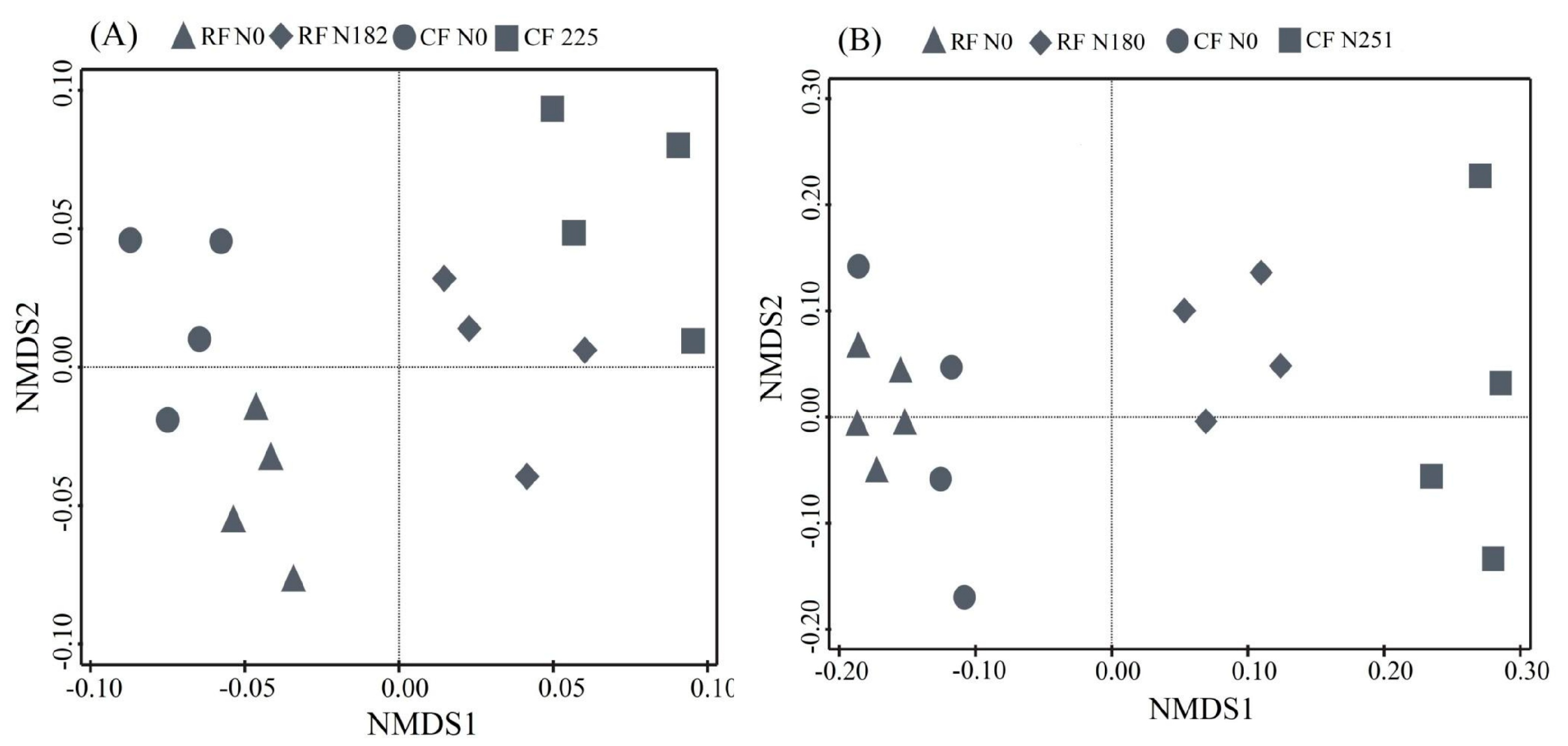
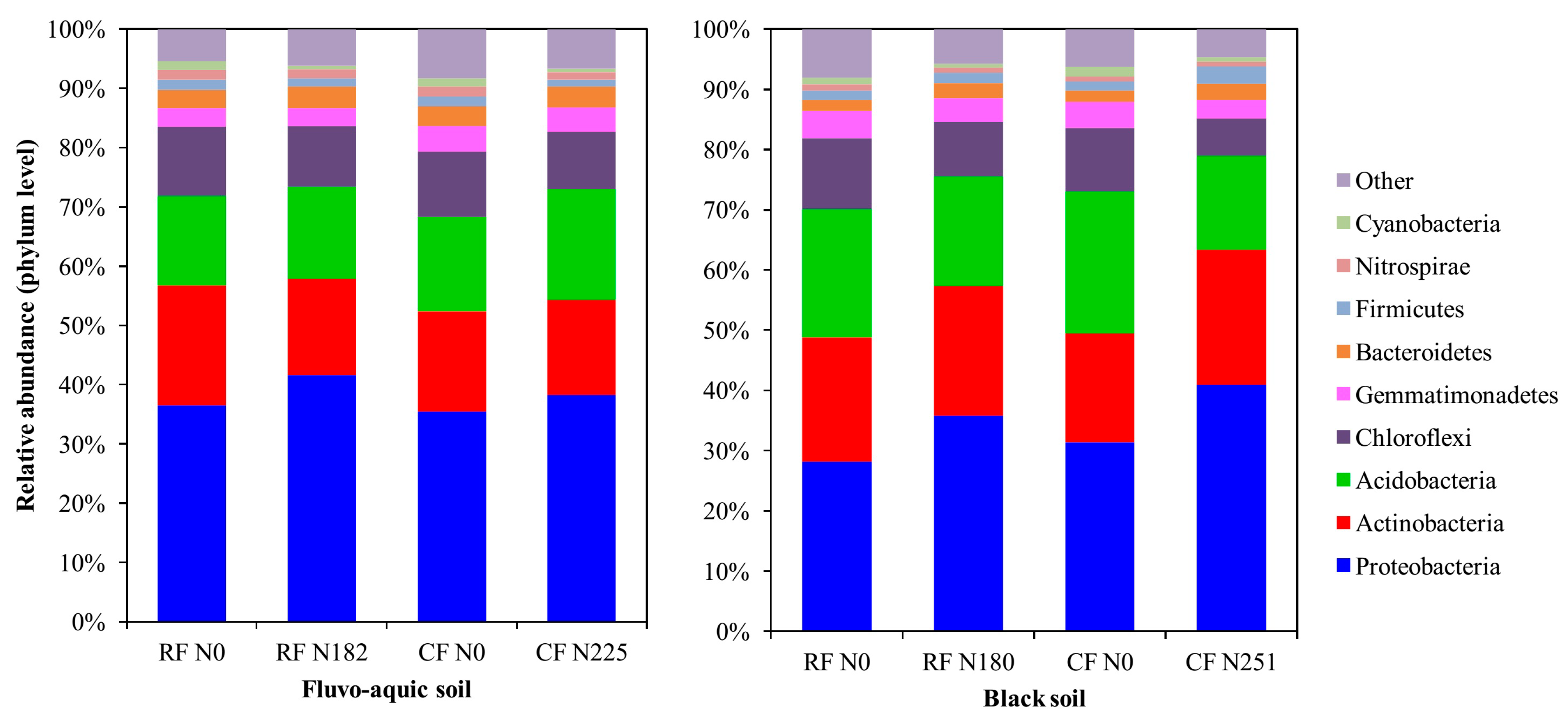
3.3. Bacterial Community Composition under Long-Term Fertilization
3.4. Bacterial Diversity and Community Composition in Relation to Soil Properties
4. Discussion
4.1. Soil Chemical Properties and Biochemical Properties in Relation to Maize Yield
4.2. Bacterial Alpha Diversity
4.3. Bacterial Community Structure
4.4. Relationship between Soil Properties versus Bacterial Diversity and Community Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Galloway, J.N.; Townsend, A.R.; Erisman, J.W.; Bekunda, M.; Cai, Z.; Freney, J.R.; Martinelli, L.A.; Seitzinger, S.P.; Sutton, M.A. Transformation of the nitrogen cycle: Recent trends, questions, and potential solutions. Science 2008, 320, 889–892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fowler, D.; Coyle, M.; Skiba, U.; Sutton, M.A.; Cape, J.N.; Reis, S.; Sheppard, L.J.; Jenkins, A.; Grizzetti, B.; Galloway, J.N. The global nitrogen cycle in the twenty-first century. Philos. Trans. R. Soc. B 2013, 368, 20130164. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Bardgett, R.D.; Smith, P.; Reay, D.S. Microorganisms and climate change: Terrestrial feedbacks and mitigation options. Nat. Rev. Microbiol. 2010, 8, 779–790. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Hättenschwiler, S.; Olander, L.; Allison, S. Nitrogen and nature. Ambio J. Hum. Environ. 2002, 31, 97–101. [Google Scholar] [CrossRef]
- Allison, S.D.; Czimczik, C.I.; Treseder, K.K. Microbial activity and soil respiration under nitrogen addition in Alaskan boreal forest. Glob. Chang. Biol. 2008, 14, 1156–1168. [Google Scholar] [CrossRef] [Green Version]
- Cusack, D.F.; Silver, W.L.; Torn, M.S.; Burton, S.D.; Firestone, M.K. Changes in microbial community characteristics and soil organic matter with nitrogen additions in two tropical forests. Ecology 2011, 92, 621–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, K.S.; Craine, J.M.; Fierer, N. Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Glob. Chang. Biol. 2012, 18, 1918–1927. [Google Scholar] [CrossRef]
- Treseder, K.K. Nitrogen additions and microbial biomass: A meta-analysis of ecosystem studies. Ecol. Lett. 2008, 11, 1111–1120. [Google Scholar] [CrossRef] [Green Version]
- Chaer, G.; Fernandes, M.; Myrold, D.; Bottomley, P. Comparative resistance and resilience of soil microbial communities and enzyme activities in adjacent native forest and agricultural soils. Microb. Ecol. 2009, 58, 414–424. [Google Scholar] [CrossRef]
- Zhou, J.; Guan, D.; Zhou, B.; Zhao, B.; Ma, M.; Qin, J.; Jiang, X.; Chen, S.; Cao, F.; Shen, D. Influence of 34-years of fertilization on bacterial communities in an intensively cultivated black soil in northeast China. Soil Biol. Biochem. 2015, 90, 42–51. [Google Scholar] [CrossRef]
- Odum, E.P.; Finn, J.T.; Franz, E.H. Perturbation theory and the subsidy-stress gradient. Bioscience 1979, 29, 349–352. [Google Scholar] [CrossRef]
- Odum, E.P. Trends expected in stressed ecosystems. Bioscience 1985, 35, 419–422. [Google Scholar] [CrossRef]
- Allison, S.D.; Hanson, C.A.; Treseder, K.K. Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biol. Biochem. 2007, 39, 1878–1887. [Google Scholar] [CrossRef] [Green Version]
- Ai, C.; Zhang, S.; Zhang, X.; Guo, D.; Zhou, W.; Huang, S. Distinct responses of soil bacterial and fungal communities to changes in fertilization regime and crop rotation. Geoderma 2018, 319, 156–166. [Google Scholar] [CrossRef]
- Zeng, J.; Liu, X.; Song, L.; Lin, X.; Zhang, H.; Shen, C.; Chu, H. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 2016, 92, 41–49. [Google Scholar] [CrossRef]
- Ling, N.; Chen, D.; Guo, H.; Wei, J.; Bai, Y.; Shen, Q.; Hu, S. Differential responses of soil bacterial communities to long-term N and P inputs in a semi-arid steppe. Geoderma 2017, 292, 25–33. [Google Scholar] [CrossRef]
- Wang, Q.; Wang, C.; Yu, W.; Turak, A.; Chen, D.; Huang, Y.; Ao, J.; Jiang, Y.; Huang, Z. Effects of nitrogen and phosphorus inputs on soil bacterial abundance, diversity, and community composition in Chinese fir plantations. Front. Microbiol. 2018, 9, 1543. [Google Scholar] [CrossRef]
- Ullah, S.; Ai, C.; Huang, S.; Zhang, J.; Jia, L.; Ma, J.; Zhou, W.; He, P. The responses of extracellular enzyme activities and microbial community composition under nitrogen addition in an upland soil. PLoS ONE 2019, 14, e0223026. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, D.; Bai, E. Decreasing soil microbial diversity is associated with decreasing microbial biomass under nitrogen addition. Soil Biol. Biochem. 2018, 120, 126–133. [Google Scholar] [CrossRef]
- Staley, C.; Breuillin-Sessoms, F.; Wang, P.; Kaiser, T.; Venterea, R.T.; Sadowsky, M.J. Urea amendment decreases microbial diversity and selects for specific nitrifying strains in eight contrasting agricultural soils. Front. Microbiol. 2018, 9, 634. [Google Scholar] [CrossRef] [Green Version]
- Bremner, J.M.; Mulvaney, C. Nitrogen—Total. In Methods of Soil Analysis. Part 2. Chemical and Microbiological Properties; Page, A.L., Miller, R.H., Keeney, D.R., Eds.; American Society of Agronomy, Soil Science Society of America: Madison, WI, USA, 1982; pp. 595–624. [Google Scholar]
- Kalembasa, S.J.; Jenkinson, D.S. A comparative study of titrimetric and gravimetric methods for the determination of organic carbon in soil. J. Sci. Food Agric. 1973, 24, 1085–1090. [Google Scholar] [CrossRef]
- Olsen, S.; Sommers, L.; Page, A. Methods of soil analysis. Part 2. Chem. Microbiol. Prop. Phosphorus. Asa Monogr. 1982, 9, 403–430. [Google Scholar]
- Jenkinson, D. Determination of microbial biomass carbon and nitrogen in soil. Adv. Nitrogen Cycl. 1988, 368–386. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De’Ath, G. Boosted trees for ecological modeling and prediction. Ecology 2007, 88, 243–251. [Google Scholar] [CrossRef]
- Tenenhaus, M.; Esposito Vinzi, V.; Chatelin, Y.; Lauro, C. PLS Path Modeling. Comput. Stat. Data Anal. 2005, 48, 159–205. [Google Scholar] [CrossRef]
- Barberán, A.; Ramirez, K.S.; Leff, J.W.; Bradford, M.A.; Wall, D.H.; Fierer, N. Why are some microbes more ubiquitous than others? Predicting the habitat breadth of soil bacteria. Ecol. Lett. 2014, 17, 794–802. [Google Scholar] [CrossRef] [PubMed]
- Shannon, C. A mathematical theoryof communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- Chao, A. Nonparametric estimation of the number of classes in a population. Scand. J. Stat. 1984, 11, 265–270. [Google Scholar]
- Jagadamma, S.; Lal, R.; Hoeft, R.G.; Nafziger, E.D.; Adee, E.A. Nitrogen fertilization and cropping systems effects on soil organic carbon and total nitrogen pools under chisel-plow tillage in Illinois. Soil Tillage Res. 2007, 95, 348–356. [Google Scholar] [CrossRef]
- Schroder, J.L.; Zhang, H.; Girma, K.; Raun, W.R.; Penn, C.J.; Payton, M.E. Soil acidification from long-term use of nitrogen fertilizers on winter wheat. Soil Sci. Soc. Am. J. 2011, 75, 957–964. [Google Scholar] [CrossRef]
- Guo, J.; Liu, X.; Zhang, Y.; Shen, J.; Han, W.; Zhang, W.; Christie, P.; Goulding, K.; Vitousek, P.; Zhang, F. Significant acidification in major Chinese croplands. Science 2010, 327, 1008–1010. [Google Scholar] [CrossRef] [Green Version]
- Zeglin, L.H.; Stursova, M.; Sinsabaugh, R.L.; Collins, S.L. Microbial responses to nitrogen addition in three contrasting grassland ecosystems. Oecologia 2007, 154, 349–359. [Google Scholar] [CrossRef]
- Yang, H.; Li, Y.; Wu, M.; Zhang, Z.; Li, L.; Wan, S. Plant community responses to nitrogen addition and increased precipitation: The importance of water availability and species traits. Glob. Chang. Biol. 2011, 17, 2936–2944. [Google Scholar] [CrossRef]
- Cusack, D.F.; Karpman, J.; Ashdown, D.; Cao, Q.; Ciochina, M.; Halterman, S.; Lydon, S.; Neupane, A. Global change effects on humid tropical forests: Evidence for biogeochemical and biodiversity shifts at an ecosystem scale. Rev. Geophys. 2016, 54, 523–610. [Google Scholar] [CrossRef] [Green Version]
- Yu, H.; Ling, N.; Wang, T.; Zhu, C.; Wang, Y.; Wang, S.; Gao, Q. Responses of soil biological traits and bacterial communities to nitrogen fertilization mediate maize yields across three soil types. Soil Tillage Res. 2019, 185, 61–69. [Google Scholar] [CrossRef]
- Marinari, S.; Lagomarsino, A.; Moscatelli, M.; Di Tizio, A.; Campiglia, E. Soil carbon and nitrogen mineralization kinetics in organic and conventional three-year cropping systems. Soil Tillage Res. 2010, 109, 161–168. [Google Scholar] [CrossRef]
- Chang, S.X.; Shi, Z.; Thomas, B.R. Soil respiration and its temperature sensitivity in agricultural and afforested poplar plantation systems in northern Alberta. Biol. Fertil. Soils 2016, 52, 629–641. [Google Scholar] [CrossRef]
- Bender, S.F.; Wagg, C.; Van Der Heijden, M.G. An underground revolution: Biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef]
- Kuzyakov, Y. Priming effects: Interactions between living and dead organic matter. Soil Biol. Biochem. 2010, 42, 1363–1371. [Google Scholar] [CrossRef]
- Canarini, A.; Kaiser, C.; Merchant, A.; Richter, A.; Wanek, W. Root exudation of primary metabolites: Mechanisms and their roles in plant responses to environmental stimuli. Front. Plant Sci. 2019, 10, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansa, J.; Bukovská, P.; Gryndler, M. Mycorrhizal hyphae as ecological niche for highly specialized hypersymbionts–or just soil free-riders? Front. Plant Sci. 2013, 4, 134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Trivedi, P.; Osanai, Y.; Liu, Y.R.; Hamonts, K.; Jeffries, T.C.; Singh, B.K. Carbon content and climate variability drive global soil bacterial diversity patterns. Ecol. Monogr. 2016, 86, 373–390. [Google Scholar] [CrossRef]
- Elser, J.J.; Bracken, M.E.; Cleland, E.E.; Gruner, D.S.; Harpole, W.S.; Hillebrand, H.; Ngai, J.T.; Seabloom, E.W.; Shurin, J.B.; Smith, J.E. Global analysis of nitrogen and phosphorus limitation of primary producers in freshwater, marine and terrestrial ecosystems. Ecol. Lett. 2007, 10, 1135–1142. [Google Scholar] [CrossRef] [Green Version]
- Six, J.; Frey, S.; Thiet, R.; Batten, K. Bacterial and fungal contributions to carbon sequestration in agroecosystems. Soil Sci. Soc. Am. J. 2006, 70, 555–569. [Google Scholar] [CrossRef]
- Berg, G.; Smalla, K. Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol. Ecol. 2009, 68, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Li, J.; Yang, F.; Yaoyao, E.; Raza, W.; Huang, Q.; Shen, Q. Application of bioorganic fertilizer significantly increased apple yields and shaped bacterial community structure in orchard soil. Microb. Ecol. 2017, 73, 404–416. [Google Scholar] [CrossRef]
- Eo, J.; Park, K.-C. Long-term effects of imbalanced fertilization on the composition and diversity of soil bacterial community. Agric. Ecosyst. Environ. 2016, 231, 176–182. [Google Scholar] [CrossRef]
- Raza, W.; Faheem, M.; Yousaf, S.; Rajer, F.U.; Yamin, M. Volatile and non-volatile antifungal compounds produced by Trichoderma harzianum SQR-T037 suppressed the growth of Fusarium oxysporum f. sp. niveum. Sci. Lett. 2013, 1, 21–24. [Google Scholar]
- Montañez, A.; Sicardi, M. Effects of inoculation on growth promotion and biological nitrogen fixation in maize (Zea mays L.) under greenhouse and field conditions. Basic Res. J. Agric. Sci. Rev. 2013, 2, 102–110. [Google Scholar]
- Ji, X.; Lu, G.; Gai, Y.; Zheng, C.; Mu, Z. Biological control against bacterial wilt and colonization of mulberry by an endophytic Bacillus subtilis strain. FEMS Microbiol. Ecol. 2008, 65, 565–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, C.; Song, Z.; Zhuang, D.; Wang, J.; Xie, S.; Liu, G. Urea fertilization decreases soil bacterial diversity, but improves microbial biomass, respiration, and N-cycling potential in a semiarid grassland. Biol. Fertil. Soils 2019, 55, 229–242. [Google Scholar] [CrossRef]
- Finkmann, W.; Altendorf, K.; Stackebrandt, E.; Lipski, A. Characterization of N2O-producing Xanthomonas-like isolates from biofilters as Stenotrophomonas nitritireducens sp. nov., Luteimonas mephitis gen. nov., sp. nov. and Pseudoxanthomonas broegbernensis gen. nov., sp. nov. Int. J. Syst. Evol. Microbiol. 2000, 50, 273–282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLuca, T.; Drinkwater, L.; Wiefling, B.; DeNicola, D. Free-living nitrogen-fixing bacteria in temperate cropping systems: Influence of nitrogen source. Biol. Fertil. Soils 1996, 23, 140–144. [Google Scholar] [CrossRef]
- Ge, Y.; Zhang, J.-B.; Zhang, L.-M.; Yang, M.; He, J.-Z. Long-term fertilization regimes affect bacterial community structure and diversity of an agricultural soil in northern China. J. Soils Sediments 2008, 8, 43–50. [Google Scholar] [CrossRef]
- Thirukkumaran, C.M.; Parkinson, D. Microbial respiration, biomass, metabolic quotient and litter decomposition in a lodgepole pine forest floor amended with nitrogen and phosphorous fertilizers. Soil Biol. Biochem. 2000, 32, 59–66. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, T.; Gilliam, F.S.; Gundersen, P.; Zhang, W.; Chen, H.; Mo, J. Interactive effects of nitrogen and phosphorus on soil microbial communities in a tropical forest. PLoS ONE 2013, 8, e61188. [Google Scholar] [CrossRef] [Green Version]
- Ullah, S.; Ai, C.; Huang, S.; Song, D.; Abbas, T.; Zhang, J.; Zhou, W.; He, P. Substituting ecological intensification of agriculture for conventional agricultural practices increased yield and decreased nitrogen losses in North China. Appl. Soil Ecol. 2020, 147, 103395. [Google Scholar] [CrossRef]




| Fertilization | N-P-K | N-P-K | N-P-K | N-P-K |
|---|---|---|---|---|
| Fluvo-Aquic Soil | RF N0 | RF N182 | CF N0 | CF N225 |
| 0–73–70 | 182–73–70 | 0–120–50 | 225–120–50 | |
| Black Soil | RF N0 | RF N180 | CF N0 | CF N251 |
| 0–75–90 | 200–75–90 | 0–145–100 | 251–145–100 |
| Treatments | TN | SOC | AP | NO3− | NH4+ | MBN | MBC | pH | C:N |
|---|---|---|---|---|---|---|---|---|---|
| Fluvo-Aquic Soil | |||||||||
| RF N0 | 1.18 b | 20.08 a | 16.61 b | 6.03 c | 1.38 a | 271 a | 74.17 b | 8.05 ab | 16.90 a |
| RF N182 | 1.28 a | 21.19 a | 14.87 b | 26.50 b | 1.48 a | 329 a | 88.81 a | 8.00 bc | 16.46 a |
| CF N0 | 1.22 ab | 20.60 a | 24.40 a | 11.44 c | 1.37 a | 273 a | 77.64 ab | 8.11 a | 16.80 a |
| CF N225 | 1.27 ab | 21.07 a | 18.80 b | 34.39 a | 1.50 a | 299 a | 79.15 ab | 7.97 c | 16.55 a |
| Significance | * | ns | * | *** | ns | ns | ns | ** | ns |
| Black Soil | |||||||||
| RF N0 | 1.00 b | 22.76 a | 53.81 b | 2.51 c | 1.28 bc | 62 a | 13.30 a | 5.82 a | 22.55 a |
| RF N180 | 1.21 a | 23.99 a | 43.02 c | 13.44 a | 3.04 a | 35 b | 10.43 ab | 5.56 ab | 19.80 b |
| CF N0 | 1.01 b | 23.79 a | 62.43 a | 2.38 c | 1.10 c | 77 a | 9.98 b | 5.91 a | 23.54 a |
| CF N251 | 1.22 a | 24.18 a | 52.80 b | 9.19 b | 2.49 ab | 21 b | 5.15 c | 5.23 b | 19.73 b |
| Significance | *** | ns | ** | *** | ** | ** | * | ** | *** |
| Treatments | TN | SOC | AP | NO3− | NH4+ | MBN | MBC | pH | C:N | Yield | Chao1 Index | Shannon Index |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fertilization | ** | ns | ** | ** | ** | ns | ns | ** | ** | ** | ** | ** |
| Soil Type | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** | ** |
| Interaction | ** | ns | ns | ** | ** | * | ** | ** | ** | ** | ** | ** |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ullah, S.; He, P.; Ai, C.; Zhao, S.; Ding, W.; Song, D.; Zhang, J.; Huang, S.; Abbas, T.; Zhou, W. How Do Soil Bacterial Diversity and Community Composition Respond under Recommended and Conventional Nitrogen Fertilization Regimes? Microorganisms 2020, 8, 1193. https://doi.org/10.3390/microorganisms8081193
Ullah S, He P, Ai C, Zhao S, Ding W, Song D, Zhang J, Huang S, Abbas T, Zhou W. How Do Soil Bacterial Diversity and Community Composition Respond under Recommended and Conventional Nitrogen Fertilization Regimes? Microorganisms. 2020; 8(8):1193. https://doi.org/10.3390/microorganisms8081193
Chicago/Turabian StyleUllah, Sami, Ping He, Chao Ai, Shicheng Zhao, Wencheng Ding, Dali Song, Jiajia Zhang, Shaohui Huang, Tanveer Abbas, and Wei Zhou. 2020. "How Do Soil Bacterial Diversity and Community Composition Respond under Recommended and Conventional Nitrogen Fertilization Regimes?" Microorganisms 8, no. 8: 1193. https://doi.org/10.3390/microorganisms8081193
APA StyleUllah, S., He, P., Ai, C., Zhao, S., Ding, W., Song, D., Zhang, J., Huang, S., Abbas, T., & Zhou, W. (2020). How Do Soil Bacterial Diversity and Community Composition Respond under Recommended and Conventional Nitrogen Fertilization Regimes? Microorganisms, 8(8), 1193. https://doi.org/10.3390/microorganisms8081193





