What Drives the Diversity of the Most Abundant Terrestrial Cercozoan Family (Rhogostomidae, Cercozoa, Rhizaria)?
Abstract
:1. Introduction
- Rhogostomidae are exceptionally species-rich.
- Rhogostomidae are ubiquitously distributed, including marine, freshwater, and terrestrial habitats.
- Rhogostoma derived from aquatic relatives and colonized terrestrial habitats.
- Rhogostoma adapted to terrestrial environments with a deeper invagination of the aperture and a less elongated, but more spherical, cell shape to reduce water loss.
2. Material and Methods
2.1. Sampling and Culturing
2.2. Microscopic Observation
2.3. Phylogenetic Analyses
2.4. Statistical Analyses
3. Results
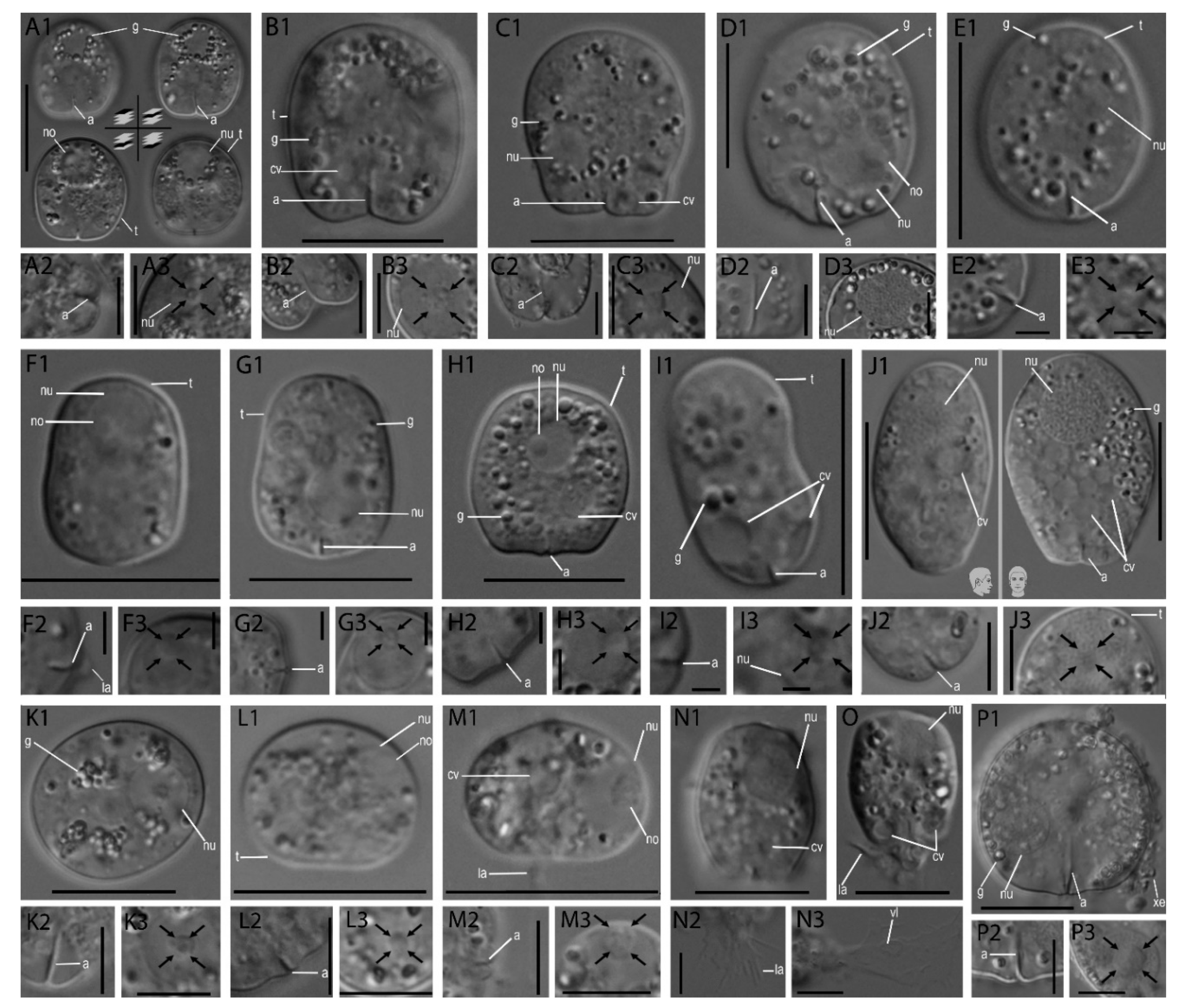
3.1. Morphological Analysis
3.2. Phylogeny
The Phylogenetic Position of Capsellina
3.3. Environmental Drivers of Species Turnover in Rhogostomidae
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Taxonomic Acts
References
- Bates, S.T.; Clemente, J.C.; Flores, G.E.; Walters, W.A.; Parfrey, L.W.; Knight, R.; Fierer, N. Global biogeography of highly diverse protistan communities in soil. ISME J. 2012, 7, 652–659. [Google Scholar] [CrossRef]
- Grossmann, L.; Jensen, M.; Heider, D.; Jost, S.; Glücksman, E.; Hartikainen, H.; Mahamdallie, S.S.; Gardner, M.; Hoffmann, D.; Bass, D.; et al. Protistan community analysis: Key findings of a large-scale molecular sampling. ISME J. 2016, 10, 2269–2279. [Google Scholar] [CrossRef] [Green Version]
- Lentendu, G.; Wubet, T.; Chatzinotas, A.; Wilhem, C.; Buscot, F.; Schlegel, M. Effects of long-term differential fertilization on eukaryotic microbial communities in an arable soil: A multiple barcoding approach. Mol. Ecol. 2014, 23, 3341–3355. [Google Scholar] [CrossRef]
- Bugge Harder, C.; Rønn, R.; Brejnrod, A.; Bass, D.; Abu Al-Soud, W.; Ekelund, F. Local diversity of heathland Cercozoa explored by in-depth sequencing Local diversity of heathland Cercozoa explored by in-depth sequencing. ISME J. 2016, 10, 2488–2497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiore-Donno, A.M.; Rixen, C.; Rippin, M.; Glaser, K.; Samolov, E.; Karsten, U.; Becker, B.; Bonkowski, M. New barcoded primers for efficient retrieval of cercozoan sequences in high-throughput environmental diversity surveys, with emphasis on worldwide biological soil crusts. Mol. Ecol. Resour. 2017, 18, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Fiore-Donno, A.M.; Richter-Heitmann, T.; Degrune, F.; Dumack, K.; Regan, K.M.; Marhan, S.; Boeddinghaus, R.S.; Rillig, M.C.; Friedrich, M.W.; Kandeler, E.; et al. Functional traits and spatio-temporal structure of a major group of soil protists (Rhizaria: Cercozoa) in a temperate grassland. Front. Microbiol. 2019, 10, 1332. [Google Scholar] [CrossRef] [Green Version]
- Matsunaga, K.; Kubota, K.; Harada, H. Molecular diversity of eukaryotes in municipal wastewater treatment processes as revealed by 18S rRNA gene analysis. Microbes Environ. 2014, 29, 401–407. [Google Scholar] [CrossRef] [Green Version]
- Remmas, N.; Melidis, P.; Paschos, G.; Statiris, E.; Ntougias, S. Protozoan indicators and extracellular polymeric substances alterations in an intermittently aerated membrane bioreactor treating mature landfill leachate. Environ. Technol. 2016, 3330, 53–64. [Google Scholar] [CrossRef] [PubMed]
- Degrune, F.; Dumack, K.; Fiore-Donno, A.M.; Bonkowski, M. Distinct communities of Cercozoa at different soil depths in a temperate agricultural field. FEMS Microbiol. Ecol. 2019, 95, fiz041. [Google Scholar] [CrossRef]
- Seppey, C.V.W.; Singer, D.; Fournier, B.; Mitchell, E.A.D.; Lara, E. Distribution patterns of soil microbial eukaryotes suggests widespread algivory by phagotrophic protists as an alternative pathway for nutrient cycling. Soil Biol. Biochem. 2017, 112, 68–76. [Google Scholar] [CrossRef]
- Dumack, K.; Flues, S.; Hermanns, K.; Bonkowski, M. Rhogostomidae (Cercozoa) from soils, roots and plant leaves (Arabidopsis thaliana): Description of Rhogostoma epiphylla sp. nov. and R. cylindrica sp. nov. Eur. J. Protistol. 2017, 60, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Dumack, K.; Öztoprak, H.; Rüger, L.; Bonkowski, M. Shedding light on the polyphyletic thecate amoeba genus plagiophrys: Transition of some of its species to rhizaspis (Tectofilosida, Thecofilosea, Cercozoa) and the establishment of sacciforma gen. nov. and rhogostomidae fam. nov. Cryomonadida, Thecofilos. Protist 2017, 168, 92–108. [Google Scholar] [CrossRef] [PubMed]
- Howe, A.T.; Bass, D.; Scoble, J.M.; Lewis, R.; Vickerman, K.; Arndt, H.; Cavalier-Smith, T. Novel cultured protists identify deep-branching environmental DNA clades of cercozoa: New genera tremula, micrometopion, minimassisteria, nudifila, peregrinia. Protist 2011, 162, 332–372. [Google Scholar] [CrossRef] [PubMed]
- Dobzhansky, T. Nothing in biology makes sense except in the light of evolution. Am. Biol. Teach. 1973, 35, 125–129. [Google Scholar] [CrossRef]
- Dumack, K.; Fiore-Donno, A.M.; Bass, D.; Bonkowski, M. Making sense of environmental sequencing data: Ecologically important functional traits of the protistan groups Cercozoa and Endomyxa (Rhizaria). Mol. Ecol. Resour. 2019, 20, 398–403. [Google Scholar] [CrossRef] [Green Version]
- Boenigk, J.; Ereshefsky, M.; Hoef-Emden, K.; Mallet, J.; Bass, D. Concepts in protistology: Species definitions and boundaries. Eur. J. Protistol. 2012, 48, 96–102. [Google Scholar] [CrossRef]
- Schlegel, M.; Meisterfeld, R. The species problem in protozoa revisited. Eur. J. Protistol. 2003, 39, 349–355. [Google Scholar] [CrossRef] [Green Version]
- Dayrat, B. Towards integrative taxonomy. Biol. J. Linn. Soc. 2005, 85, 407–417. [Google Scholar] [CrossRef]
- Will, K.; Mishler, B.; Wheeler, Q. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 2005, 54, 844–851. [Google Scholar] [CrossRef]
- Kosakyan, A.; Gomaa, F.; Lara, E.; Lahr, D.J.G. Current and future perspectives on the systematics, taxonomy and nomenclature of testate amoebae. Eur. J. Protistol. 2016, 55, 1–13. [Google Scholar] [CrossRef]
- Dumack, K.; Bonkowski, M.; Clauß, S.; Völcker, E. Phylogeny and redescription of the testate amoeba Diaphoropodon archeri (Chlamydophryidae, Thecofilosea, Cercozoa), De Saedeleer 1934, and annotations on the polyphyly of testate amoebae with agglutinated tests in the Cercozoa. J. Eukaryot. Microbiol. 2018, 65, 308–314. [Google Scholar] [CrossRef] [PubMed]
- Dumack, K.; Siemensma, F. Shell colour in Cercozoa; a simple trait to distinguish Thecofilosea from Imbricatea? Protist 2020, 171, 125718. [Google Scholar] [CrossRef] [PubMed]
- Belar, K. Untersuchungen über thecamöben der clamydophrys-gruppe. Arch Protistenkd 1921, 43, 287–354. [Google Scholar]
- Jauss, R.; Walden, S.; Fiore-Donno, A.; Dumack, K.; Schaffer, S.; Wolf, R.; Schlegel, M.; Bonkowski, M. From forest soil to the canopy: Increased habitat diversity does not increase species richness of Cercozoa and Oomycota in tree canopies. Mol. Ecol. 2020. [Google Scholar] [CrossRef]
- Heger, T.J.; Giesbrecht, I.J.W.; Gustavsen, J.; del Campo, J.; Kellogg, C.T.E.; Hoffman, K.M.; Lertzman, K.; Mohn, W.W.; Keeling, P.J. High-throughput environmental sequencing reveals high diversity of litter and moss associated protist communities along a gradient of drainage and tree productivity. Environ. Microbiol. 2018, 20, 1185–1203. [Google Scholar] [CrossRef] [PubMed]
- Mcfadden, G.; Melkonian, M. Use of Hepes buffer for micro algal culture media and fixation for electron microscopy. Phycologia 1986, 25, 551–557. [Google Scholar] [CrossRef]
- Bonkowski, M. Microcosm Approaches to Investigate Multitrophic Interactions between Microbial Communities in the Rhizosphere of Plants. In Methods in Rhizosphere Biology Research; Reinhardt, D., Sharma, A.K., Eds.; Springer: Singapore, 2019; pp. 255–270. [Google Scholar]
- Medlin, L.; Elwood, H.J.; Stickel, S.; Sogin, M.L. The characterization of enzymatically amplified eukaryotic 16S-like rRNA-coding regions. Gene 1988, 71, 491–499. [Google Scholar] [CrossRef] [Green Version]
- Quintela-Alonso, P.; Nitsche, F.; Arndt, H. Molecular characterization and revised systematics of Microdiaphanosoma arcuatum (Ciliophora, Colpodea). J. Eukaryot. Microbiol. 2011, 58, 114–119. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in Performance and usability article fast track. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Gouy, M.; Guindon, S.; Gascuel, O. Sea view version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2019. Available online: http://www.R-project.org/ (accessed on 1 March 2020).
- Borcard, D.; Gillet, F.; Legendre, P. Numerical Ecology with R; Springer: Berlin/Heidelberg, Germany, 2011. [Google Scholar]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package Version 2.5-6. Available online: http://CRAN.R-project.org/package=vegan (accessed on 1 March 2020).
- Schurko, A.M.; Logsdon, J. Using a Meiosis Detection Toolkit to Investigate Ancient Asexual ‘Scandals’ and the Evolution of Sex. Bioessays 2008, 30, 579–589. [Google Scholar] [CrossRef] [PubMed]
- Hofstatter, P.G.; Lahr, D.J.G. All Eukaryotes are sexual, unless proven otherwise. BioEssays 2019, 41, 1800246. [Google Scholar] [CrossRef]
- Chi, J.; Parrow, M.W.; Dunthorn, M. Cryptic sex in Symbiodinium (alveolata, dinoflagellata) is supported by an inventory of meiotic genes. J. Eukaryot. Microbiol. 2014, 61, 322–327. [Google Scholar] [CrossRef]
- Engelen, S.; Hingamp, P.; Sieracki, M.; Vargas, C.; Audic, S.; Henry, N.; Decelle, J.; Mahé, F.; Logares, R.; Lara, E.; et al. Eukaryotic plankton diversity in the sunlit ocean. Science (80-) 2015, 348, 1261605. [Google Scholar]
- Bork, P.; Bowler, C.; De Vargas, C.; Gorsky, G.; Karsenti, E.; Wincker, P. Tara Oceans studies plankton at Planetary scale. Science (80-) 2015, 348, 873. [Google Scholar] [CrossRef] [Green Version]
- Mahé, F.; de Vargas, C.; Bass, D.; Czech, L.; Stamatakis, A.; Lara, E.; Singer, D.; Mayor, J.; Bunge, J.; Sernaker, S.; et al. Parasites dominate hyperdiverse soil protist communities in Neotropical rainforests. Nat. Ecol. Evol. 2017, 1, 91. [Google Scholar] [CrossRef] [Green Version]
- Singer, D.; Mitchell, E.A.D.; Payne, R.J.; Blandenier, Q.; Duckert, C.; Fernández, L.D.; Fournier, B.; Hernández, C.E.; Granath, G.; Rydin, H.; et al. Dispersal limitations and historical factors determine the biogeography of specialized terrestrial protists. Mol. Ecol. 2019, 28, 3089–3100. [Google Scholar] [CrossRef]
- Singer, D.; Kosakyan, A.; Seppey, C.V.W.; Pillonel, A.; Fernández, L.D.; Fontaneto, D.; Mitchell, E.A.D.; Lara, E. Environmental filtering and phylogenetic clustering correlate with the distribution patterns of cryptic protist species. Ecology 2018, 99, 904–914. [Google Scholar] [CrossRef] [Green Version]
- Dumack, K.; Pundt, J.; Bonkowski, M. Food Choice experiments indicate selective fungivorous predation in Fisculla terrestris (Thecofilosea, Cercozoa). J. Eukaryot. Microbiol. 2018, 66, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Simonin, M.; Dasilva, C.; Terzi, V.; Ngonkeu, E.L.M.; Diouf, D.; Kane, A.; Béna, G.; Moulin, L. Influence of plant genotype and soil on the wheat rhizosphere microbiome: Evidences for a core microbiome across eight African and European soils. FEMS Microbiol. Ecol. 2020, 96, fiaa067. [Google Scholar] [CrossRef] [PubMed]
- Rossmann, M.; Pérez-Jaramillo, J.E.; Kavamura, V.N.; Chiaramonte, J.B.; Dumack, K.; Fiore-Donno, A.M.; Mendes, L.W.; Ferreira, M.C.; Bonkowski, M.; Raaijmakers, J.M.; et al. Multitrophic interactions in the rhizosphere microbiome of wheat: From bacteria and fungi to protists. FEMS Microbiol. Ecol. 2020, 96, fiaa032. [Google Scholar] [CrossRef]
- Ploch, S.; Rose, L.E.; Bass, D.; Bonkowski, M. High diversity revealed in leaf-associated protists (Rhizaria: Cercozoa) of brassicaceae. J. Eukaryot. Microbiol. 2016, 63, 635–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Strain | CCAP Reference | Sampling Spots | Coordinates | Isolation Date | Habitat | Used Primers | Sequence Length (nt) |
|---|---|---|---|---|---|---|---|
| WM | 1966/8 | Germany, Rostock, Warnemünde | 54.180267, 12.080450 | December 2018 | Sand beach, close to the Baltic Sea, algae-rich soil crust sample | EukA, 616F, 963R, EukB | 1718 |
| il-I | 1966/13 | Germany, Inden | 50.875, 6.325 | May 2019 | Monoculture, bur clover (Medicago sativa) | EukA, 590F, 1300R, EukB | 1627 |
| IGS | 1966/12 | Germany, Inden | 50.886, 6.317 | May 2019 | Monoculture, Barley shoot | EukA, 590F, 1300R, EukB | 1767 |
| 3A | 1966/11 | Germany, Rostock, Warnemünde | 54.180267, 12.080450 | February 2019 | Sand beach, close to the Baltic Sea, soil crust sample | EukA, cercomix, 963R, EukB | 1619 |
| B10 | 1966/6 | Germany, Cologne | 50.927186, 6.935997 | December 2018 | Courtyard of the Cologne Biocenter, leaf samples, phyllosphere | EukA, cercomix, 963R, EukB | 1376 |
| K8 | 1966/14 | Germany, Cologne | 51.025008, 6.751871 | April 2019 | Puddle with moss | EukA, cercomix, 963R, EukB | 1606 |
| K9 | 1966/15 | Germany, Cologne | 51.025041, 6.751496 | April 2019 | Litter from an Atlas cedar (Cedrus atlantica) | EukA, cercomix, 963R, EukB | 1686 |
| W2 | 1996/18 | Austria, Vienna | 48.200537, 16.370177 | February 2019 | Charles‘ Square, litter | EukA, 616F, 963R, EukB | 1719 |
| RC | 1966/5 | Germany, Cologne | 50.927186, 6.935997 | October 2017 | Courtyard of the Cologne Biocenter, leaf samples, phyllosphere | EukA, cercomix, 963R, EukB | 1720 |
| 1A | 1966/9 | Hilversum, Netherlands | 52.247196, 5.167029 | March 2019 | Zanderij Crailoo, large pond, freshwater sample | EukA, cercomix, 963R, EukB | 1708 |
| WH4 | 1944/20 | Germany, Wendershagen | 50.899719, 7.735686 | May 2019 | Lichen from apple tree trunk | EukA, 590F, 1300R, EukB | 1386 |
| W3 | 1966/19 | Austria, Vienna | 48.207667, 16.366056 | February 2019 | Hofburg, lichen from cut branches | EukA, 616F, 963R, EukB | 1721 |
| 1B | 1966/10 | Hilversum, Netherlands | 52.247196, 5.167029 | March 2019 | Zanderij Crailoo, large pond, freshwater sample | EukA, cercomix, 963R, EukB | 1719 |
| TG4.2-II | 1966/16 | Germany, Cologne | 50.920378, 7.105548 | December 2018 | Königsforst, forest litter | EukA, 590F, 1300R, EukB | 909 |
| TG4.2-IV | 1966/17 | Germany, Cologne | 50.920378, 7.105548 | December 2018 | Königsforst, forest litter | EukA, cercomix, 963R, EukB | 1721 |
| B14 | 1966/7 | Germany, Cologne | 50.927186, 6.935997 | December 2018 | Courtyard of the Cologne Biocenter, leaf samples, phyllosphere | EukA, 590F, 1300R, EukB | 1761 |
| Used Primer | Primer Sequence |
|---|---|
| EukA [28] | general eukaryotic primer 5′-CCGAATTCGTCGACAACCTGGTTGATCCTGCCAGT-3′ |
| EukB [28] | general eukaryotic primer 5′-CCCGGGATCCAAGCTTGATCCTTCTGCAGGTTCACCTAC-3′ |
| 590F [29] | general eukaryotic primer 5′-CGGTAATTCCAGCTCCAATAGC-3′ |
| 1300R [29] | general eukaryotic primer 5′-CACCAACTAAGAACGGCCATGC-3′ |
| S963R_Cerco [5] | Cercozoa specific primer 5′- CAACTTTCGTTCTTGATTAAA-3′ |
| S616F [5] * | Cercozoa specific primer 5′-TTAAAAAGCTCGTAGTTG-3′ |
| S616F_Eocer [5] * | Cercozoa specific primer 5′-TTAAAAAGCGCGTAGTTG-3′ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Öztoprak, H.; Walden, S.; Heger, T.; Bonkowski, M.; Dumack, K. What Drives the Diversity of the Most Abundant Terrestrial Cercozoan Family (Rhogostomidae, Cercozoa, Rhizaria)? Microorganisms 2020, 8, 1123. https://doi.org/10.3390/microorganisms8081123
Öztoprak H, Walden S, Heger T, Bonkowski M, Dumack K. What Drives the Diversity of the Most Abundant Terrestrial Cercozoan Family (Rhogostomidae, Cercozoa, Rhizaria)? Microorganisms. 2020; 8(8):1123. https://doi.org/10.3390/microorganisms8081123
Chicago/Turabian StyleÖztoprak, Hüsna, Susanne Walden, Thierry Heger, Michael Bonkowski, and Kenneth Dumack. 2020. "What Drives the Diversity of the Most Abundant Terrestrial Cercozoan Family (Rhogostomidae, Cercozoa, Rhizaria)?" Microorganisms 8, no. 8: 1123. https://doi.org/10.3390/microorganisms8081123
APA StyleÖztoprak, H., Walden, S., Heger, T., Bonkowski, M., & Dumack, K. (2020). What Drives the Diversity of the Most Abundant Terrestrial Cercozoan Family (Rhogostomidae, Cercozoa, Rhizaria)? Microorganisms, 8(8), 1123. https://doi.org/10.3390/microorganisms8081123






