Deciphering Additional Roles for the EF-Tu, l-Asparaginase II and OmpT Proteins of Shiga Toxin-Producing Escherichia coli
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Construction of Isogenic Mutants
2.3. Cloning and Expression of the ansB and ompT Genes
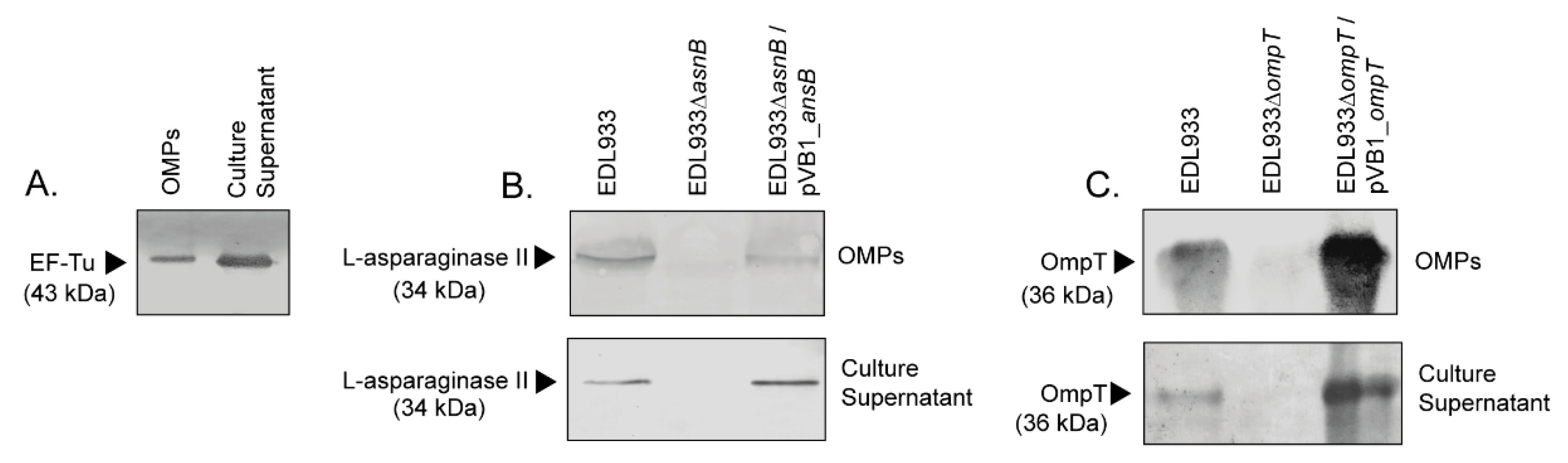
2.4. Western Blotting
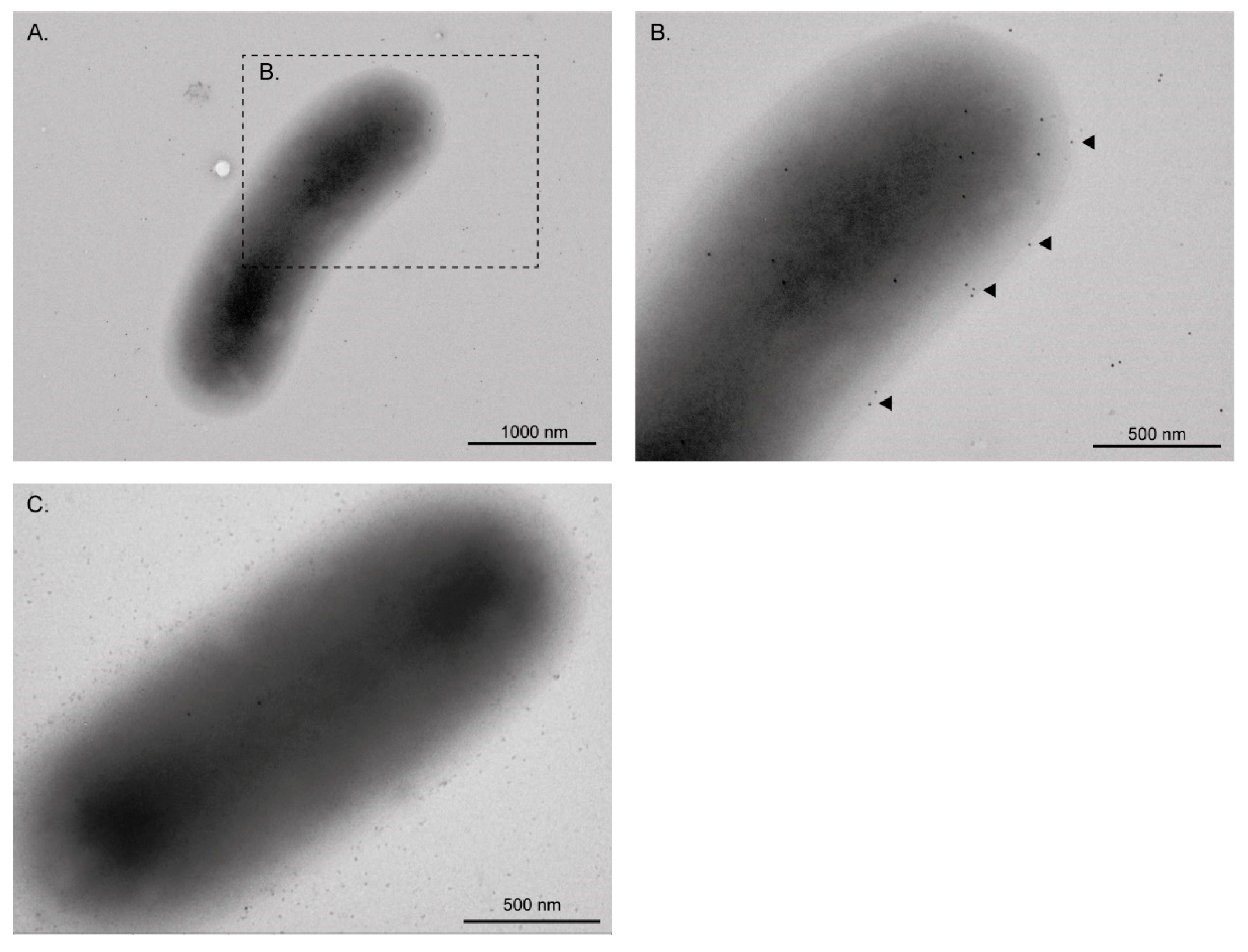
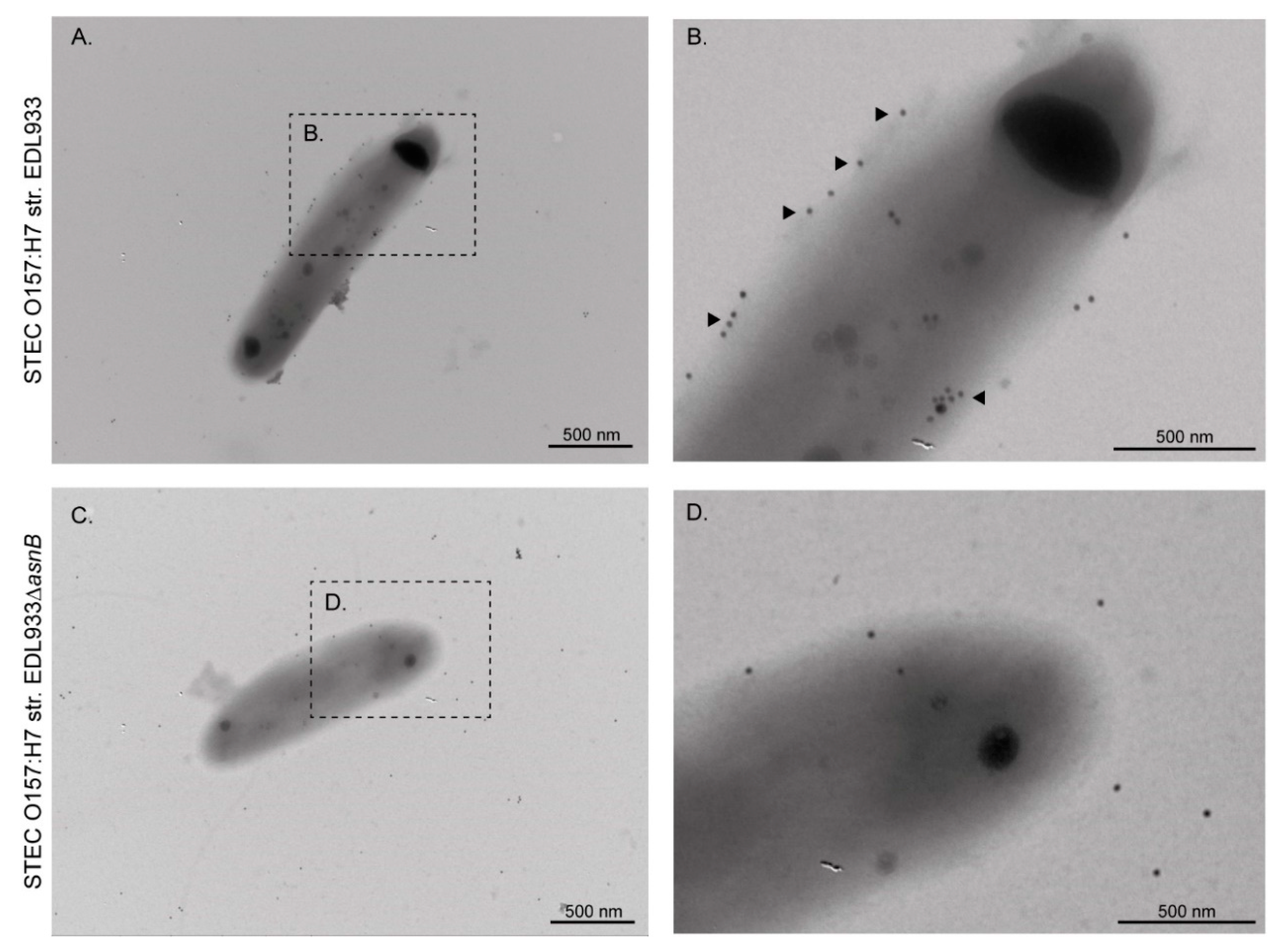
2.5. Transmission Electron Microscopy (TEM) and Immunogold Labeling
2.6. Proteomic Analysis of Outer Membrane Vesicles (OMVs)
2.7. Bacterial Adhesion Assays
2.8. Proliferation Assay
2.9. Statistical Analysis
3. Results
3.1. EF-Tu and l-Asparaginase II Are Secreted and Associated with the Surface of STEC O157:H7
3.2. The Monoclonal Anti-EF-Tu Antibody (mAb 900) Does Not Affect the Adhesion of STEC O157:H7 str. EDL933 to HT-29 Epithelial Cells, Fibronectin and Laminin
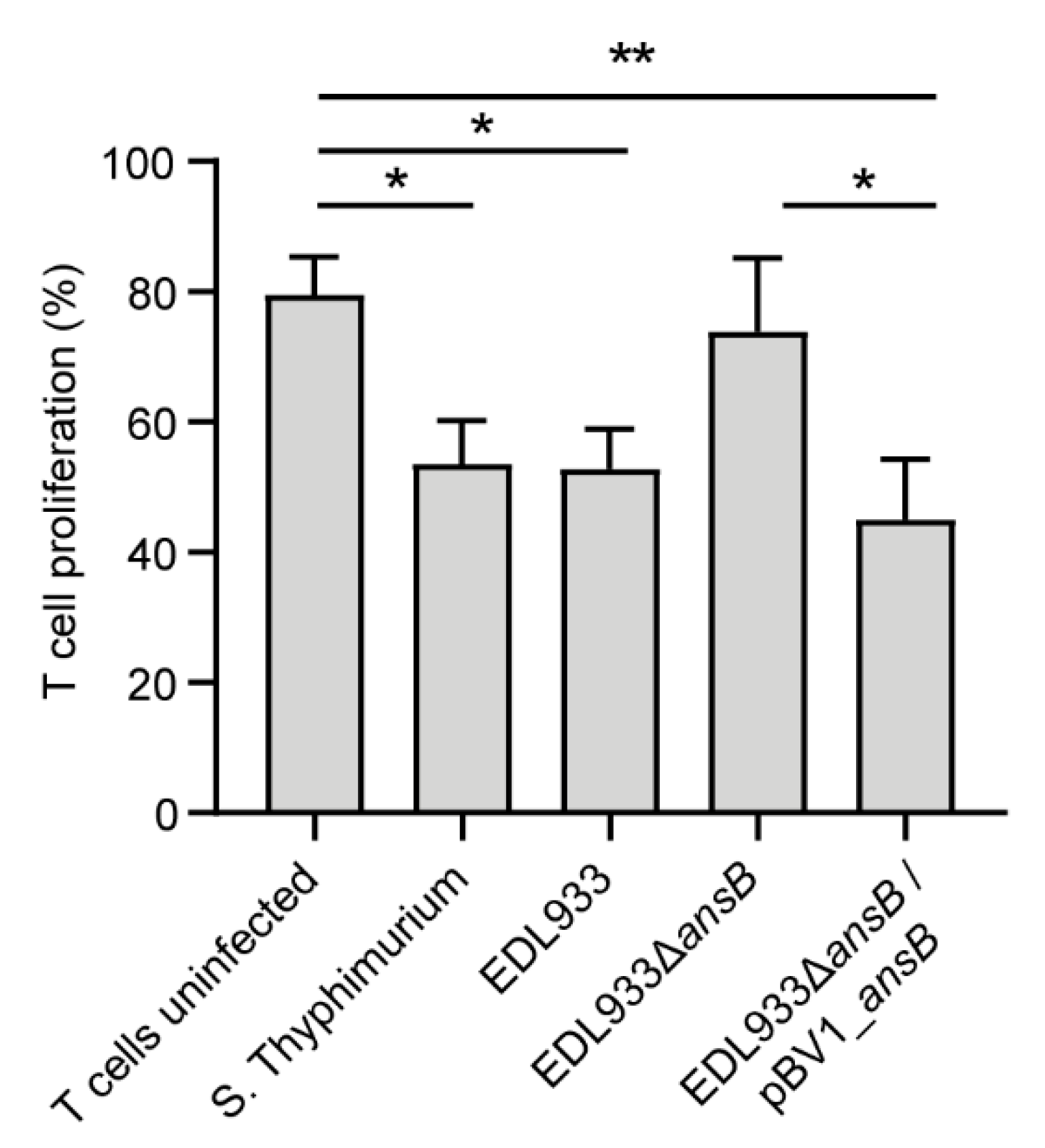
3.3. l-Asparaginase II Produced by STEC O157:H7 str. EDL933 Inhibits T Cell Proliferation
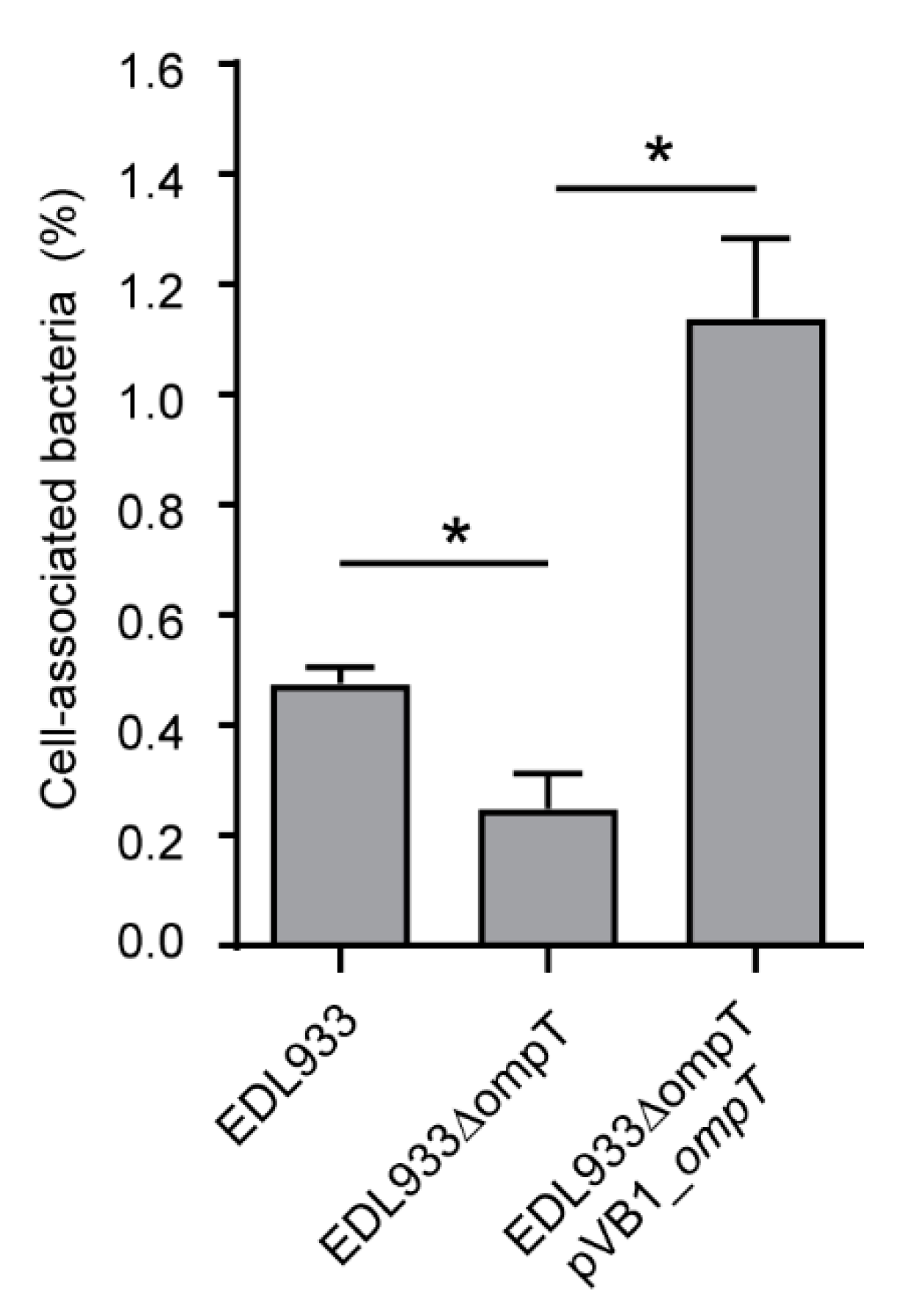
3.4. Deletion of The ompT Gene Affects The Adhesion of STEC O157:H7 str. EDL933 to HT-29 Cells
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Jeffery, C. Moonlighting proteins. Trends Biochem. Sci. 1999, 24, 8–11. [Google Scholar] [CrossRef]
- Jeffery, C. Moonlighting proteins: Old proteins learning new tricks. Trends Genet. 2003, 19, 415–417. [Google Scholar] [CrossRef]
- Henderson, B.; Martin, A. Bacterial virulence in the moonlight: Multitasking bacterial moonlighting proteins are virulence determinants in infectious disease. Infect. Immun. 2011, 79, 3476–3491. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Xia, Y.; Cui, J.; Gu, Z.; Song, Y.; Chen, Y.Q.; Chen, H.; Zhang, H.; Chen, W. The roles of moonlighting proteins in bacteria. Curr. Issues Mol. Biol. 2014, 16, 15–22. [Google Scholar]
- Harvey, K.L.; Jarocki, V.M.; Charles, I.G.; Djordjevic, S.P. The diverse functional roles of elongation factor tu (Ef-tu) in microbial pathogenesis. Front. Microbiol. 2019, 10, 1–19. [Google Scholar] [CrossRef]
- Kainulainen, V.; Korhonen, T.K. Dancing to another tune-adhesive moonlighting proteins in bacteria. Biology 2014, 3, 178–204. [Google Scholar] [CrossRef] [Green Version]
- O’Ryan, M.; Vidal, R.; del Canto, F.; Carlos Salazar, J.; Montero, D. Vaccines for viral and bacterial pathogens causing acute gastroenteritis: Part II: Vaccines for Shigella, Salmonella, enterotoxigenic E. coli (ETEC) enterohemorragic E. coli (EHEC) and Campylobacter jejuni. Hum. Vaccin. Immunother. 2015, 11, 601–619. [Google Scholar] [CrossRef] [Green Version]
- Egea, L.; Aguilera, L.; Giménez, R.; Sorolla, M.A.; Aguilar, J.; Badía, J.; Baldoma, L. Role of secreted glyceraldehyde-3-phosphate dehydrogenase in the infection mechanism of enterohemorrhagic and enteropathogenic Escherichia coli: Interaction of the extracellular enzyme with human plasminogen and fibrinogen. Int. J. Biochem. Cell Biol. 2007, 39, 1190–1203. [Google Scholar] [CrossRef]
- Da Silva, W.M.; Bei, J.; Amigo, N.; Valacco, M.P.; Amadio, A.; Zhang, Q.; Wu, X.; Yu, T.; Larzabal, M.; Chen, Z.; et al. Quantification of enterohemorrhagic Escherichia coli O157:H7 protein abundance by high-throughput proteome. PLoS ONE 2018, 13, e0208520. [Google Scholar] [CrossRef]
- Montero, D.; Orellana, P.; Gutiérrez, D.; Araya, D.; Salazar, J.C.; Prado, V.; Oñate, A.; Del Canto, F.; Vidal, R. Immunoproteomic analysis to identify Shiga toxin-producing Escherichia coli outer membrane proteins expressed during human infection. Infect. Immun. 2014, 82, 4767–4777. [Google Scholar] [CrossRef] [Green Version]
- Beck, B.D.; Arscott, P.G.; Jacobson, A. Novel properties of bacterial elongation factor Tu. Proc. Natl. Acad. Sci. USA 1978, 75, 1250–1254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soufo, H.J.D.; Reimold, C.; Breddermann, H.; Mannherz, H.G.; Graumann, P.L. Translation elongation factor EF-Tu modulates filament formation of actin-like MreB protein in vitro. J. Mol. Biol. 2015, 427, 1715–1727. [Google Scholar] [CrossRef] [Green Version]
- Soufo, H.J.D.; Reimold, C.; Linne, U.; Knust, T.; Gescher, J.; Graumann, P.L. Bacterial translation elongation factor EF-Tu interacts and colocalizes with actin-like MreB protein. Proc. Natl. Acad. Sci. USA 2010, 107, 3163–3168. [Google Scholar] [CrossRef] [Green Version]
- Caldas, T.D.; El Yaagoubi, A.; Richarme, G. Chaperone properties of bacterial elongation factor EF-Tu. J. Biol. Chem. 1998, 273, 11478–11482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kunert, A.; Losse, J.; Gruszin, C.; Kaendler, K.; Mikkat, S.; Volke, D.; Jokiranta, T.S.; Seeberger, H.; Hellwage, J.; Zipfel, P.F.; et al. Pseudomonas aeruginosa: Elongation Factor Tuf Is a Factor H and Plasminogen Binding Protein. J. Immunol. 2013, 179, 29792988. [Google Scholar]
- Wolff, D.G.; Castiblanco-Valencia, M.M.; Abe, C.M.; Monaris, D.; Morais, Z.M.; Souza, G.O.; Vasconcellos, S.A.; Isaac, L.; Abreu, P.A.E.; Barbosa, A.S. Interaction of Leptospira elongation factor Tu with plasminogen and complement factor H: A metabolic leptospiral protein with moonlighting activities. PLoS ONE 2013, 8, e81818. [Google Scholar] [CrossRef]
- Montes-García, J.F.; Chincoya Martinez, D.A.; Vaca Pacheco, S.; Vázquez Cruz, C.; Sanchez Alonso, P.; Xicohtencatl Cortes, J.; Trujillo-Ruiz, H.; Negrete-Abascal, E. Identification of two adhesins of Actinobacillus seminis. Small Rumin. Res. 2018, 167, 100–103. [Google Scholar] [CrossRef]
- Dallo, S.F.; Kannan, T.R.; Blaylock, M.W.; Baseman, J.B. Elongation factor Tu and E1 beta subunit of pyruvate dehydrogenase complex act as fibronectin binding proteins in Mycoplasma pneumoniae. Mol. Microbiol. 2002, 46, 1041–1051. [Google Scholar] [CrossRef]
- Dallo, S.F.; Zhang, B.; Denno, J.; Hong, S.; Tsai, A.; Haskins, W.; Ye, J.Y.; Weitao, T. Association of Acinetobacter baumannii EF-Tu with cell surface, outer membrane vesicles, and fibronectin. Sci. World J. 2012, 2012, 128705. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Wang, H.; Wang, J.; Feng, Z.; Wu, M.; Liu, B.; Xin, J.; Xiong, Q.; Liu, M.; Shao, G. Elongation factor thermo unstable (EF-Tu) moonlights as an adhesin on the surface of Mycoplasma hyopneumoniae by binding to fibronectin. Front. Microbiol. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Muñoz-Provencio, D.; Pérez-Martínez, G.; Monedero, V. Identification of Surface Proteins from Lactobacillus casei BL23 Able to Bind Fibronectin and Collagen. Probiotics Antimicrob. Proteins 2011, 3, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Kannan, T.R.; Baseman, J.B. The surface-exposed carboxyl region of Mycoplasma pneumoniae elongation factor Tu interacts with fibronectin. Infect. Immun. 2008, 76, 3116–3123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Liu, H.; Du, D.; Yu, Y.; Ma, C.; Jiao, F.; Yao, H.; Lu, C.; Zhang, W. Identification of Novel Laminin- and Fibronectin-binding Proteins by Far-Western Blot: Capturing the Adhesins of Streptococcus suis Type 2. Front. Cell. Infect. Microbiol. 2015, 5, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Koenigs, A.; Zipfel, P.F.; Kraiczy, P. Translation elongation factor Tuf of Acinetobacter baumannii is a plasminogen-binding protein. PLoS ONE 2015, 10, e0134418. [Google Scholar]
- Widjaja, M.; Harvey, K.L.; Hagemann, L.; Berry, I.J.; Jarocki, V.M.; Raymond, B.B.A.; Tacchi, J.L.; Gründel, A.; Steele, J.R.; Padula, M.P.; et al. Elongation factor Tu is a multifunctional and processed moonlighting protein. Sci. Rep. 2017, 7, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Mohan, S.; Hertweck, C.; Dudda, A.; Hammerschmidt, S.; Skerka, C.; Hallström, T.; Zipfel, P.F. Tuf of Streptococcus pneumoniae is a surface displayed human complement regulator binding protein. Mol. Immunol. 2014, 62, 249–264. [Google Scholar] [CrossRef]
- Granato, D.; Bergonzelli, G.E.; Pridmore, R.D.; Marvin, L.; Rouvet, M.; Corthésy-Theulaz, I.E. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 2004, 72, 2160–2169. [Google Scholar] [CrossRef] [Green Version]
- Dhanani, A.S.; Bagchi, T. The expression of adhesin EF-Tu in response to mucin and its role in Lactobacillus adhesion and competitive inhibition of enteropathogens to mucin. J. Appl. Microbiol. 2013, 115, 546–554. [Google Scholar] [CrossRef]
- Kesimer, M.; Kiliç, N.; Mehrotra, R.; Thornton, D.J.; Sheehan, J.K. Identification of salivary mucin MUC7 binding proteins from Streptococcus gordonii. BMC Microbiol. 2009, 9, 1–10. [Google Scholar] [CrossRef]
- Amimanan, P.; Tavichakorntrakool, R.; Fong-Ngern, K.; Sribenjalux, P.; Lulitanond, A.; Prasongwatana, V.; Wongkham, C.; Boonsiri, P.; Welbat, J.U.; Thongboonkerd, V. Elongation factor Tu on Escherichia coli isolated from urine of kidney stone patients promotes calcium oxalate crystal growth and aggregation. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Kullas, A.L.; McClelland, M.; Yang, H.J.; Tam, J.W.; Torres, A.; Porwollik, S.; Mena, P.; McPhee, J.B.; Bogomolnaya, L.; Andrews-Polymenis, H.; et al. l-asparaginase II produced by salmonella typhimurium inhibits t cell responses and mediates virulence. Cell Host Microbe 2012, 12, 791–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Batool, T.; Makky, E.A.; Jalal, M.; Yusoff, M.M. A Comprehensive Review on l-Asparaginase and Its Applications. Appl. Biochem. Biotechnol. 2016, 178, 900–923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vimal, A.; Kumar, A. The morpheein model of allosterism: A remedial step for targeting virulent l-asparaginase. Drug Discov. Today 2017, 22, 814–822. [Google Scholar] [CrossRef] [PubMed]
- Torres, A.; Luke, J.D.; Kullas, A.L.; Kapilashrami, K.; Botbol, Y.; Koller, A.; Tonge, P.J.; Chen, E.I.; Macian, F.; van der Velden, A.W.M. Asparagine deprivation mediated by Salmonella asparaginase causes suppression of activation-induced T cell metabolic reprogramming. J. Leukoc. Biol. 2016, 99, 387–398. [Google Scholar] [CrossRef] [Green Version]
- Stumpe, S.; Schmid, R.; Stephens, D.L.; Georgiou, G.; Bakker, E.P. Identification of OmpT as the protease that hydrolyzes the antimicrobial peptide protamine before it enters growing cells of Escherichia coli. J. Bacteriol. 1998, 180, 4002–4006. [Google Scholar] [CrossRef] [Green Version]
- Thomassin, J.-L.; Brannon, J.R.; Gibbs, B.F.; Gruenheid, S.; Le Moual, H. OmpT outer membrane proteases of enterohemorrhagic and enteropathogenic Escherichia coli contribute differently to the degradation of human LL-37. Infect. Immun. 2012, 80, 483–492. [Google Scholar] [CrossRef] [Green Version]
- Brannon, J.R.; Thomassin, J.-L.; Desloges, I.; Gruenheid, S.; Le Moual, H. Role of uropathogenic Escherichia coli OmpT in the resistance against human cathelicidin LL-37. FEMS Microbiol. Lett. 2013, 345, 64–71. [Google Scholar] [CrossRef] [Green Version]
- Premjani, V.; Tilley, D.; Gruenheid, S.; Le Moual, H.; Samis, J.A. Enterohemorrhagic Escherichia coli OmpT regulates outer membrane vesicle biogenesis. FEMS Microbiol. Lett. 2014, 355, 185–192. [Google Scholar] [CrossRef] [Green Version]
- He, X.L.; Wang, Q.; Peng, L.; Qu, Y.R.; Puthiyakunnon, S.; Liu, X.L.; Hui, C.Y.; Boddu, S.; Cao, H.; Huang, S.H. Role of uropathogenic Escherichia coli outer membrane protein T in pathogenesis of urinary tract infection. Pathog. Dis. 2015, 73, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Wan, L.; Guo, Y.; Hui, C.Y.; Liu, X.L.; Zhang, W.B.; Cao, H. The surface protease OmpT serves as Escherichia coli K1 adhesion in binding to human brain micro vascular endothelial cells. Pak. J. Pharm. Sci. 2014, 27, 617–624. [Google Scholar]
- Hejair, H.M.A.; Ma, J.; Zhu, Y.; Sun, M.; Dong, W.; Zhang, Y.; Pan, Z.; Zhang, W.; Yao, H. Role of outer membrane protein T in pathogenicity of avian pathogenic Escherichia coli. Res. Vet. Sci. 2017, 115, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Riley, L.W.; Remis, R.S.; Helgerson, S.D.; Mcgee, H.B.; Wells, J.G.; Davis, B.R.; Hebert, R.J.; Olcott, E.S.; Johnson, L.M.; Hargrett, N.T.; et al. Hemorrhagic Colitis Associated with a Rare Escherichia coli Serotype. N. Engl. J. Med. 1983, 308, 681–685. [Google Scholar] [CrossRef] [PubMed]
- Montero, D.A.; Del Canto, F.; Velasco, J.; Colello, R.; Padola, N.L.; Salazar, J.C.; Martin, C.S.; Oñate, A.; Blanco, J.; Rasko, D.A.; et al. Cumulative acquisition of pathogenicity islands has shaped virulence potential and contributed to the emergence of LEE-negative Shiga toxin-producing Escherichia coli strains. Emerg. Microbes Infect. 2019, 8, 486–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Céspedes, S.; Saitz, W.; Del Canto, F.; De la Fuente, M.; Quera, R.; Hermoso, M.; Muñoz, R.; Ginard, D.; Khorrami, S.; Girón, J.; et al. Genetic Diversity and Virulence Determinants of Escherichia coli Strains Isolated from Patients with Crohn’s Disease in Spain and Chile. Front. Microbiol. 2017, 8, 639. [Google Scholar] [CrossRef] [PubMed]
- Datsenko, K.; Wanner, B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 2000, 97, 6640–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutiérrez, D.; Pardo, M.; Montero, D.; Oñate, A.; Farfán, M.J.; Ruiz-Pérez, F.; Del Canto, F.; Vidal, R. TleA, a Tsh-Like Autotransporter Identified in a Human Enterotoxigenic Escherichia coli Strain. Infect. Immun. 2015, 83, 1893–1903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero, D.A.; Velasco, J.; Del Canto, F.; Puente, J.L.; Padola, N.L.; Rasko, D.A.; Farfán, M.; Salazar, J.C.; Vidal, R. Locus of Adhesion and Autoaggregation (LAA), a pathogenicity island present in emerging Shiga Toxin-producing Escherichia coli strains. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Farfan, M.J.; Cantero, L.; Vidal, R.; Botkin, D.J.; Torres, A.G. Long Polar Fimbriae of Enterohemorrhagic Escherichia coli O157:H7 Bind to Extracellular Matrix Proteins. Infect. Immun. 2011, 79, 3744–3750. [Google Scholar] [CrossRef] [Green Version]
- Falcón-Beas, C.; Tittarelli, A.; Mora-Bau, G.; Tempio, F.; Pérez, C.; Hevia, D.; Behrens, C.; Flores, I.; Falcón-Beas, F.; Garrido, P.; et al. Dexamethasone turns tumor antigen-presenting cells into tolerogenic dendritic cells with T cell inhibitory functions. Immunobiology 2019, 224, 697–705. [Google Scholar] [CrossRef]
- Del Canto, F.; Botkin, D.J.; Valenzuela, P.; Popov, V.; Ruiz-Perez, F.; Nataro, J.P.; Levine, M.M.; Stine, O.C.; Pop, M.; Torres, A.G.; et al. Identification of coli surface antigen 23, a novel adhesin of enterotoxigenic Escherichia coli. Infect. Immun. 2012, 80, 2791–2801. [Google Scholar] [CrossRef] [Green Version]
- Zuurmond, A.M.; Rundlöf, A.K.; Kraal, B. Either of the chromosomal tuf genes of E. coli K-12 can be deleted without loss of cell viability. Mol. Gen. Genet. 1999, 260, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Scotti, C.; Sommi, P.; Pasquetto, M.V.; Cappelletti, D.; Stivala, S.; Mignosi, P.; Savio, M.; Chiarelli, L.R.; Valentini, G.; Bolanos-Garcia, V.M.; et al. Cell-cycle inhibition by Helicobacter pylori l-asparaginase. PLoS ONE 2010, 5, e13892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hofreuter, D.; Novik, V.; Galán, J.E. Metabolic Diversity in Campylobacter jejuni Enhances Specific Tissue Colonization. Cell Host Microbe 2008, 4, 425–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, G.R.; Rosenbusch, J.P. Abundance and membrane association of elongation factor Tu in E. coli. Nature 1976, 261, 23–26. [Google Scholar] [CrossRef]
- Dombou, M.; Bhide, S.V.; Mizushima, S. Appearance of elongation factor Tu in the outer membrane of sucrose-dependent spectinomycin-resistant mutants of Escherichia coli. Eur. J. Biochem. 1981, 113, 397–403. [Google Scholar] [CrossRef]
- Cordwell, S.J.; Nouwens, A.S.; Walsh, B.J. Comparative proteomics of bacterial pathogens. Proteomics 2001, 1, 461–472. [Google Scholar] [CrossRef]
- Mujahid, S.; Pechan, T.; Wang, C. Improved solubilization of surface proteins from Listeria monocytogenes for 2-DE. Electrophoresis 2007, 28, 3998–4007. [Google Scholar] [CrossRef]
- Ying, T.; Wang, H.; Li, M.; Wang, J.; Wang, J.; Shi, Z.; Feng, E.; Liu, X.; Su, G.; Wei, K.; et al. Immunoproteomics of outer membrane proteins and extracellular proteins of Shigella flexneri 2a 2457T. Proteomics 2005, 5, 4777–4793. [Google Scholar] [CrossRef]
- George, D.T.; Mathesius, U.; Behm, C.A.; Verma, N.K. The periplasmic enzyme, AnsB, of Shigella flexneri modulates bacterial adherence to host epithelial cells. PLoS ONE 2014, 9, e94954. [Google Scholar] [CrossRef]
- Kukkonen, M.; Korhonen, T.K. The omptin family of enterobacterial surface proteases/adhesins: From housekeeping in Escherichia coli to systemic spread of Yersinia pestis. Int. J. Med. Microbiol. 2004, 294, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Feodorova, V.A.; Lyapina, A.M.; Zaitsev, S.S.; Khizhnyakova, M.A.; Sayapina, L.V.; Ulianova, O.V.; Ulyanov, S.S.; Motin, V.L. New promising targets for synthetic omptin-based peptide vaccine against gram-negative pathogens. Vaccines 2019, 7, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero, D.A.; Del Canto, F.; Salazar, J.C.; Céspedes, S.; Cádiz, L.; Arenas-Salinas, M.; Reyes, J.; Oñate, Á.; Vidal, R.M. Immunization of mice with chimeric antigens displaying selected epitopes confers protection against intestinal colonization and renal damage caused by Shiga toxin-producing Escherichia coli. npj Vaccines 2020, 5, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Primer Name | Sequence 5′ to 3′ * | Protocol | Source |
|---|---|---|---|
| d-ompT_F | ATCATTAAAACGATTGAATGGAGACCTTTTATGCGGGCGAAACTT CTGGGAATAGTCCTGACAACCCCTAGTGTAGGCTGGAGCTGCTTC | Deletion of the ompT gene | This work |
| d-ompT _R | CCAACCGGGGAGAAAATCTGGTTAACTTCGTTAAAAGGTGTACTT AAGACCAGCAGTAGTGATGAAGTTACATATGAATATCCTCCTTAG | Deletion of the ompT gene | This work |
| ompT-e _F | TCCAGCATATTAAACCACAACAATAATA | Confirmation of the ompT deletion | This work |
| ompTc_F | CATATGCGGGCGAAACTTCT | Cloning of the ompT gene | This work |
| ompTc_R | GGATCCTTAAAAGGTGTACTTA | Cloning of the ompT gene | This work |
| d-ansB _F | AAGGGATAATGCGTAGCGTTCACGTAACTGGAGGAATGAAATGGA GTTTTTCAAAAAGACGGTGTAGGCTGGAGCTGCTTC | Deletion of the ansB gene | This work |
| d-ansB _R | TGAAGTGAAAAAGCCCCGGCGCGATACCGGGGCGAAATGATTAGT ACTGATTGAAGATCTGCATATGAATATCCTCCTTAG | Deletion of the ansB gene | This work |
| ansB-e _F | CCTCAATCCAAACCGAGTGGAAA | Confirmation of the ansB deletion | This work |
| ansBc_F | CATATGATGGAGTTTTTCAAAAAGACGGCAC | Cloning of the ompT gene | This work |
| ansBC_R | GGATCCTTAGTACTGATTGAAGATCTGCTGG | Cloning of the ompT gene | This work |
| kanR_R | GCCATGATGGATACTTTCT | Confirmation of allelic replacement | [50] |
| Protein Name | Description | Coverage Percentage | No. of Peptides | Protein Score * | Protein Intensity Values | ||
|---|---|---|---|---|---|---|---|
| STEC O157:H7 | STEC O91:H21 | AIEC NRG857c | |||||
| A0A0H3EMH4 | EF-Tu | 69.04 | 27 | 1184.8 | 4.5 × 1014 | 1.6 × 1015 | 1.0 × 1015 |
| A0A0H3EPM6 | EF-Tu | 69.04 | 27 | 1184.8 | 4.5 × 1014 | 1.6 × 1015 | 1.0 × 1015 |
| A0A0H3ERC0 | OmpT | 14.20 | 6 | 124 | 1.6 × 1013 | 0 | 3.8 × 1013 |
| Experiment | HT-29 Cells | Fibronectin | Laminin | |||
|---|---|---|---|---|---|---|
| % Bacteria ± SD | p Value * | % Bacteria ± SD | p Value * | % Bacteria ± SD | p Value * | |
| No antibody | 100 ± 9.9 | - | 100 ± 7.3 | - | 100 ± 10.4 | |
| Anti-EF-Tu (1:10) | 107.7 ± 6.0 | >0.05 | 118 ± 23.3 | >0.05 | 135.2 ± 3.9 | >0.05 |
| Anti-EF-Tu (1:100) | 105.4 ± 17.2 | >0.05 | Not evaluated | - | Not evaluated | - |
| Mouse IgG (1:10) ** | 113.6 ± 5.6 | >0.05 | 131.9 ± 10.1 | >0.05 | 133.8 ± 20.6 | >0.05 |
| Mouse IgG (1:100) ** | 121.1 ± 10.6 | >0.05 | Not evaluated | - | Not evaluated | - |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres, A.N.; Chamorro-Veloso, N.; Costa, P.; Cádiz, L.; Del Canto, F.; Venegas, S.A.; López Nitsche, M.; Coloma-Rivero, R.F.; Montero, D.A.; Vidal, R.M. Deciphering Additional Roles for the EF-Tu, l-Asparaginase II and OmpT Proteins of Shiga Toxin-Producing Escherichia coli. Microorganisms 2020, 8, 1184. https://doi.org/10.3390/microorganisms8081184
Torres AN, Chamorro-Veloso N, Costa P, Cádiz L, Del Canto F, Venegas SA, López Nitsche M, Coloma-Rivero RF, Montero DA, Vidal RM. Deciphering Additional Roles for the EF-Tu, l-Asparaginase II and OmpT Proteins of Shiga Toxin-Producing Escherichia coli. Microorganisms. 2020; 8(8):1184. https://doi.org/10.3390/microorganisms8081184
Chicago/Turabian StyleTorres, Alexia N., Nayaret Chamorro-Veloso, Priscila Costa, Leandro Cádiz, Felipe Del Canto, Sebastián A. Venegas, Mercedes López Nitsche, Roberto F. Coloma-Rivero, David A. Montero, and Roberto M. Vidal. 2020. "Deciphering Additional Roles for the EF-Tu, l-Asparaginase II and OmpT Proteins of Shiga Toxin-Producing Escherichia coli" Microorganisms 8, no. 8: 1184. https://doi.org/10.3390/microorganisms8081184
APA StyleTorres, A. N., Chamorro-Veloso, N., Costa, P., Cádiz, L., Del Canto, F., Venegas, S. A., López Nitsche, M., Coloma-Rivero, R. F., Montero, D. A., & Vidal, R. M. (2020). Deciphering Additional Roles for the EF-Tu, l-Asparaginase II and OmpT Proteins of Shiga Toxin-Producing Escherichia coli. Microorganisms, 8(8), 1184. https://doi.org/10.3390/microorganisms8081184






