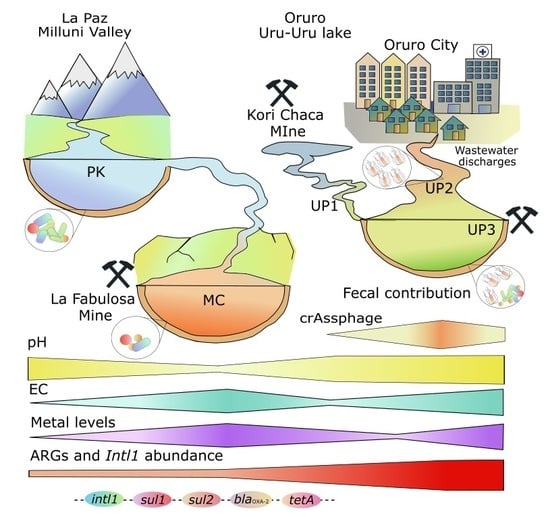
Fecal Pollution Drives Antibiotic Resistance and Class 1 Integron Abundance in Aquatic Environments of the Bolivian Andes Impacted by Mining and Wastewater
Abstract
1. Introduction
2. Materials and Methods
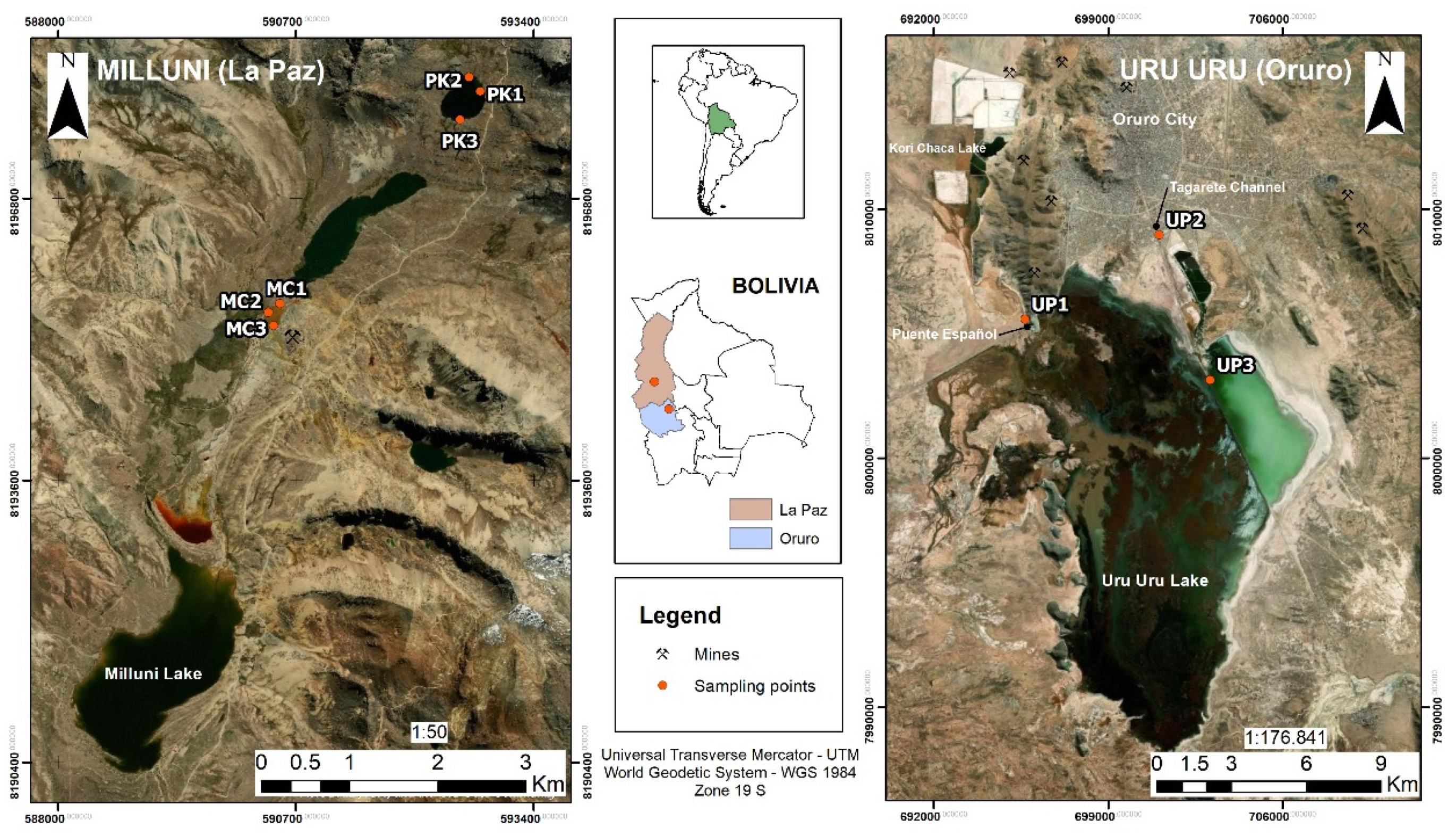
2.1. Sampling Sites
2.1.1. The Milluni Valley Lakes
2.1.2. Uru Uru Lake
2.1.3. Sample Collection and Processing
2.1.4. Quantification of Metals
2.1.5. DNA Extraction
2.1.6. Quantitative PCR
2.1.7. Statistical Analysis
3. Results
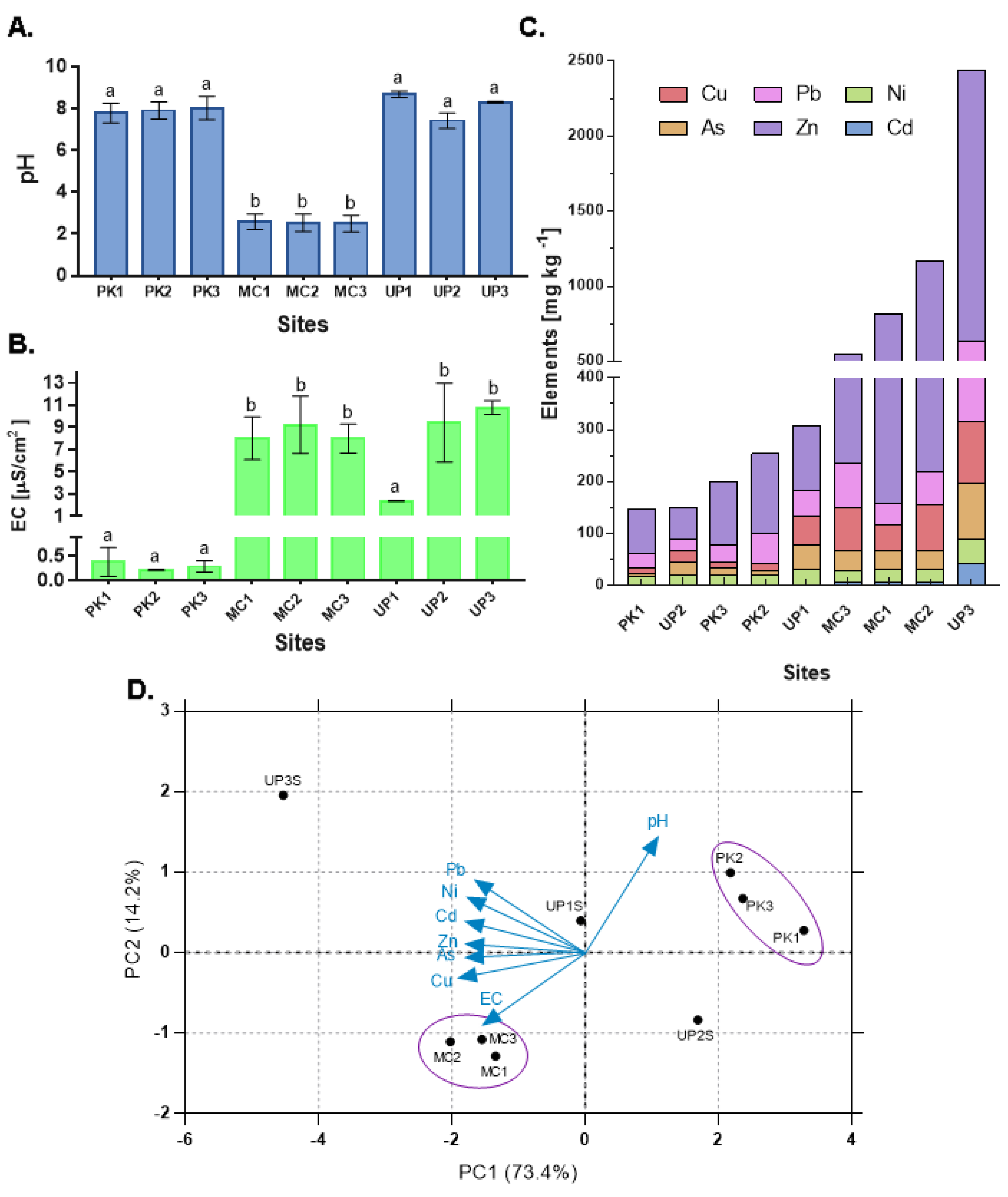
3.1. Physicochemical Conditions and Metal Levels
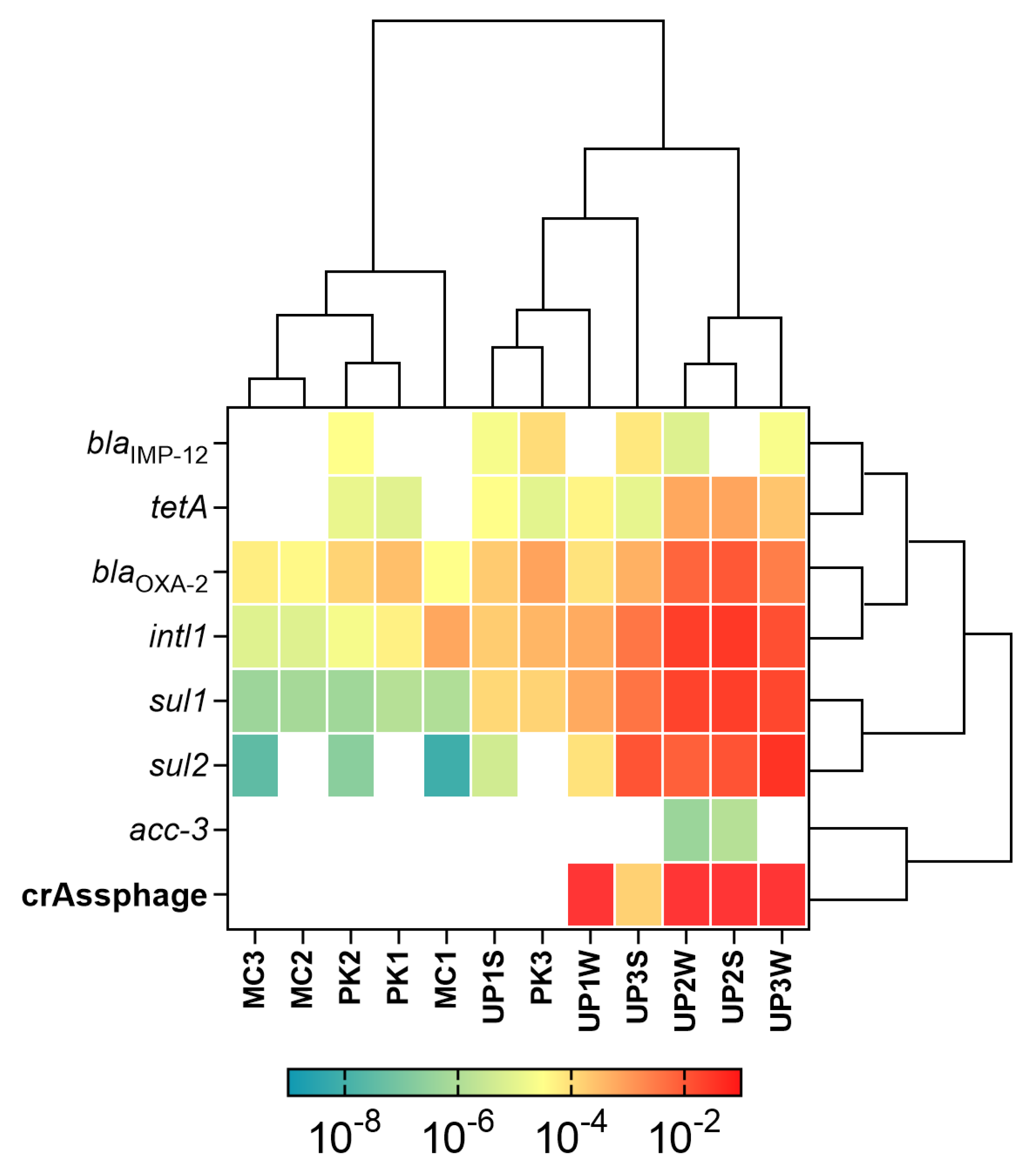
3.2. Detection and Quantification of ARGs and MGEs
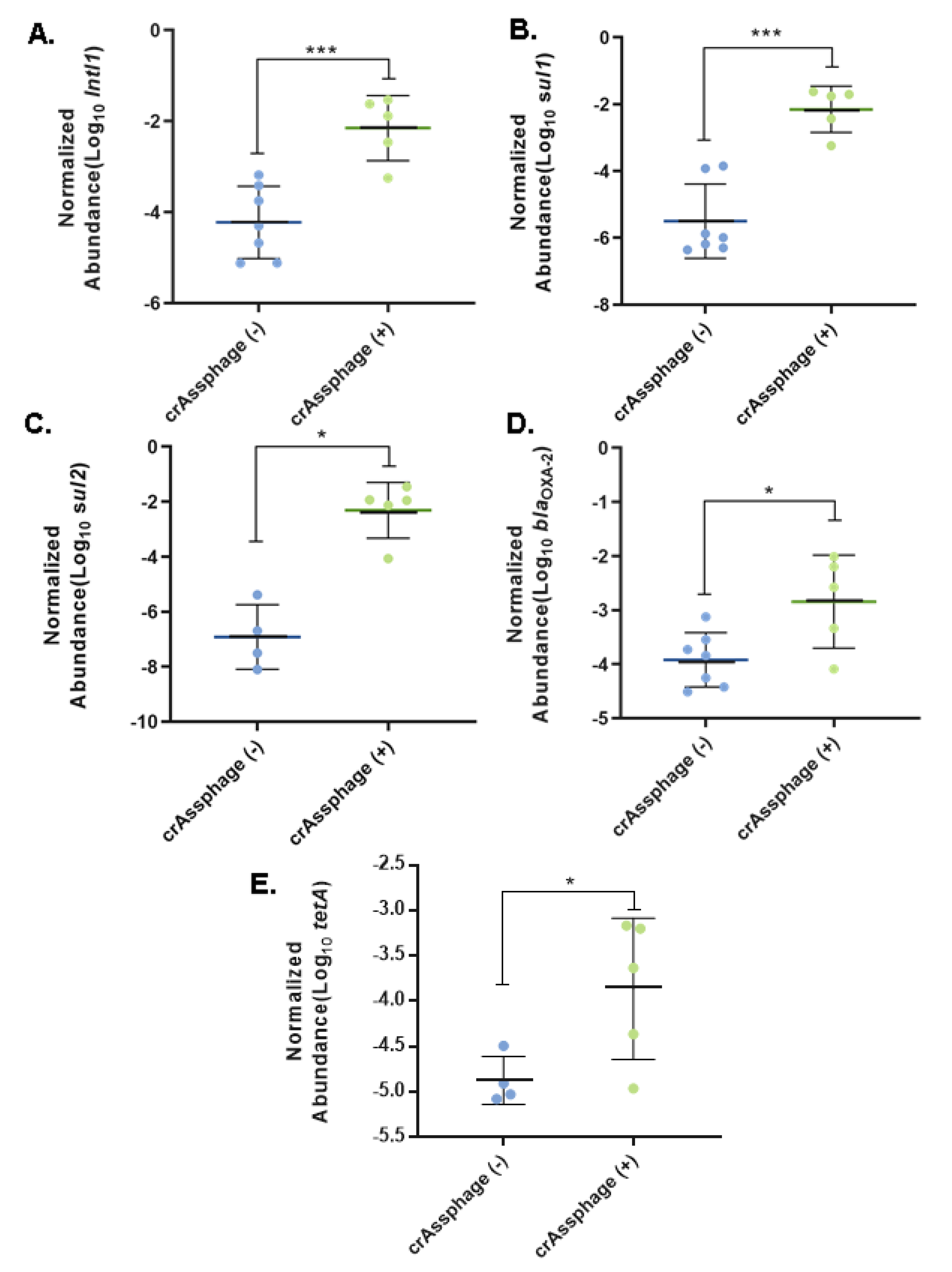
3.3. Fecal Pollution and Antibiotic Resistance
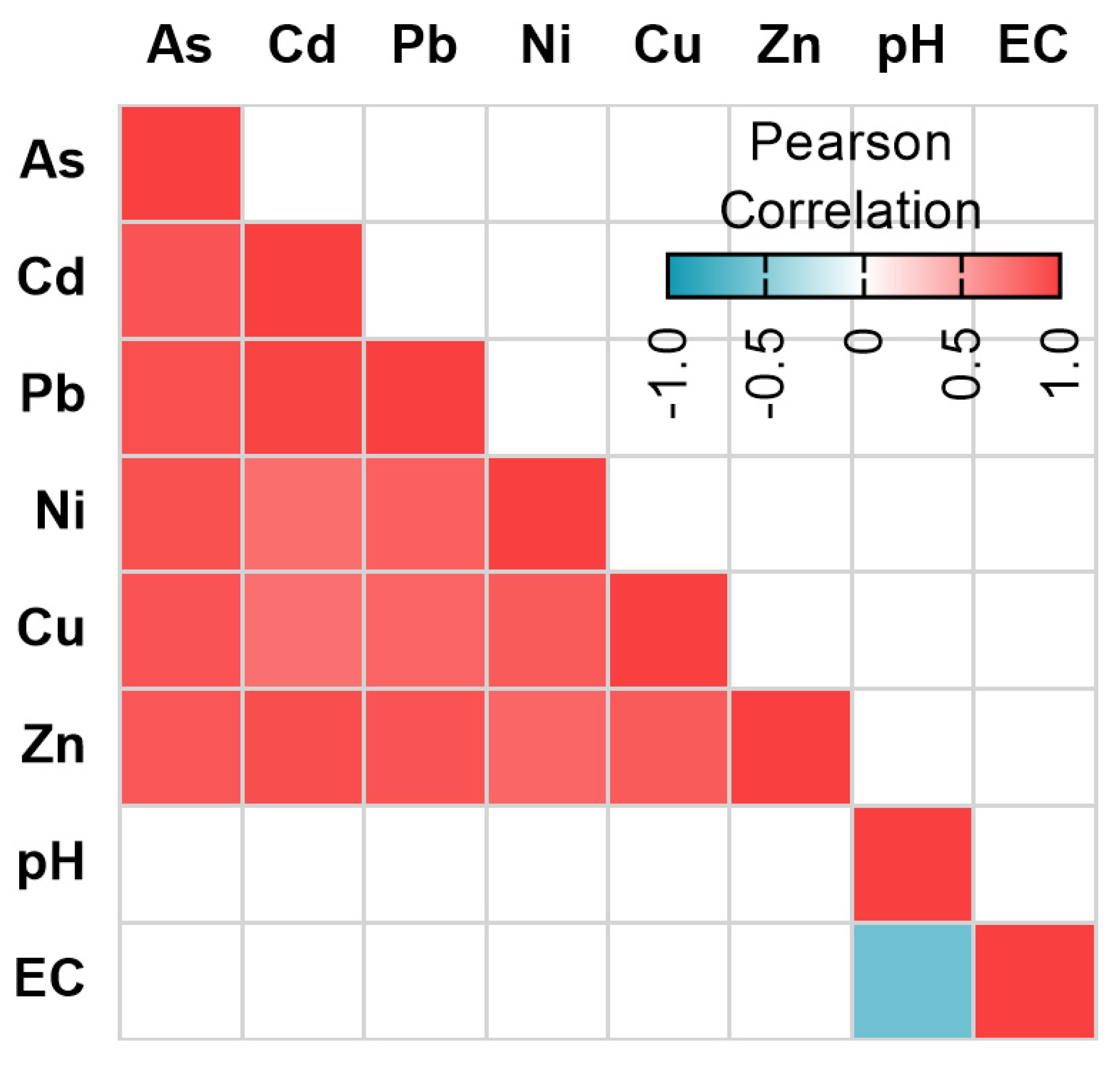
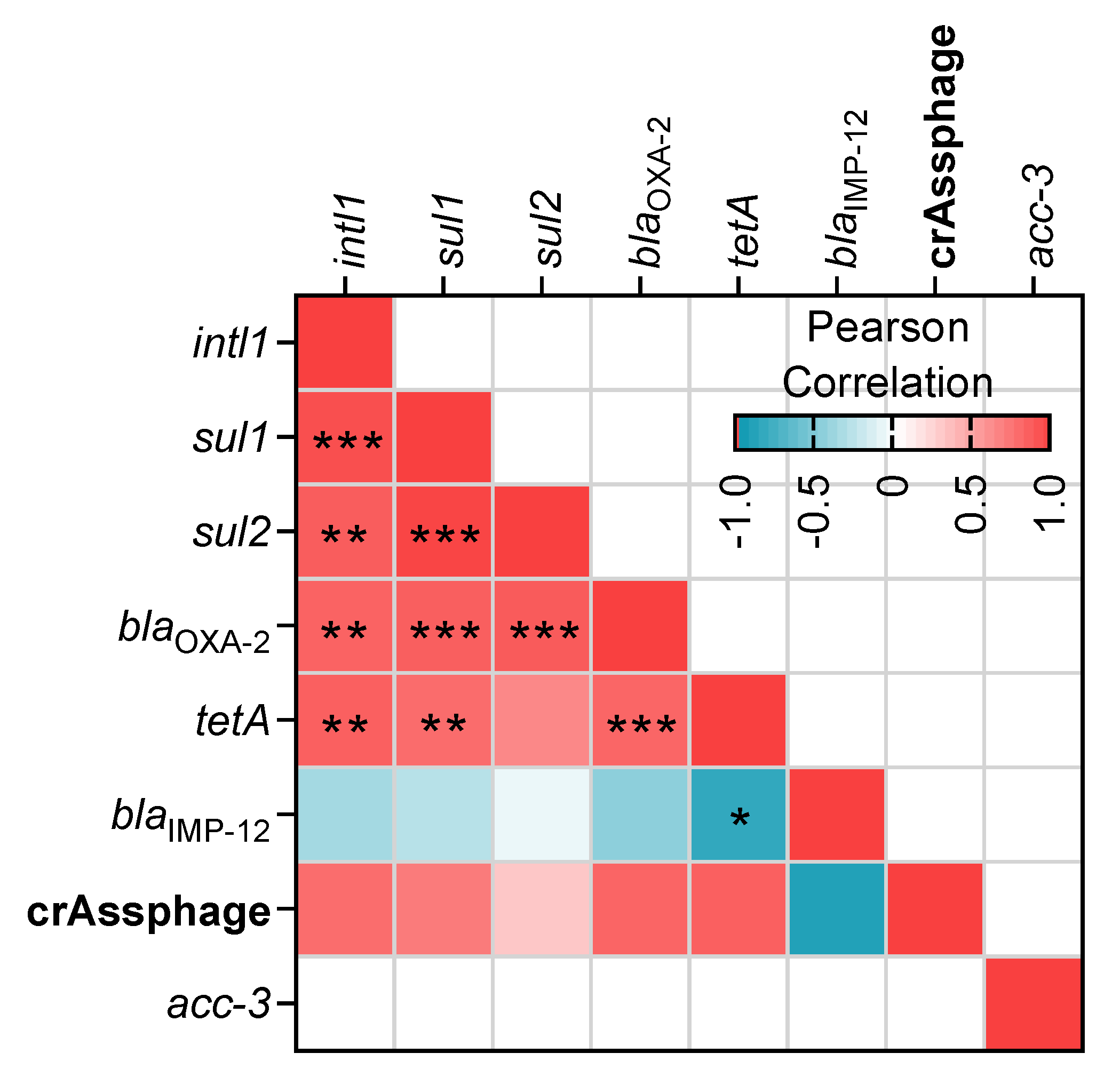
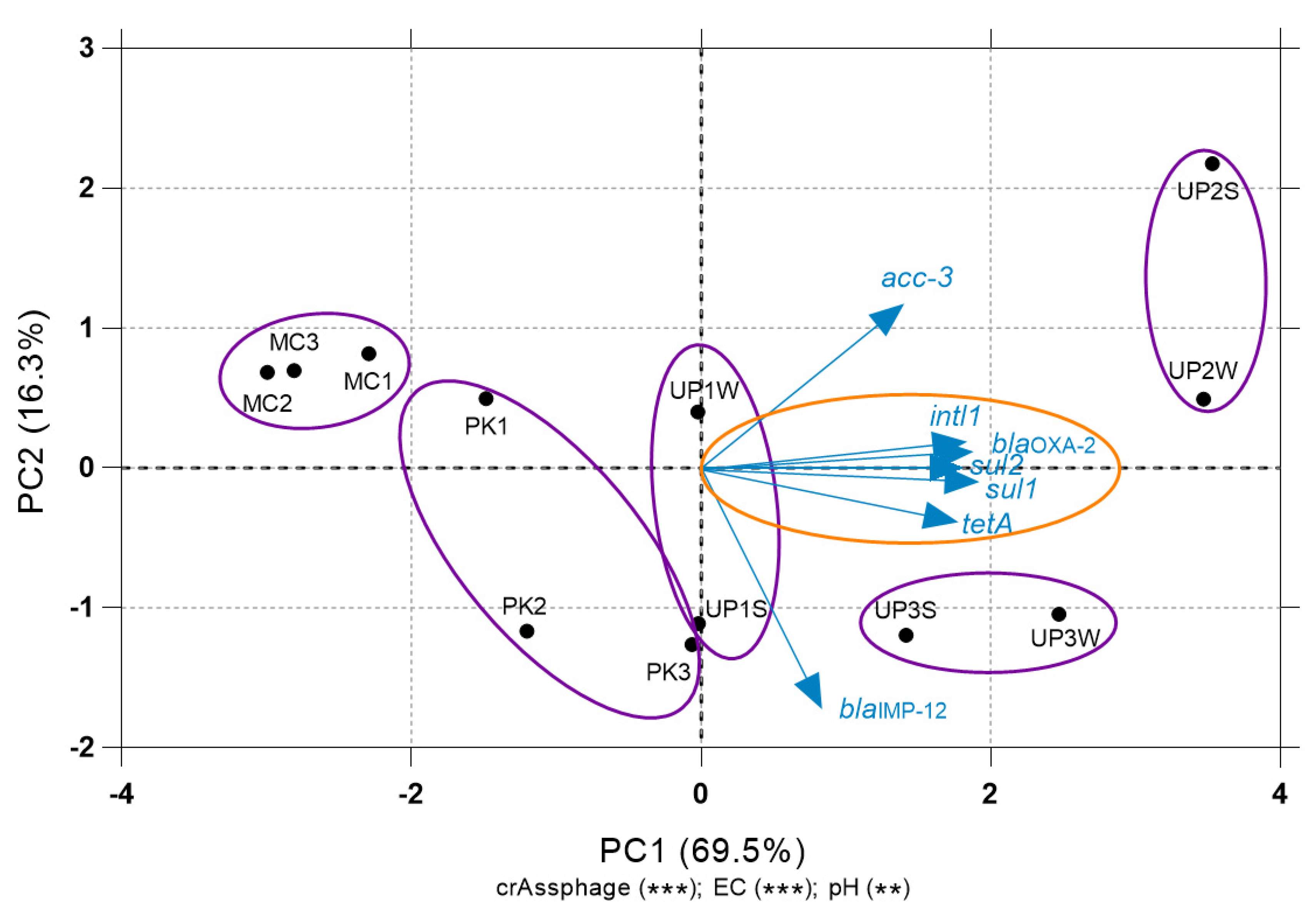
3.4. Relationship between ARGs and Environmental Variables
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Llor, C.; Bjerrum, L. Antimicrobial resistance: Risk associated with antibiotic overuse and initiatives to reduce the problem. Ther. Adv. Drug Saf. 2014, 5, 229–241. [Google Scholar] [CrossRef] [PubMed]
- Berendonk, T.U.; Manaia, C.M.; Merlin, C.; Fatta-Kassinos, D.; Cytryn, E.; Walsh, F.; Burgmann, H.; Sorum, H.; Norstrom, M.; Pons, M.N.; et al. Tackling antibiotic resistance: The environmental framework. Nat. Rev. Microbiol. 2015, 13, 310–317. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R.; Westoby, M.; Ghaly, T.M. Pollutants That Replicate: Xenogenetic DNAs. Trends Microbiol. 2018, 26, 975–977. [Google Scholar] [CrossRef] [PubMed]
- Marti, E.; Variatza, E.; Balcazar, J.L. The role of aquatic ecosystems as reservoirs of antibiotic resistance. Trends Microbiol. 2014, 22, 36–41. [Google Scholar] [CrossRef] [PubMed]
- Chu, B.T.T.; Petrovich, M.L.; Chaudhary, A.; Wright, D.; Murphy, B.; Wells, G.; Poretsky, R. Metagenomics Reveals the Impact of Wastewater Treatment Plants on the Dispersal of Microorganisms and Genes in Aquatic Sediments. Appl. Environ. Microbiol. 2018, 84. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Zhao, Y.; Li, B.; Huang, C.-L.; Zhang, S.-Y.; Yu, S.; Chen, Y.-S.; Zhang, T.; Gillings, M.R.; Su, J.-Q. Continental-scale pollution of estuaries with antibiotic resistance genes. Nat. Microbiol. 2017, 2, 16270. [Google Scholar] [CrossRef]
- Jutkina, J.; Marathe, N.P.; Flach, C.F.; Larsson, D.G.J. Antibiotics and common antibacterial biocides stimulate horizontal transfer of resistance at low concentrations. Sci. Total Environ. 2018, 616–617, 172–178. [Google Scholar] [CrossRef]
- Nesme, J.; Cecillon, S.; Delmont, T.O.; Monier, J.M.; Vogel, T.M.; Simonet, P. Large-scale metagenomic-based study of antibiotic resistance in the environment. Curr. Biol. 2014, 24, 1096–1100. [Google Scholar] [CrossRef]
- Navarro, M.C.; Pérez-Sirvent, C.; Martínez-Sánchez, M.J.; Vidal, J.; Tovar, P.J.; Bech, J. Abandoned mine sites as a source of contamination by heavy metals: A case study in a semi-arid zone. J. Geochem. Explor. 2008, 96, 183–193. [Google Scholar] [CrossRef]
- Liu, J.; Yin, M.; Luo, X.; Xiao, T.; Wu, Z.; Li, N.; Wang, J.; Zhang, W.; Lippold, H.; Belshaw, N.S.; et al. The mobility of thallium in sediments and source apportionment by lead isotopes. Chemosphere 2019, 219, 864–874. [Google Scholar] [CrossRef]
- Baker-Austin, C.; Wright, M.S.; Stepanauskas, R.; McArthur, J.V. Co-selection of antibiotic and metal resistance. Trends Microbiol. 2006, 14, 176–182. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Cocerva, T.; Cox, S.; Tardif, S.; Su, J.-Q.; Zhu, Y.-G.; Brandt, K.K. Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci. Total Environ. 2019, 656, 512–520. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Rensing, C.; Holm, P.E.; Virta, M.; Brandt, K.K. Comparison of Metals and Tetracycline as Selective Agents for Development of Tetracycline Resistant Bacterial Communities in Agricultural Soil. Environ. Sci. Technol. 2017, 51, 3040–3047. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Wang, J.; Han, Y.; Chen, J.; Liu, G.; Lu, H.; Yan, B.; Chen, S. Nutrients, heavy metals and microbial communities co-driven distribution of antibiotic resistance genes in adjacent environment of mariculture. Environ. Pollut. 2017, 220, 909–918. [Google Scholar] [CrossRef]
- Gao, P.; He, S.; Huang, S.; Li, K.; Liu, Z.; Xue, G.; Sun, W. Impacts of coexisting antibiotics, antibacterial residues, and heavy metals on the occurrence of erythromycin resistance genes in urban wastewater. Appl. Microbiol. Biotechnol. 2015, 99, 3971–3980. [Google Scholar] [CrossRef]
- Taylor, N.G.; Verner-Jeffreys, D.W.; Baker-Austin, C. Aquatic systems: Maintaining, mixing and mobilising antimicrobial resistance? Trends Ecol. Evol. 2011, 26, 278–284. [Google Scholar] [CrossRef]
- Baquero, F.; Martínez, J.-L.; Cantón, R. Antibiotics and antibiotic resistance in water environments. Curr. Opin. Biotechnol. 2008, 19, 260–265. [Google Scholar] [CrossRef]
- Karkman, A.; Pärnänen, K.; Larsson, D.G.J. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat. Commun. 2019, 10, 80. [Google Scholar] [CrossRef]
- Dutilh, B.E.; Cassman, N.; McNair, K.; Sanchez, S.E.; Silva, G.G.; Boling, L.; Barr, J.J.; Speth, D.R.; Seguritan, V.; Aziz, R.K.; et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat. Commun. 2014, 5, 4498. [Google Scholar] [CrossRef]
- García-Aljaro, C.; Ballesté, E.; Muniesa, M.; Jofre, J. Determination of crAssphage in water samples and applicability for tracking human faecal pollution. Microb. Biotechnol. 2017, 10, 1775–1780. [Google Scholar] [CrossRef]
- Garrido, A.E.; Strosnider, W.H.; Wilson, R.T.; Condori, J.; Nairn, R.W. Metal-contaminated potato crops and potential human health risk in Bolivian mining highlands. Environ. Geochem. Health 2017, 39, 681–700. [Google Scholar] [CrossRef] [PubMed]
- Quaghebeur, W.; Mulhern, R.E.; Ronsse, S.; Heylen, S.; Blommaert, H.; Potemans, S.; Valdivia Mendizábal, C.; Terrazas García, J. Arsenic contamination in rainwater harvesting tanks around Lake Poopó in Oruro, Bolivia: An unrecognized health risk. Sci. Total Environ. 2019, 688, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Satgé, F.; Hussain, Y.; Xavier, A.; Zolá, R.P.; Salles, L.; Timouk, F.; Seyler, F.; Garnier, J.; Frappart, F.; Bonnet, M.-P. Unraveling the impacts of droughts and agricultural intensification on the Altiplano water resources. Agric. For. Meteorol. 2019, 279, 107710. [Google Scholar] [CrossRef]
- Poma, V.; Mamani, N.; Iñiguez, V. Impact of urban contamination of the La Paz River basin on thermotolerant coliform density and occurrence of multiple antibiotic resistant enteric pathogens in river water, irrigated soil and fresh vegetables. SpringerPlus 2016, 5, 499. [Google Scholar] [CrossRef]
- Guzman-Otazo, J.; Gonzales-Siles, L.; Poma, V.; Bengtsson-Palme, J.; Thorell, K.; Flach, C.F.; Iniguez, V.; Sjoling, A. Diarrheal bacterial pathogens and multi-resistant enterobacteria in the Choqueyapu River in La Paz, Bolivia. PLoS ONE 2019, 14, e0210735. [Google Scholar] [CrossRef] [PubMed]
- Archundia, D.; Duwig, C.; Lehembre, F.; Chiron, S.; Morel, M.C.; Prado, B.; Bourdat-Deschamps, M.; Vince, E.; Aviles, G.F.; Martins, J.M. Antibiotic pollution in the Katari subcatchment of the Titicaca Lake: Major transformation products and occurrence of resistance genes. Sci. Total Environ. 2017, 576, 671–682. [Google Scholar] [CrossRef]
- Stalder, T.; Barraud, O.; Casellas, M.; Dagot, C.; Ploy, M.-C. Integron Involvement in Environmental Spread of Antibiotic Resistance. Front. Microbiol. 2012, 3, 119. [Google Scholar] [CrossRef]
- Gillings, M.R.; Gaze, W.H.; Pruden, A.; Smalla, K.; Tiedje, J.M.; Zhu, Y.-G. Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution. ISME J. 2015, 9, 1269–1279. [Google Scholar] [CrossRef]
- Salvarredy-Aranguren, M.M.; Probst, A.; Roulet, M.; Isaure, M.-P. Contamination of surface waters by mining wastes in the Milluni Valley (Cordillera Real, Bolivia): Mineralogical and hydrological influences. Appl. Geochem. 2008, 23, 1299–1324. [Google Scholar] [CrossRef]
- Alanoca, L.; Guedron, S.; Amouroux, D.; Audry, S.; Monperrus, M.; Tessier, E.; Goix, S.; Acha, D.; Seyler, P.; Point, D. Synergistic effects of mining and urban effluents on the level and distribution of methylmercury in a shallow aquatic ecosystem of the Bolivian Altiplano. Environ. Sci. Process. Impacts 2016, 18, 1550–1560. [Google Scholar] [CrossRef]
- Stepanauskas, R.; Glenn, T.C.; Jagoe, C.H.; Tuckfield, R.C.; Lindell, A.H.; King, C.J.; McArthur, J.V. Coselection for microbial resistance to metals and antibiotics in freshwater microcosms. Environ. Microbiol. 2006, 8, 1510–1514. [Google Scholar] [CrossRef] [PubMed]
- Knapp, C.W.; McCluskey, S.M.; Singh, B.K.; Campbell, C.D.; Hudson, G.; Graham, D.W. Antibiotic Resistance Gene Abundances Correlate with Metal and Geochemical Conditions in Archived Scottish Soils. PLoS ONE 2011, 6, e27300. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.W.; Olivares-Rieumont, S.; Knapp, C.W.; Lima, L.; Werner, D.; Bowen, E. Antibiotic Resistance Gene Abundances Associated with Waste Discharges to the Almendares River near Havana, Cuba. Environ. Sci. Technol. 2011, 45, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Gullberg, E.; Albrecht, L.M.; Karlsson, C.; Sandegren, L.; Andersson, D.I. Selection of a Multidrug Resistance Plasmid by Sublethal Levels of Antibiotics and Heavy Metals. mBio 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Kang, W.; Zhang, Y.J.; Shi, X.; He, J.Z.; Hu, H.W. Short-term copper exposure as a selection pressure for antibiotic resistance and metal resistance in an agricultural soil. Environ. Sci. Pollut. Res. Int. 2018, 25, 29314–29324. [Google Scholar] [CrossRef]
- Driesen, K. Contamination of Surface Waters by the Former Mining Industry in the Milluni Valley (Cordillera Real, Bolivia) and the Application of the Water Planning Model WEAP. Bachelor’s Thesis, University College Ghent, Ghent, Bélgica, 2012. [Google Scholar]
- Zamora Echenique, G.; Mata Parello, J.; Quezada, R. Propuesta de “Geoparque Oruro-Bolivia” Alternativa para la preservación del patrimonio geológico, minero y sociocultural. Rev. MAMYM 2018, 5, 19–33. [Google Scholar]
- Guedron, S.; Point, D.; Acha, D.; Bouchet, S.; Baya, P.A.; Tessier, E.; Monperrus, M.; Molina, C.I.; Groleau, A.; Chauvaud, L.; et al. Mercury contamination level and speciation inventory in Lakes Titicaca & Uru-Uru (Bolivia): Current status and future trends. Environ. Pollut. 2017, 231, 262–270. [Google Scholar] [CrossRef]
- Fang, Y.; Xu, M.; Chen, X.; Sun, G.; Guo, J.; Wu, W.; Liu, X. Modified pretreatment method for total microbial DNA extraction from contaminated river sediment. Front. Environ. Sci. Eng. 2015, 9, 444–452. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.Y.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.V.; Cheng, A.A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef]
- Nolvak, H.; Truu, M.; Truu, J. Evaluation of quantitative real-time PCR workflow modifications on 16S rRNA and tetA gene quantification in environmental samples. Sci. Total Environ. 2012, 426, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.-G.; Johnson, T.A.; Su, J.-Q.; Qiao, M.; Guo, G.-X.; Stedtfeld, R.D.; Hashsham, S.A.; Tiedje, J.M. Diverse and abundant antibiotic resistance genes in Chinese swine farms. Proc. Natl. Acad. Sci. USA 2013, 110, 3435–3440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Qu, Y.; Li, S.; Feng, K.; Wang, S.; Cai, W.; Liang, Y.; Li, H.; Xu, M.; Yin, H.; et al. Soil bacterial quantification approaches coupling with relative abundances reflecting the changes of taxa. Sci. Rep. 2017, 7, 4837. [Google Scholar] [CrossRef] [PubMed]
- Gray, N.F. Field assessment of acid mine drainage contamination in surface and ground water. Environ. Geol. 1996, 27, 358–361. [Google Scholar] [CrossRef]
- Rios, C. Estudio de la Contaminación Ambiental por las Descargas Mineras de Comsur en la Represa Milluni; Universidad Mayor de San Andres: La Paz, Bolivia, 1985. [Google Scholar]
- Lucassen, R.; Rehberg, L.; Heyden, M.; Bockmühl, D. Strong correlation of total phenotypic resistance of samples from household environments and the prevalence of class 1 integrons suggests for the use of the relative prevalence of intI1 as a screening tool for multi-resistance. PLoS ONE 2019, 14, e0218277. [Google Scholar] [CrossRef]
- Uyaguari-Díaz, M.I.; Croxen, M.A.; Luo, Z.; Cronin, K.I.; Chan, M.; Baticados, W.N.; Nesbitt, M.J.; Li, S.; Miller, K.M.; Dooley, D.; et al. Human Activity Determines the Presence of Integron-Associated and Antibiotic Resistance Genes in Southwestern British Columbia. Front. Microbiol. 2018, 9, 852. [Google Scholar] [CrossRef]
- Storteboom, H.; Arabi, M.; Davis, J.G.; Crimi, B.; Pruden, A. Tracking Antibiotic Resistance Genes in the South Platte River Basin Using Molecular Signatures of Urban, Agricultural, And Pristine Sources. Environ. Sci. Technol. 2010, 44, 7397–7404. [Google Scholar] [CrossRef]
- Ouyang, W.-Y.; Huang, F.-Y.; Zhao, Y.; Li, H.; Su, J.-Q. Increased levels of antibiotic resistance in urban stream of Jiulongjiang River, China. Appl. Microbiol. Biotechnol. 2015, 99, 5697–5707. [Google Scholar] [CrossRef]
- Van Goethem, M.W.; Pierneef, R.; Bezuidt, O.K.I.; Van De Peer, Y.; Cowan, D.A.; Makhalanyane, T.P. A reservoir of ‘historical’ antibiotic resistance genes in remote pristine Antarctic soils. Microbiome 2018, 6, 40. [Google Scholar] [CrossRef]
- Seiler, C.; Berendonk, T. Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 2012, 3, 399. [Google Scholar] [CrossRef]
- Stachler, E.; Crank, K.; Bibby, K. Co-Occurrence of crAssphage with Antibiotic Resistance Genes in an Impacted Urban Watershed. Environ. Sci. Technol. Lett. 2019, 6, 216–221. [Google Scholar] [CrossRef]
- Chen, H.; Bai, X.; Li, Y.; Jing, L.; Chen, R.; Teng, Y. Source identification of antibiotic resistance genes in a peri-urban river using novel crAssphage marker genes and metagenomic signatures. Water Res. 2019, 167, 115098. [Google Scholar] [CrossRef] [PubMed]
- Girlich, D.; Naas, T.; Bellais, S.; Poirel, L.; Karim, A.; Nordmann, P. Biochemical-genetic characterization and regulation of expression of an ACC-1-like chromosome-borne cephalosporinase from Hafnia alvei. Antimicrob. Agents Chemother. 2000, 44, 1470–1478. [Google Scholar] [CrossRef] [PubMed]
- Romanowska, E. Immunochemical aspects of Hafnia alvei O antigens. FEMS Immunol. Med. Microbiol. 2000, 27, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Stachler, E.; Akyon, B.; de Carvalho, N.A.; Ference, C.; Bibby, K. Correlation of crAssphage qPCR Markers with Culturable and Molecular Indicators of Human Fecal Pollution in an Impacted Urban Watershed. Environ. Sci. Technol. 2018, 52, 7505–7512. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Samal, N.; Roy, P.; Roy, M.B. Electrical Conductivity of Lake Water as Environmental Monitoring—A Case study of Rudra sagar Lake. J. Environ. Sci. Toxicol. Food Technol. 2015, 9, 66–71. [Google Scholar]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-Based Assessment of Soil pH as a Predictor of Soil Bacterial Community Structure at the Continental Scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Bahram, M.; Hildebrand, F.; Forslund, S.K.; Anderson, J.L.; Soudzilovskaia, N.A.; Bodegom, P.M.; Bengtsson-Palme, J.; Anslan, S.; Coelho, L.P.; Harend, H.; et al. Structure and function of the global topsoil microbiome. Nature 2018, 560, 233–237. [Google Scholar] [CrossRef]
- Forsberg, K.J.; Patel, S.; Gibson, M.K.; Lauber, C.L.; Knight, R.; Fierer, N.; Dantas, G. Bacterial phylogeny structures soil resistomes across habitats. Nature 2014, 509, 612–616. [Google Scholar] [CrossRef]
- Herlihy, A.T.; Mills, A.L. The pH regime of sediments underlying acidified waters. Biogeochemistry 1986, 2, 95–99. [Google Scholar] [CrossRef]
- Chen, Y.; Jiang, Y.; Huang, H.; Mou, L.; Ru, J.; Zhao, J.; Xiao, S. Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Sci. Total Environ. 2018, 637–638, 1400–1412. [Google Scholar] [CrossRef] [PubMed]
| Gene | Forward (5′ → 3′) | Reverse (5′ → 3′) | Size (bp) | Ref. |
|---|---|---|---|---|
| sul1 | GGATTTTTCTTGAGCCCCGC | CACCGAGACCAATAGCGGAA | 99 | This study |
| sul2 | TCATCTGCCAAACTCGTCGT | CAAAGAACGCCGCAATGTGA | 103 | This study |
| blaIMP-5 | CTTGGTTTGTGGAACGCGG | TAAGCCACTCTATTCCGCCC | 87 | This study |
| blaIMP-2 | GAGCGCGTTTGCCTGATTTA | AGAAACAACACCCCAACCGT | 95 | This study |
| blaIMP-12 | TGAAGAGGTTAGCGGTTGGG | CGCCCTACAAACCAAGCAAC | 132 | This study |
| blaOXA-2 | GGTAGGATGGGTTGAGTGGC | ATAGAGCGAAGGATTGCCCG | 120 | This study |
| acc-3 | GTTGCTACGCCGATTGTTCC | GCGATGTAGGCACCAAAACC | 92 | This study |
| tetA | TCAATTTCCTGACGGGCTG | GAAGCGAGCGGGTTGAGAG | 91 | [42] |
| tetB | AGTGCGCTTTGGATGCTGTA | GCTGAGGTGGTATCGGCAAT | 98 | This study |
| msrA | CTGCTAACACAAGTACGATTCCAAAT | TCAAGTAAAGTTGTCTTACCTACACCATT | 89 | [43] |
| mecA | GGTTACGGACAAGGTGAAATACTGAT | TGTCTTTTAATAAGTGAGGTGCGTTAATA | 106 | [43] |
| QnrB-1 | GCGGCACTGAATTTATCGGC | GGCATCTTTCAGCATCGCAC | 86 | This study |
| QnrB-5 | CGGGGTGTTGATTTACAAGGC | GCCAATAATCGCGATGCCAA | 84 | This study |
| intl1 | CAGCACCTTGCCGTAGAAGA | GAGGCATTTCTGTCCTGGCT | 99 | This study |
| crAss- phage | AGGAGAAAGTGAACGTGGAAACA | AACGAGCACCAATTTTAAGCTTTA | 78 | Modified from [20] |
| 16S rRNA | CCTACGGGAGGCAGCAG | TTACCGCGGCTGCTGGCAC | 192 | [44] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Agramont, J.; Gutiérrez-Cortez, S.; Joffré, E.; Sjöling, Å.; Calderon Toledo, C. Fecal Pollution Drives Antibiotic Resistance and Class 1 Integron Abundance in Aquatic Environments of the Bolivian Andes Impacted by Mining and Wastewater. Microorganisms 2020, 8, 1122. https://doi.org/10.3390/microorganisms8081122
Agramont J, Gutiérrez-Cortez S, Joffré E, Sjöling Å, Calderon Toledo C. Fecal Pollution Drives Antibiotic Resistance and Class 1 Integron Abundance in Aquatic Environments of the Bolivian Andes Impacted by Mining and Wastewater. Microorganisms. 2020; 8(8):1122. https://doi.org/10.3390/microorganisms8081122
Chicago/Turabian StyleAgramont, Jorge, Sergio Gutiérrez-Cortez, Enrique Joffré, Åsa Sjöling, and Carla Calderon Toledo. 2020. "Fecal Pollution Drives Antibiotic Resistance and Class 1 Integron Abundance in Aquatic Environments of the Bolivian Andes Impacted by Mining and Wastewater" Microorganisms 8, no. 8: 1122. https://doi.org/10.3390/microorganisms8081122
APA StyleAgramont, J., Gutiérrez-Cortez, S., Joffré, E., Sjöling, Å., & Calderon Toledo, C. (2020). Fecal Pollution Drives Antibiotic Resistance and Class 1 Integron Abundance in Aquatic Environments of the Bolivian Andes Impacted by Mining and Wastewater. Microorganisms, 8(8), 1122. https://doi.org/10.3390/microorganisms8081122