Effects of Maresin 1 (MaR1) on Colonic Inflammation and Gut Dysbiosis in Diet-Induced Obese Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Models and Experimental Design
2.2. DNA Extraction Microbiome
2.3. Metagenomic Analysis
2.4. Gene Expression in Colonic Mucosa by qRT-PCR
2.5. Statistics Analysis
3. Results
3.1. Effects of Maresin 1 (MaR1) Treatment on the Expression of Inflammatory Genes in Colon Mucosa of Diet-Induced Obese (DIO) Mice
3.2. Effects of MaR1 on Gut Microbiota Composition in DIO Mice
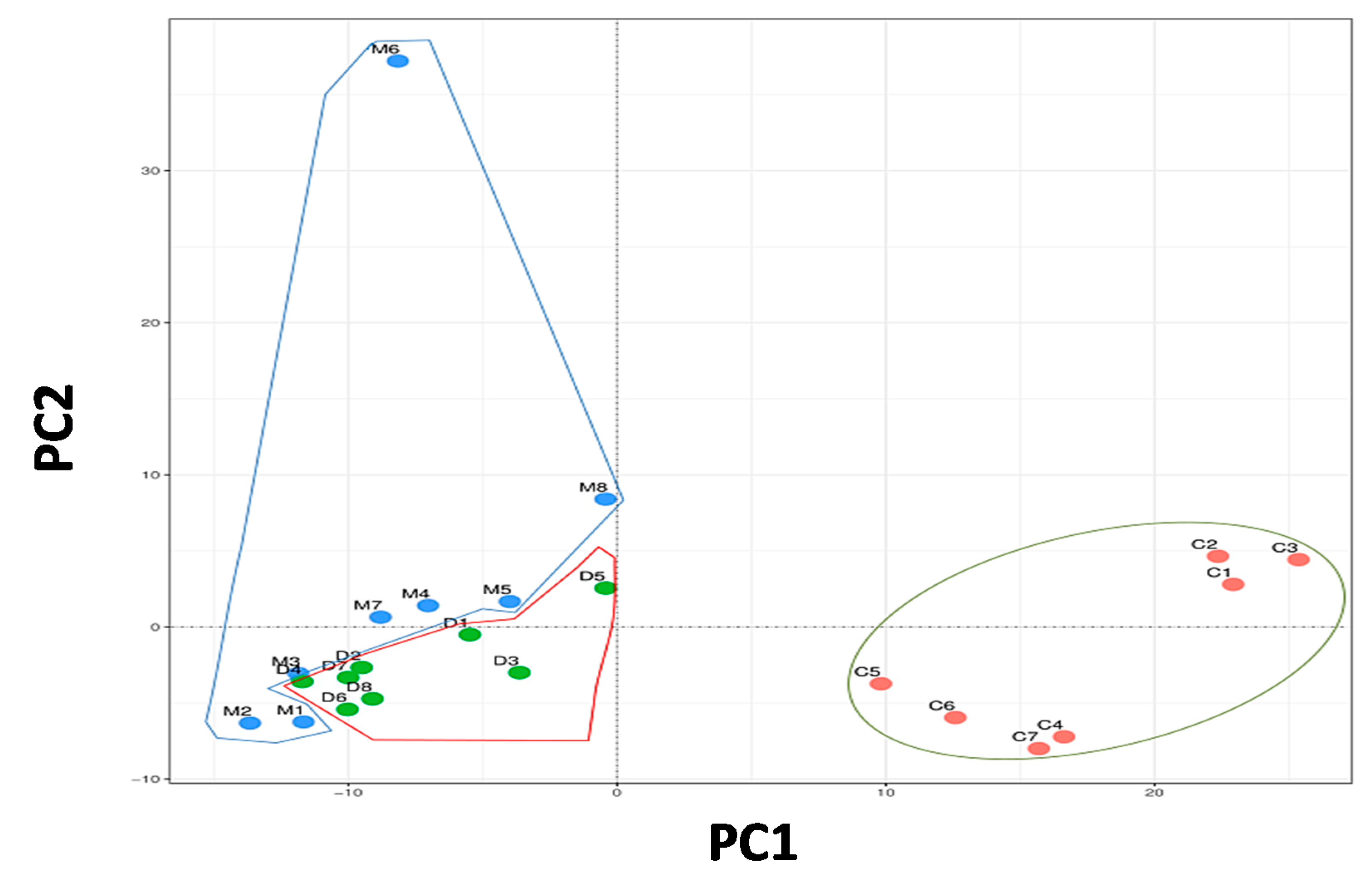
3.2.1. Principal Component Analysis
3.2.2. Taxonomic Changes in Faecal Microbiome
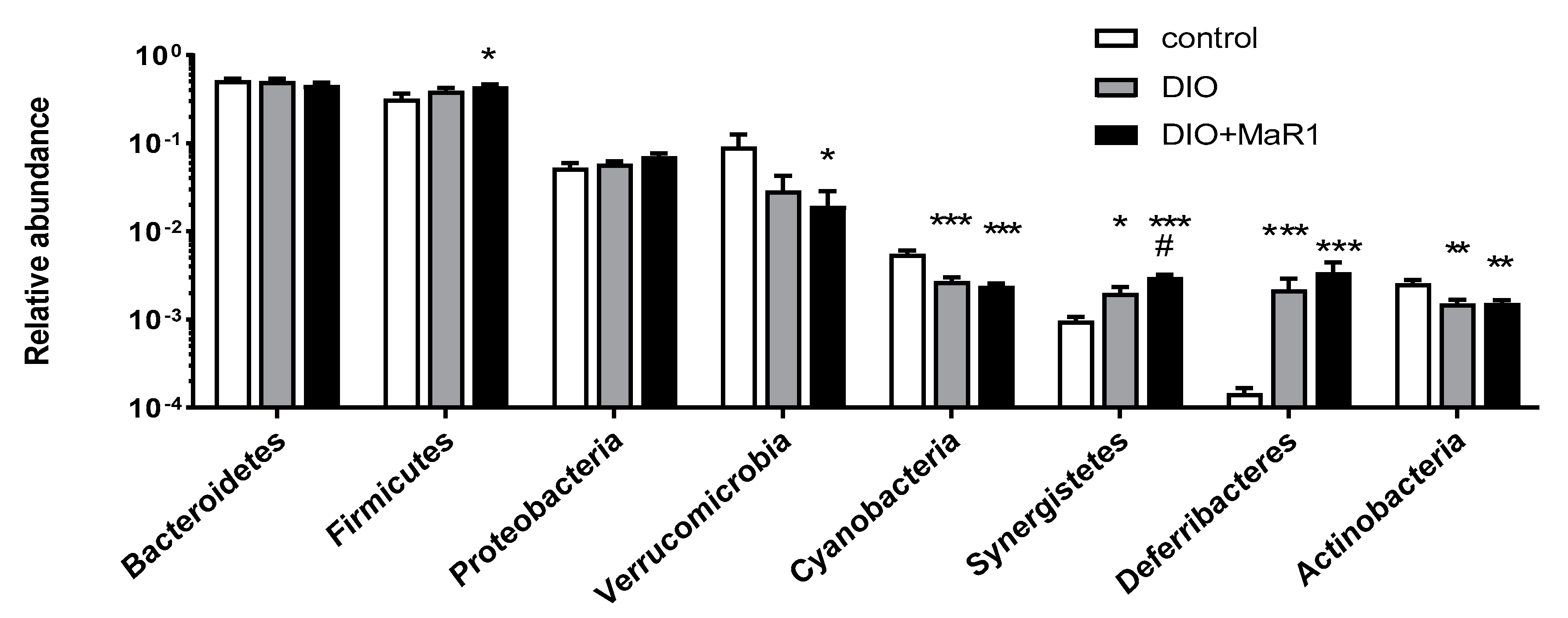
Analysis of Taxonomic Changes at the Phylum Level
Analysis of Taxonomic Changes at the Genus Level
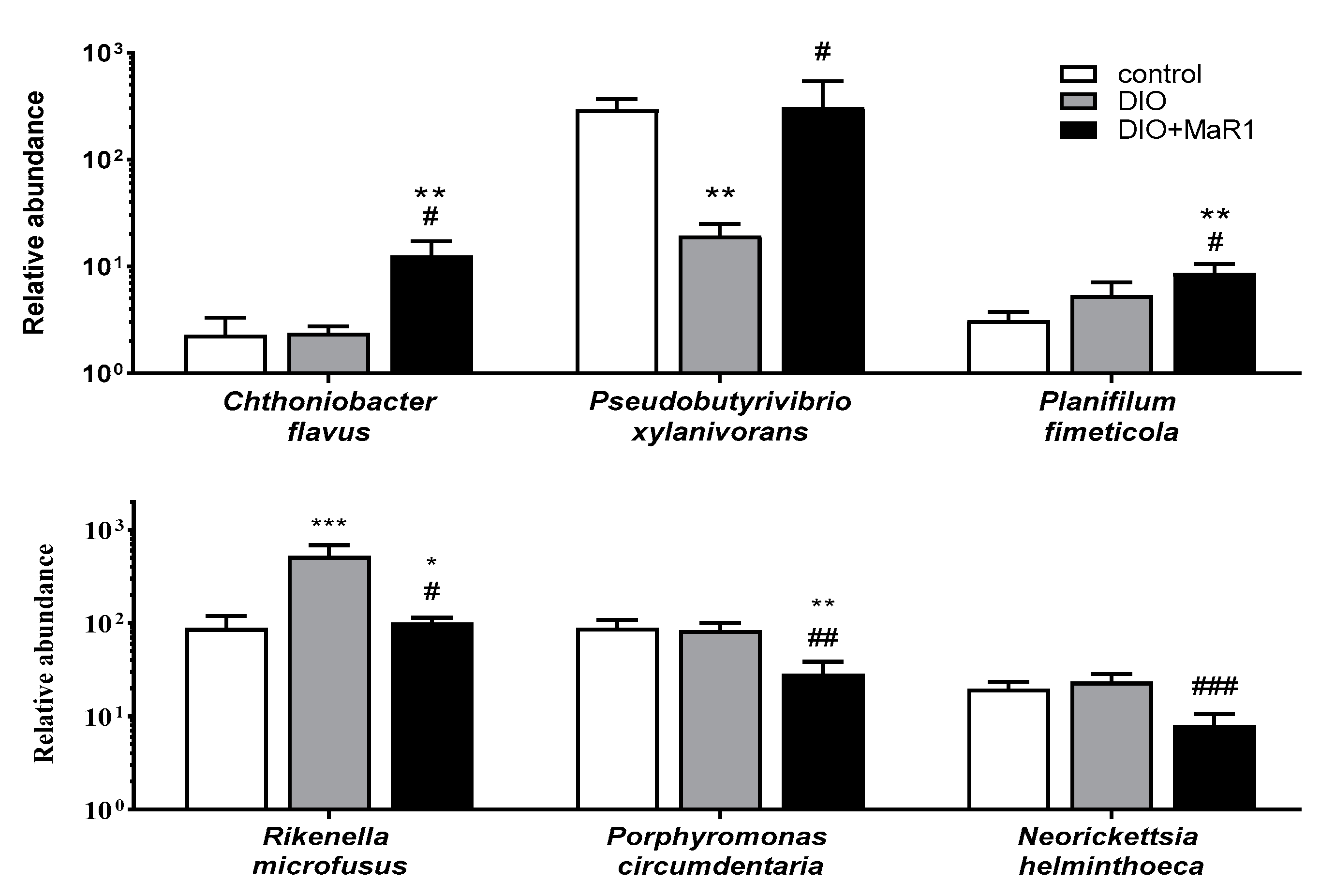
Analysis of Taxonomic Changes at the Species Level
3.3. Correlation between Specific Inflammatory Genes in Colonic Mucosa and the Gut Microbiota Composition at the Phylum Level
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Angelakis, E.; Armougom, F.; Million, M.; Raoult, D. The relationship between gut microbiota and weight gain in humans. Future Microbiol. 2012, 7, 91–109. [Google Scholar] [CrossRef] [PubMed]
- Neseer, M.I.; Bibi, F.; Alqahtani, M.H.; Chaudhary, A.G.; Azhar, E.I.; Kamal, M.A.; Yasir, M. Role of gut microbiota in obesity, type 2 diabetes and Alzheimer’s disease. CNS Neurol. Disord. Drug Targets 2014, 13, 305–311. [Google Scholar] [CrossRef]
- Wood, N.J. Microbiota: Dysbiosis driven by inflammasome deficiency exacerbates hepatic steatosis and governs rate of NAFLD progression. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 123. [Google Scholar] [CrossRef]
- Rinninella, E.; Raoul, P.; Cintoni, M.; Franceschi, F.; Miggiano, G.; Gasbarrini, A.; Mele, M. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms 2019, 7, 14. [Google Scholar] [CrossRef]
- Febbraio, M.A.; Reibe, S.; Shalapour, S.; Ooi, G.; Watt, M.J.; Karin, M. Preclinical models for studying NASH-driven HCC: How useful are they? Cell Metab. 2020, 29, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Palm, N.W.; de Zoete, M.R.; Flavell, R.A. Immune-microbiota interactions in health and disease. Clin. Immunol. 2015, 159, 122–127. [Google Scholar] [CrossRef]
- Ding, S.; Chi, M.M.; Scull, B.P.; Rigby, R.; Schwerbrock, N.M.J.; Magness, S.; Jobin, C.; Lund, P.K. High-fat diet: Bacteria interactions promote intestinal inflammation which precedes and correlates with obesity and insulin resistance in mouse. PLoS ONE 2010, 5, e12191. [Google Scholar] [CrossRef]
- Gil-Cardoso, K.; Ginés, I.; Pinent, M.; Ardévol, A.; Blay, M.; Terra, X. Effects of flavonoids on intestinal inflammation, barrier integrity and changes in gut microbiota during diet-induced obesity. Nutr. Res. Rev. 2016, 29, 234–248. [Google Scholar] [CrossRef]
- Ni, J.; Wu, G.D.; Albenberg, L.; Tomov, V.T. Gut microbiota and IBD: Causation or correlation? Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 573–584. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chen, W.D.; Wang, Y.D. The relationship between gut microbiota and inflammatory diseases: The role of macrophages. Front. Microbiol. 2020, 11, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Pavelock, N.; Masood, U.; Minchenberg, S.; Heisig, D. Effects of obesity on the course of inflammatory bowel disease. Baylor Univ. Med. Cent. Proc. 2019, 32, 14–17. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zhou, Q.; Li, H. Gut microbiota and nonalcoholic fatty liver disease: Insights on mechanisms and therapy. Nutrients 2017, 9, 1124. [Google Scholar] [CrossRef] [PubMed]
- Rosso, C.; Caviglia, G.P.; Younes, R.; Foglia, B.; Blanco, M.J.G.; Ribaldone, D.G.; Bugianesi, E. Circulating Zonulin is related to hepatic necroinflammation in patients with non alcoholic fatty liver disease. Clin. Lab. 2020, 66, 705–708. [Google Scholar] [CrossRef]
- Le Roy, T.; Llopis, M.; Lepage, P.; Bruneau, A.; Rabot, S.; Bevilacqua, C.; Martin, P.; Philippe, C.; Walker, F.; Bado, A.; et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 2013, 62, 1787–1794. [Google Scholar] [CrossRef]
- McGuckin, M.A.; Eri, R.; Simms, L.A.; Florin, T.H.J.; Radford-Smith, G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm. Bowel Dis. 2009, 15, 100–113. [Google Scholar] [CrossRef]
- Martínez-Fernández, L.; Laiglesia, L.M.; Huerta, A.E.; Martínez, J.A.; Moreno-Aliaga, M.J. Omega-3 fatty acids and adipose tissue function in obesity and metabolic syndrome. Prostaglandins Other Lipid Mediat. 2015, 121, 24–41. [Google Scholar] [CrossRef]
- Serhan, C.; Chiang, N.; Van Dyke, T.E. Resolving inflammation: Dual anti-inflammatory and pro-resolution lipid mediators. Nat. Rev. Immunol. 2008, 8, 349–361. [Google Scholar] [CrossRef]
- Neuhofer, A.; Zeyda, M.; Mascher, D.; Itariu, B.K.; Murano, I.; Leitner, L.; Hochbrugger, E.E.; Fraisl, P.; Cinti, S.; Serhan, C.N.; et al. Impaired local production of proresolving lipid mediators in obesity and 17-HDHA as a potential treatment for obesity-associated inflammation. Diabetes 2013, 62, 1945–1956. [Google Scholar] [CrossRef]
- Spite, M.; Clària, J.; Serhan, C.N. Resolvins, specialized proresolving lipid mediators, and their potential roles in metabolic diseases. Cell Metab. 2014, 19, 21–36. [Google Scholar] [CrossRef]
- Martínez-Fernández, L.; González-Muniesa, P.; Laiglesia, L.M.; Sáinz, N.; Prieto-Hontoria, P.L.; Escoté, X.; Odriozola, L.; Corrales, F.J.; Arbones-Mainar, J.M.; Martínez, J.A.; et al. Maresin 1 improves insulin sensitivity and attenuates adipose tissue inflammation in ob/ob and diet-induced obese mice. FASEB J. 2017, 31, 2135–2145. [Google Scholar] [CrossRef] [PubMed]
- Bento, A.F.; Claudino, R.F.; Dutra, R.C.; Marcon, R.; Calixto, J.B. Omega-3 fatty acid-derived mediators 17(R)-hydroxy docosahexaenoic acid, aspirin-triggered Resolvin D1 and Resolvin D2 prevent experimental colitis in mice. J. Immunol. 2011, 187, 1957–1969. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Yang, R.; Martinod, K.; Kasuga, K.; Pillai, P.S.; Porter, T.F.; Oh, S.F.; Spite, M. Maresins: Novel macrophage mediators with potent antiinflammatory and proresolving actions. J. Exp. Med. 2009, 206, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Serhan, C.N.; Dalli, J.; Karamnov, S.; Choi, A.; Park, C.-K.; Xu, Z.-Z.; Ji, R.-R.; Zhu, M.; Petasis, N.A. Macrophage proresolving mediator maresin 1 stimulates tissue regeneration and controls pain. FASEB J. 2012, 26, 1755–1765. [Google Scholar] [CrossRef]
- Laiglesia, L.M.; Lorente-Cebrián, S.; López-Yoldi, M.; Lanas, R.; Sáinz, N.; Martínez, J.A.; Moreno-Aliaga, M.J. Maresin 1 inhibits TNF-alpha-induced lipolysis and autophagy in 3T3-L1 adipocytes. J. Cell. Physiol. 2018, 233, 2238–2246. [Google Scholar] [CrossRef]
- Laiglesia, L.M.; Lorente-Cebrián, S.; Martínez-Fernández, L.; Sáinz, N.; Prieto-Hontoria, P.L.; Burrell, M.A.; Rodríguez-Ortigosa, C.M.; Martínez, J.A.; Moreno-Aliaga, M.J. Maresin 1 mitigates liver steatosis in ob/ob and diet-induced obese mice. Int. J. Obes. 2018, 42, 572–579. [Google Scholar] [CrossRef]
- Rius, B.; Duran-Güell, M.; Flores-Costa, R.; López-Vicario, C.; Lopategi, A.; Alcaraz-Quiles, J.; Casulleras, M.; José Lozano, J.; Titos, E.; Clària, J. The specialized proresolving lipid mediator maresin 1 protects hepatocytes from lipotoxic and hypoxia-induced endoplasmic reticulum stress. FASEB J. 2017, 31, 5384–5398. [Google Scholar] [CrossRef]
- Martínez-Fernández, L.; González-Muniesa, P.; Sáinz, N.; Laiglesia, L.M.; Escoté, X.; Martínez, J.A.; Moreno-Aliaga, M.J. Maresin 1 regulates hepatic FGF21 in diet-induced obese mice and in cultured hepatocytes. Mol. Nutr. Food Res. 2019, 63, e1900358. [Google Scholar] [CrossRef]
- Jung, T.W.; Kim, H.C.; El-Aty, A.M.A.; Jeong, J.H. Maresin 1 attenuates NAFLD by suppression of endoplasmic reticulum stress via AMPK-SERCA2b pathway. J. Biol. Chem. 2018, 293, 3981–3988. [Google Scholar] [CrossRef]
- Han, Y.H.; Shin, K.O.; Kim, J.Y.; Khadka, D.B.; Kim, H.J.; Lee, Y.M.; Cho, W.J.; Cha, J.Y.; Lee, B.J.; Lee, M.O. A maresin 1/RORα/12-lipoxygenase autoregulatory circuit prevents inflammation and progression of nonalcoholic steatohepatitis. J. Clin. Investig. 2019, 129, 1684–1698. [Google Scholar] [CrossRef]
- Marcon, R.; Bento, A.F.; Dutra, R.C.; Bicca, M.A.; Leite, D.F.P.; Calixto, J.B. Maresin 1, a proresolving lipid mediator derived from Omega-3 polyunsaturated fatty acids, exerts protective actions in murine models of colitis. J. Immunol. 2013, 191, 4288–4298. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Li, P.; Zhao, H.; Li, X. Maresin 1 alleviates dextran sulfate sodium-induced ulcerative colitis by regulating NRF2 and TLR4/NF-kB signaling pathway. Int. Immunopharmacol. 2020, 78, 106018. [Google Scholar] [CrossRef] [PubMed]
- Castilla-Madrigal, R.; Gil-Iturbe, E.; López de Calle, M.; Moreno-Aliaga, M.J.; Lostao, M.P. DHA and its derived lipid mediators MaR1, RvD1 and RvD2 block TNF-α inhibition of intestinal sugar and glutamine uptake in Caco-2 cells. J. Nutr. Biochem. 2020, 76, 108264. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Aliaga, M.J.; Pérez-Echarri, N.; Marcos-Gómez, B.; Larequi, E.; Gil-Bea, F.J.; Viollet, B.; Gimenez, I.; Martínez, J.A.; Prieto, J.; Bustos, M. Cardiotrophin-1 is a key regulator of glucose and lipid metabolism. Cell Metab. 2011, 14, 242–253. [Google Scholar] [CrossRef] [PubMed]
- Nagy-Szakal, D.; Mir, S.A.V.; Harris, R.A.; Dowd, S.E.; Yamada, T.; Lacorazza, H.D.; Tatevian, N.; Smith, C.W.; De Zoeten, E.F.; Klein, J.; et al. Loss of n-6 fatty acid induced pediatric obesity protects against acute murine colitis. FASEB J. 2015, 29, 3151–3159. [Google Scholar] [CrossRef]
- Etxeberria, U.; Arias, N.; Boqué, N.; Romo-Hualde, A.; Macarulla, M.T.; Portillo, M.P.; Milagro, F.I.; Martínez, J.A. Metabolic faecal fingerprinting of trans-resveratrol and quercetin following a high-fat sucrose dietary model using liquid chromatography coupled to high-resolution mass spectrometry. Food Funct. 2015, 6, 2758–2767. [Google Scholar] [CrossRef]
- Yang, Y.W.; Chen, M.K.; Yang, B.Y.; Huang, X.J.; Zhang, X.R.; He, L.Q.; Zhang, J.; Hua, Z.C. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl. Environ. Microbiol. 2015, 81, 6749–6756. [Google Scholar] [CrossRef]
- Hildebrand, F.; Tadeo, R.; Voigt, A.Y.; Bork, P.; Raes, J. LotuS: An efficient and user-friendly OTU processing pipeline. Microbiome 2014, 2, 1–7. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Edgar, R.C. UCHIME2: Improved chimera prediction for amplicon sequencing. BioRxiv 2016, 074252. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Meola, M.; Rifa, E.; Shani, N.; Delbès, C.; Berthoud, H.; Chassard, C. DAIRYdb: A manually curated reference database for improved taxonomy annotation of 16S rRNA gene sequences from dairy products. BMC Genom. 2019, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Chiang, N.; Libreros, S.; Norris, P.C.; De Rosa, X.; Serhan, C.N. Maresin 1 activates LGR6 receptor promoting phagocyte immunoresolvent functions Graphical abstract Find the latest version: Maresin 1 activates LGR6 receptor promoting phagocyte immunoresolvent functions. J. Clin. Investig. 2019, 129, 5294–5311. [Google Scholar] [CrossRef] [PubMed]
- Arora, T.; Bäckhed, F. The gut microbiota and metabolic disease: Current understanding and future perspectives. J. Intern. Med. 2016, 280, 339–349. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Ding, S.; Lund, P.K. Role of intestinal inflammation as an early event in obesity and insulin resistance. Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 328–333. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Tang, R.; Zhang, G.; Zeng, H.; Wood, R.J.; Liu, Z. High fat diet alters gut microbiota and the expression of paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediat. Inflamm. 2017, 2017, 9474896. [Google Scholar] [CrossRef]
- Warner, D.R.; Warner, J.B.; Hardesty, J.E.; Song, Y.L.; King, T.N.; Kang, J.X.; Chen, C.; Xie, S.; Yuan, F.; Prodhan, A.I.; et al. Decreased ω-6: ω-3 PUFA ratio attenuates ethanol-induced alterations in intestinal homeostasis, microbiota, and liver injury. J. Lipid Res. 2019, 60, 2034–2049. [Google Scholar] [CrossRef]
- Wilk, J.N.; Bilsborough, J.; Viney, J.L. The mdr1a-/- mouse model of spontaneous colitis: A relevant and appropriate animal model to study inflammatory bowel disease. Immunol. Res. 2005, 31, 151–159. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Gou, Y.K.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Hildebrandt, M.A.; Hoffmann, C.; Sherrill-Mix, S.A.; Keilbaugh, S.A.; Hamady, M.; Chen, Y.Y.; Knight, R.; Ahima, R.S.; Bushman, F.; Wu, G.D. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009, 137, 1716–1724.e2. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Sonne, S.B.; Feng, Q.; Chen, N.; Xia, Z.; Li, X.; Fang, Z.; Zhang, D.; Fjaere, E.; Midtbø, L.K.; et al. High-fat feeding rather than obesity drives taxonomical and functional changes in the gut microbiota in mice. Microbiome 2017, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Lobley, G.E.; Holtrop, G.; Ince, J.; Johnstone, A.M.; Louis, P.; Flint, H.J. Human colonic microbiota associated with diet, obesity and weight loss. Int. J. Obes. 2008, 32, 1720–1724. [Google Scholar] [CrossRef]
- Fernandes, J.; Su, W.; Rahat-Rozenbloom, S.; Wolever, T.M.S.; Comelli, E.M. Adiposity, gut microbiota and faecal short chain fatty acids are linked in adult humans. Nutr. Diabetes 2014, 4, e121. [Google Scholar] [CrossRef]
- Lam, Y.Y.; Ha, C.W.Y.; Hoffmann, J.M.A.; Oscarsson, J.; Dinudom, A.; Mather, T.J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Effects of dietary fat profile on gut permeability and microbiota and their relationships with metabolic changes in mice. Obesity 2015, 23, 1429–1439. [Google Scholar] [CrossRef]
- Chávez-Carbajal, A.; Pizano-Zárate, M.L.; Hernández-Quiroz, F.; Ortiz-Luna, G.F.; Morales-Hernández, R.M.; De Sales-Millán, A.; Hernández-Trejo, M.; García-Vite, A.; Beltrán-Lagunes, L.; Hoyo-Vadillo, C.; et al. Characterization of the gut microbiota of individuals at different T2D stages reveals a complex relationship with the host. Microorganisms 2020, 8, 94. [Google Scholar] [CrossRef]
- Shin, N.R.; Bose, S.; Wang, J.H.; Ansari, A.Z.; Lim, S.K.; Chin, Y.W.; Choi, H.S.; Kim, H. Flos Lonicera combined with metformin ameliorates hepatosteatosis and glucose intolerance in association with gut microbiota modulation. Front. Microbiol. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Walker, A.; Pfitzner, B.; Neschen, S.; Kahle, M.; Harir, M.; Lucio, M.; Moritz, F.; Tziotis, D.; Witting, M.; Rothballer, M.; et al. Distinct signatures of host-microbial meta-metabolome and gut microbiome in two C57BL/6 strains under high-fat diet. ISME J. 2014, 8, 2380–2396. [Google Scholar] [CrossRef]
- Etxeberria, U.; Hijona, E.; Aguirre, L.; Milagro, F.I.; Bujanda, L.; Rimando, A.M.; Martínez, J.A.; Portillo, M.P. Pterostilbene-induced changes in gut microbiota composition in relation to obesity. Mol. Nutr. Food Res. 2017, 61, 1500906. [Google Scholar] [CrossRef] [PubMed]
- Selvanantham, T.; Lin, Q.; Guo, C.X.; Surendra, A.; Fieve, S.; Escalante, N.K.; Guttman, D.S.; Streutker, C.J.; Robertson, S.J.; Philpott, D.J.; et al. NKT cell-deficient mice harbor an altered microbiota that fuels intestinal inflammation during chemically induced colitis. J. Immunol. 2016, 197, 4464–4472. [Google Scholar] [CrossRef] [PubMed]
- Chávez-Carbajal, A.; Nirmalkar, K.; Pérez-Lizaur, A.; Hernández-Quiroz, F.; Ramírez-Del-Alto, S.; García-Mena, J.; Hernández-Guerrero, C. Gut microbiota and predicted metabolic pathways in a sample of Mexican women affected by obesity and obesity plus metabolic syndrome. Int. J. Mol. Sci. 2019, 20, 438. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Chen, Z.; Guo, H.; He, D.; Zhao, H.; Wang, Z.; Zhang, W.; Liao, L.; Zhang, C.; Ni, L. The modulatory effect of infusions of green tea, oolong tea, and black tea on gut microbiota in high-fat-induced obese mice. Food Funct. 2016, 7, 4869–4879. [Google Scholar] [CrossRef]
- Pérez-Salcedo, L.; Herrera, D.; Esteban-Saltiveri, D.; León, R.; Jeusette, I.; Torre, C.; O’Connor, A.; González, I.; González, I. Isolation and identification of Porphyromonas spp. and other putative pathogens from cats with periodontal disease. J. Vet. Dent. 2013, 30, 208–213. [Google Scholar] [CrossRef]
- Lin, M.; Bachman, K.; Cheng, Z.; Daugherty, S.C.; Nagaraj, S.; Sengamalay, N.; Ott, S.; Godinez, A.; Tallon, L.J.; Sadzewicz, L.; et al. Analysis of complete genome sequence and major surface antigens of Neorickettsia helminthoeca, causative agent of salmon poisoning disease. Microb. Biotechnol. 2017, 10, 933–957. [Google Scholar] [CrossRef]
- Cepeljnik, T.; Zorec, M.; Kostanjsek, R.; Nekrep, F.V.; Marinsek-Logar, R. Is Pseudobutyrivibrio xylanivorans strain Mz5T suitable as a probiotic? An in vitro study. Folia Microbiol. 2003, 48, 339–345. [Google Scholar] [CrossRef]
- Rivière, A.; Selak, M.; Lantin, D.; Leroy, F.; De Vuyst, L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front. Microbiol. 2016, 7, 979. [Google Scholar] [CrossRef]
- Grilli, D.J.; Mansilla, M.E.; Giménez, M.C.; Sohaefer, N.; Ruiz, M.S.; Terebiznik, M.R.; Sosa, M.; Arenas, G.N. Pseudobutyrivibrio xylanivorans adhesion to epithelial cells. Anaerobe 2019, 56, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Im, D.S. Maresin-1 resolution with RORα and LGR6. Prog. Lipid Res. 2020, 78, 10–14. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Kim, D.; Kim, K.; Boo, K.; Yu, Y.S.; Kim, I.S.; Jeon, Y.; Im, S.K.; Lee, S.H.; Lee, J.M.; et al. RORα is crucial for attenuated inflammatory response to maintain intestinal homeostasis. Proc. Natl. Acad. Sci. USA 2019, 116, 21140–21149. [Google Scholar] [CrossRef] [PubMed]

| Downregulated in DIO + MaR1 vs. DIO | Upregulated in DIO + MaR1 vs. DIO | ||||
|---|---|---|---|---|---|
| Genus | LogFC | p | Genus | LogFC | P |
| Gluconacetobacter | −2.007 | 0.001 | Pseudobutyrivibrio | 2.771 | 0.009 |
| Pseudomonas | −1.595 | 0.001 | Chthoniobacter | 1.723 | 0.010 |
| Psychrobacter | −0.841 | 0.001 | Planifilum | 1.190 | 0.022 |
| Hymenobacter | −0.776 | 0.002 | Sphaerisporangium | 1.146 | 0.026 |
| Microvirus | −1.356 | 0.004 | Dethiosulfovibrio | 1.031 | 0.033 |
| Neorickettsia | −1.068 | 0.007 | Euzebya | 0.879 | 0.044 |
| Rikenella | −1.288 | 0.011 | Actinomyces | 0.681 | 0.045 |
| Thiothrix | −0.584 | 0.011 | Faecalibacterium | 1.000 | 0.049 |
| Candidatus Amoebophilus | −0.758 | 0.012 | |||
| Emticicia | −0.899 | 0.023 | |||
| Gillisia | −0.670 | 0.032 | |||
| Tetragenococcus | −0.549 | 0.032 | |||
| Thiothrix | −0.584 | 0.011 | |||
| Enterococcus | −1.378 | 0.033 | |||
| Polaribacter | −0.792 | 0.037 | |||
| Paraprevotella | −0.933 | 0.039 | |||
| Candidatus Glomeribacter | −0.647 | 0.042 | |||
| Coraliomargarita | −1.223 | 0.049 | |||
| Downregulated in DIO + MaR1 vs. DIO | Upregulated in DIO + MaR1 vs. DIO | ||||
|---|---|---|---|---|---|
| Species | LogFC | p | Species | LogFC | p |
| Porphyromonas circumdentaria | −1.055 | 0.003 | Actinomyces naturae | 0.853 | 0.019 |
| Microvirus enterobacteria | −1.264 | 0.007 | Slackia faecicanis | 0.959 | 0.025 |
| Coraliomargarita akajimensis | −1.201 | 0.011 | Euzebyatangerine | 1.051 | 0.027 |
| Rikenella microfusus | −1.116 | 0.015 | Planifilum fimeticola | 1.349 | 0.031 |
| Neorickettsia helminthoeca | −0.928 | 0.016 | Desulfovibrio litoralis | 1.397 | 0.039 |
| Chthoniobacter flavus | 1.742 | 0.041 | |||
| Pseudobutyrivibrio xylanivorans | 2.956 | 0.045 | |||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
León, I.C.; Quesada-Vázquez, S.; Sáinz, N.; Guruceaga, E.; Escoté, X.; Moreno-Aliaga, M.J. Effects of Maresin 1 (MaR1) on Colonic Inflammation and Gut Dysbiosis in Diet-Induced Obese Mice. Microorganisms 2020, 8, 1156. https://doi.org/10.3390/microorganisms8081156
León IC, Quesada-Vázquez S, Sáinz N, Guruceaga E, Escoté X, Moreno-Aliaga MJ. Effects of Maresin 1 (MaR1) on Colonic Inflammation and Gut Dysbiosis in Diet-Induced Obese Mice. Microorganisms. 2020; 8(8):1156. https://doi.org/10.3390/microorganisms8081156
Chicago/Turabian StyleLeón, Irene C., Sergio Quesada-Vázquez, Neira Sáinz, Elizabeth Guruceaga, Xavier Escoté, and María Jesús Moreno-Aliaga. 2020. "Effects of Maresin 1 (MaR1) on Colonic Inflammation and Gut Dysbiosis in Diet-Induced Obese Mice" Microorganisms 8, no. 8: 1156. https://doi.org/10.3390/microorganisms8081156
APA StyleLeón, I. C., Quesada-Vázquez, S., Sáinz, N., Guruceaga, E., Escoté, X., & Moreno-Aliaga, M. J. (2020). Effects of Maresin 1 (MaR1) on Colonic Inflammation and Gut Dysbiosis in Diet-Induced Obese Mice. Microorganisms, 8(8), 1156. https://doi.org/10.3390/microorganisms8081156







