Production of Marine Probiotic Bacteria in a Cost-Effective Marine Media Based on Peptones Obtained from Discarded Fish By-Products
Abstract
:1. Introduction
2. Materials and Methods
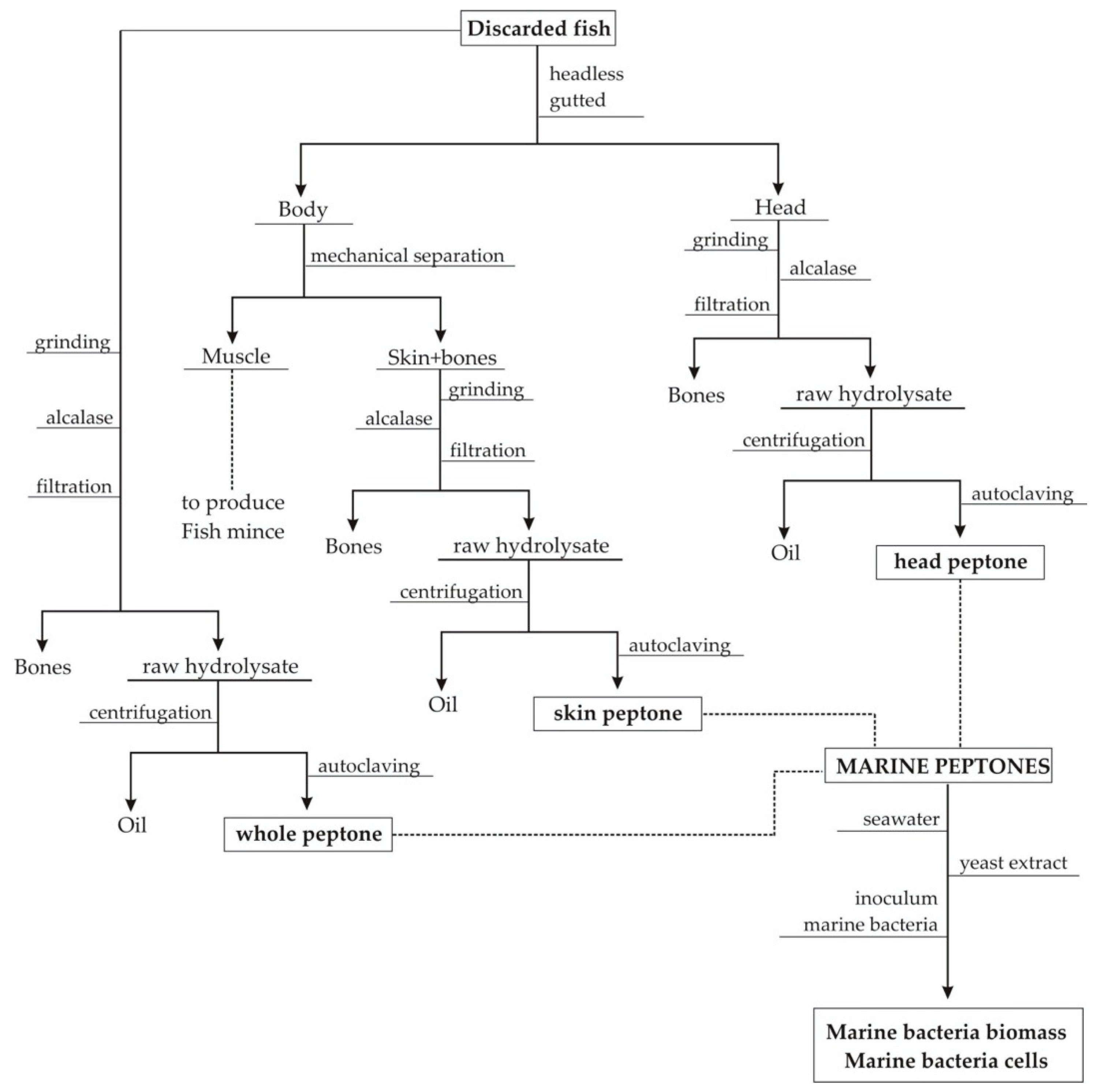
2.1. Preparation of Peptones from Fish Discard By-Products
2.2. Microbiological Methods, Culture Media, and Analytical Determinations
2.3. Bacterial Sampling and Biomass and Cell Analysis
2.4. Mathematical Equations for Cultures Modelling
2.5. Economical Assessment of MPB Growth Costs
2.6. Numerical and Statistical Analyses
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vine, N.G.; Leukes, W.D.; Kaiser, H. Probiotics in marine larviculture. FEMS Microbiol. Rev. 2006, 30, 404–427. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Defoirdt, T.; Boon, N.; Sorgeloos, P.; Verstraete, W.; Bossier, P. Alternatives to antibiotics to control bacterial infections: Luminescent vibriosis in aquaculture as an example. Trends Biotechnol. 2007, 25, 472–479. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.-B.; Li, J.-R.; Lin, J. Probiotics in aquaculture: Challenges and outlook. Aquaculture 2008, 281, 1–4. [Google Scholar] [CrossRef]
- Rasmussen, B.B.; Kalatzis, P.G.; Middelboe, M.; Gram, L. Combining probiotic Phaeobacter inhibens DSM17395 and broad-host-range vibriophage KVP40 against fish pathogenic vibrios. Aquaculture 2019, 513, 734415. [Google Scholar] [CrossRef]
- Planas, M.; Vázquez, J.A.; Marques, J.; Pérez-Lomba, R.; González, M.P.; Murado, M.A. Enhancement of rotifer (Brachionus plicatilis) growth by using terrestrial acid lactic bacteria. Aquaculture 2004, 240, 313–329. [Google Scholar] [CrossRef] [Green Version]
- Dash, G.; Raman, R.P.; Prasad, K.P.; Makesh, M.; Pradeep, M.A.; Sen, S. Evaluation of paraprobiotic applicability of Lactobacillus plantarum in improving the immune response and disease protection in giant freshwater prawn, Macrobrachium rosenbergii (de Man, 1879). Fish Shellfish Immunol. 2015, 43, 167–174. [Google Scholar] [CrossRef]
- Pintado, J.; Pérez-Lorenzo, M.; Luna-González, A.; Sotelo, C.G.; Prol, M.J.; Planas, M. Monitoring of the bioencapsulation of a probiotic Phaeobacter strain in the rotifer Brachionus plicatilis using denaturing gradient gel electrophoresis. Aquaculture 2010, 302, 182–194. [Google Scholar] [CrossRef] [Green Version]
- D’Alvise, P.W.; Lillebø, S.; Prol-Garcia, M.J.; Wergeland, H.I.; Nielsen, K.F.; Bergh, Ø.; Gram, L. Phaeobacter gallaeciensis reduces Vibrio anguillarum in cultures of microalgae and rotifers, and prevents vibriosis in cod larvae. PLoS ONE 2012, 7, e43996. [Google Scholar] [CrossRef] [Green Version]
- Grotkjær, T.; Bentzon-Tilia, M.; D’Alvise, P.; Dourala, N.; Nielsen, K.F.; Gram, L. Isolation of TDA-producing Phaeobacter strains from sea bass larval rearing units and their probiotic effect against pathogenic Vibrio spp. in Artemia cultures. Syst. Appl. Microbiol. 2016, 39, 180–188. [Google Scholar] [CrossRef] [Green Version]
- Planas, M.; Pérez-Lorenzo, M.; Hjelm, M.; Gram, L.; Fiksdal, I.U.; Bergh, Ø.; Pintado, J. Probiotic effect In Vivo of Roseobacter strain 27-4 against Vibrio (Listonella) anguillarum infections in turbot (Scophthalmus maximus L.) larvae. Aquaculture 2006, 255, 323–333. [Google Scholar] [CrossRef] [Green Version]
- D’Alvise, P.W.; Lillebø, S.; Wergeland, H.I.; Gram, L.; Bergh, Ø. Protection of cod larvae from vibriosis by Phaeobacter spp.: A comparison of strains and introduction times. Aquaculture 2013, 384–387, 82–86. [Google Scholar] [CrossRef]
- Prol-García, M.J.; Pintado, J. Effectiveness of probiotic Phaeobacter bacteria grown in biofilters against Vibrio anguillarum infections in the rearing of turbot (Psetta maxima) Larvae. Mar. Biotechnol. 2013, 15, 726–738. [Google Scholar] [CrossRef]
- Gram, L.; Løvold, T.; Nielsen, J.; Melchiorsen, J.; Spanggaard, B. In Vitro antagonism of the probiont Pseudomonas fluorescens strain AH2 against Aeromonas salmonicida does not confer protection of salmon against furunculosis. Aquaculture 2001, 199, 1–11. [Google Scholar] [CrossRef]
- D’Alvise, P.W.; Melchiorsen, J.; Porsby, C.H.; Nielsen, K.F.; Gram, L. Inactivation of Vibrio anguillarum by attached and planktonic roseobacter cells. Appl. Environ. Microbiol. 2010, 76, 2366–2370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prol-García, M.J.; Gómez, M.; Sánchez, L.; Pintado, J. Phaeobacter grown in biofilters: A new strategy for the control of Vibrionaceae in aquaculture. Aquac. Res. 2014, 45, 1012–1025. [Google Scholar] [CrossRef]
- Gram, L.; Melchiorsen, J.; Spanggaard, B.; Huber, I.; Nielsen, T.F. Inhibition of Vibrio anguillarum by Pseudomonas fluorescens AH2, a possible probiotic treatment of fish. Appl. Environ. Microbiol. 1999, 65, 969–973. [Google Scholar] [CrossRef] [Green Version]
- Porsby, C.H.; Gram, L. Phaeobacter inhibens as biocontrol agent against Vibrio vulnificus in oyster models. Food Microbiol. 2016, 57, 63–70. [Google Scholar] [CrossRef]
- Sonnenschein, E.C.; Phippen, C.B.W.; Nielsen, K.F.; Mateiu, R.V.; Melchiorsen, J.; Gram, L.; Overmann, J.; Freese, H.M. Phaeobacter piscinae sp. nov., a species of the Roseobacter group and potential aquaculture probiont. Int. J. Syst. Evol. Microbiol. 2017, 67, 4559–4564. [Google Scholar] [CrossRef]
- Safari, R.; Motamedzadegan, A.; Ovissipour, M.; Regenstein, J.M.; Gildberg, A.; Rasco, B. Use of hydrolysates from yellowfin tuna (Thunnus albacares) heads as a complex nitrogen. Food Bioprocess Technol. 2012, 5, 73–79. [Google Scholar] [CrossRef]
- Gildberg, A.; Dahl, R.; Mikkelsen, H.; Nilsen, K. Peptones from Atlantic cod stomach as nitrogen sources in growth media to marine bacteria. J. Aquat. Food Prod. Technol. 2010, 19, 75–83. [Google Scholar] [CrossRef]
- European Commission. Discarding and the Landing Obligation. 2020. Available online: https://ec.europa.eu/fisheries/cfp/fishing_rules/discards_en (accessed on 13 April 2020).
- Iñarra, B.; Bald, C.; Cebrián, M.; Antelo, L.T.; Franco-Uría, A.; Vázquez, J.A.; Pérez-Martín, R.I.; Zufía, J. What to do with unwanted catches: Valorisation options and selection strategies. In The European Landing Obligation, Reducing Discards in Complex, Multi-Species and Multi-Jurisdictional Fisheries; Uhlmann, S.S., Ulrich, C., Kennelly, S.J., Eds.; Springer Open: Berlin, Germany, 2019; ISBN 978-3-030-03307-1. [Google Scholar] [CrossRef]
- Pérez-Martín, R.I.; Antelo, L.T.; Vázquez, J.A.; Mirón, J. An on-land management and valorisation approach for biomass associated with landing obligation compliance. Mar. Policy 2020, 116, 103506. [Google Scholar] [CrossRef]
- Borderías, A.J.; Moreno, H.M. Valorization of recurrently discarded fish species in trawler fisheries in North-West Spain. J. Food Sci. Technol. 2018, 55, 4477–4484. [Google Scholar]
- Vázquez, J.A.; Fernández-Compás, A.; Blanco, M.; Rodríguez-Amado, I.; Moreno, H.; Borderías, J.; Pérez-Martín, R.I. Development of bioprocesses for the integral valorisation of fish discards. Biochem. Eng. J. 2019, 144, 198–208. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Menduíña, A.; Durán, A.I.; Nogueira, M.; Fernández-Compás, A.; Pérez-Martín, R.I.; Amado, I.R. Production of valuable compounds and bioactive metabolites from by-products of fish discards using chemical processing, enzymatic hydrolysis, and bacterial fermentation. Mar. Drugs 2019, 17, 139. [Google Scholar]
- Vázquez, J.A.; Fraguas, J.; Mirón, J.; Valcarcel, J.; Pérez-Martín, R.I.; Antelo, L.T. Valorisation of fish discards assisted by enzymatic hydrolysis and microbial bioconversion: Lab and pilot plant studies and preliminary sustainability evaluation. J. Clean. Prod. 2020, 246, 119027. [Google Scholar] [CrossRef]
- Bernfeld, P. Enzymes of starch degradation and synthesis. Adv. Enzymol. 1951, 12, 379–427. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the folin phenol reagent. J. Biol. Chem. 1951, 270, 27299–27304. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric method for determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Lorenzo, J.M.; Fuciños, P.; Franco, D. Evaluation of non-linear equations to model different animal growths with mono and bi-sigmoid profiles. J. Theor. Biol. 2012, 314, 95–105. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Caprioni, R.; Nogueira, M.; Menduiña, A.; Ramos, P.; Pérez-Martín, R.I. Valorisation of effluents obtained from chemical and enzymatic chitin production of Illex argentinus pen by-products as nutrient supplements for various bacterial fermentations. Biochem. Eng. J. 2016, 116, 34–44. [Google Scholar]
- Vázquez, J.A.; González, M.P.; Murado, M.A. A new marine medium. Use of the different fish peptones and comparative study of the growth of selected species of marine bacteria. Enzyme Microb. Technol. 2004, 35, 385–392. [Google Scholar]
- Vázquez, J.A.; Docasal, S.F.; Mirón, J.; González, M.P.; Murado, M.A. Proteases production by two Vibrio species on residuals marine media. J. Ind. Microbiol. Biotechnol. 2006, 33, 661–668. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lapeña, D.; Vuoristo, K.S.; Kosa, G.; Horn, S.J.; Eijsink, V.G.H. Comparative assessment of enzymatic hydrolysis for valorization of different protein-rich industrial byproducts. J. Agric. Food Chem. 2018, 66, 9738–9749. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Chang, S.K.C.; Meng, S. Comparing the kinetics of the hydrolysis of by-product from channel catfish (Ictalurus punctatus) fillet processing by eight proteases. LWT 2019, 111, 809–820. [Google Scholar] [CrossRef]
- Vázquez, J.A.; González, M.P.; Murado, M.A. Peptones from autohydrolysed fish viscera for nisin and pediocin production. J. Biotechnol. 2004, 112, 299–311. [Google Scholar] [CrossRef] [Green Version]
- Castro-Ceseña, A.B.; Sánchez-Saavedra, M.P.; Márquez-Rocha, F.J. Characterisation and partial purification of proteolytic enzymes from sardine by-products to obtain concentrated hydrolysates. Food Chem. 2012, 135, 583–589. [Google Scholar] [CrossRef]
- Derouiche, B.M.H.; Guadix, E.M.; Guadix, A.; Gargouri, M.; Espejo-Carpio, F.J. Valorisation of tuna viscera by endogenous enzymatic treatment. Int. J. Food Sci. Technol. 2019, 54, 1100–1108. [Google Scholar] [CrossRef]
- Deraz, S.F.; El-Fawal, G.F.; Abd-Ellatif, S.A.; Khalil, A.A. Autohydrolysed Tilapia nilotica fish viscera as a peptone source in bacteriocin production. Indian J. Microbiol. 2011, 51, 171–175. [Google Scholar] [CrossRef] [Green Version]
- Vázquez, J.A.; Amado, I.R.; Sotelo, C.G.; Sanz, N.; Pérez-Martín, R.I.; Valcarcel, J. Production, characterization, and bioactivity of fish protein hydrolysates from aquaculture turbot (Scophthalmus maximus) wastes. Biomolecules 2020, 10, 310. [Google Scholar] [CrossRef] [Green Version]
- Halim, N.R.A.; Yusof, H.M.; Sarbon, N.M. Functional and bioactive properties of fish protein hydolysates and peptides: A comprehensive review. Trends Food Sci. Technol. 2016, 51, 24–33. [Google Scholar] [CrossRef]
- Fan, B.; Sun, J.; Dong, P.; Xue, C.; Mao, X. Conversion of turbot skin wastes into valuable functional substances with an eco-friendly fermentation technology. J. Clean. Prod. 2017, 156, 367–377. [Google Scholar]
- Aspmo, S.I.; Horn, S.J.; Eijsink, V.G.H. Use of hydrolysates from Atlantic cod (Gadus morhua L.) viscera as a complex nitrogen source for lactic acid bacteria. FEMS Microbiol. Lett. 2005, 248, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Safari, R.; Saravi, H.N.; Pourgholam, R.; Motalebi, A.A.; Ghoroghi, A. Use of hydrolysates from silver carp (Hypophthalmichthys molitrix) head as peptone for Vibrio anguillarum and optimization using response surface method (RSM). J. Aquat. Food Prod. Technol. 2011, 20, 247–257. [Google Scholar] [CrossRef]
- Strathe, A.B.; Danfær, A.; Sørensen, H.; Kebreab, E. A multi level non linear mixed-effects approach to model growth in pigs. J. Anim. Sci. 2010, 88, 638–649. [Google Scholar] [CrossRef]
- Franco, D.; Rois, D.; Vázquez, J.A.; Purriños, L.; González, R.; Lorenzo, J.M. Breed effect between Mos rooster (Galician indigenous breed) and SassoT-44 line and finishing feed effect of commercial fodder or corn. Poult. Sci. 2012, 91, 487–498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhabra, A.T.; Carter, W.H.; Linton, R.H.; Cousin, M.A. A predictive model that evaluates the effect of growth conditions on the thermal resistance of Listeria monocytogenes. Int. J. Food Microbiol. 2002, 78, 235–243. [Google Scholar] [CrossRef]
- Vázquez, J.A.; Montemayor, M.I.; Fraguas, J.; Murado, M.A. High production of hyaluronic and lactic acids by Streptococcus zooepidemicus in fed-batch cultures using commercial and marine peptones from fishing by-products. Biochem. Eng. J. 2009, 44, 125–130. [Google Scholar] [CrossRef] [Green Version]
- Rutledge, R.G.; Stewart, D. A kinetic-based sigmoidal model for the polymerase chain reaction and its application to high-capacity absolute quantitative real-time PCR. BMC Biotechnol. 2008, 8, 47. [Google Scholar] [CrossRef] [Green Version]
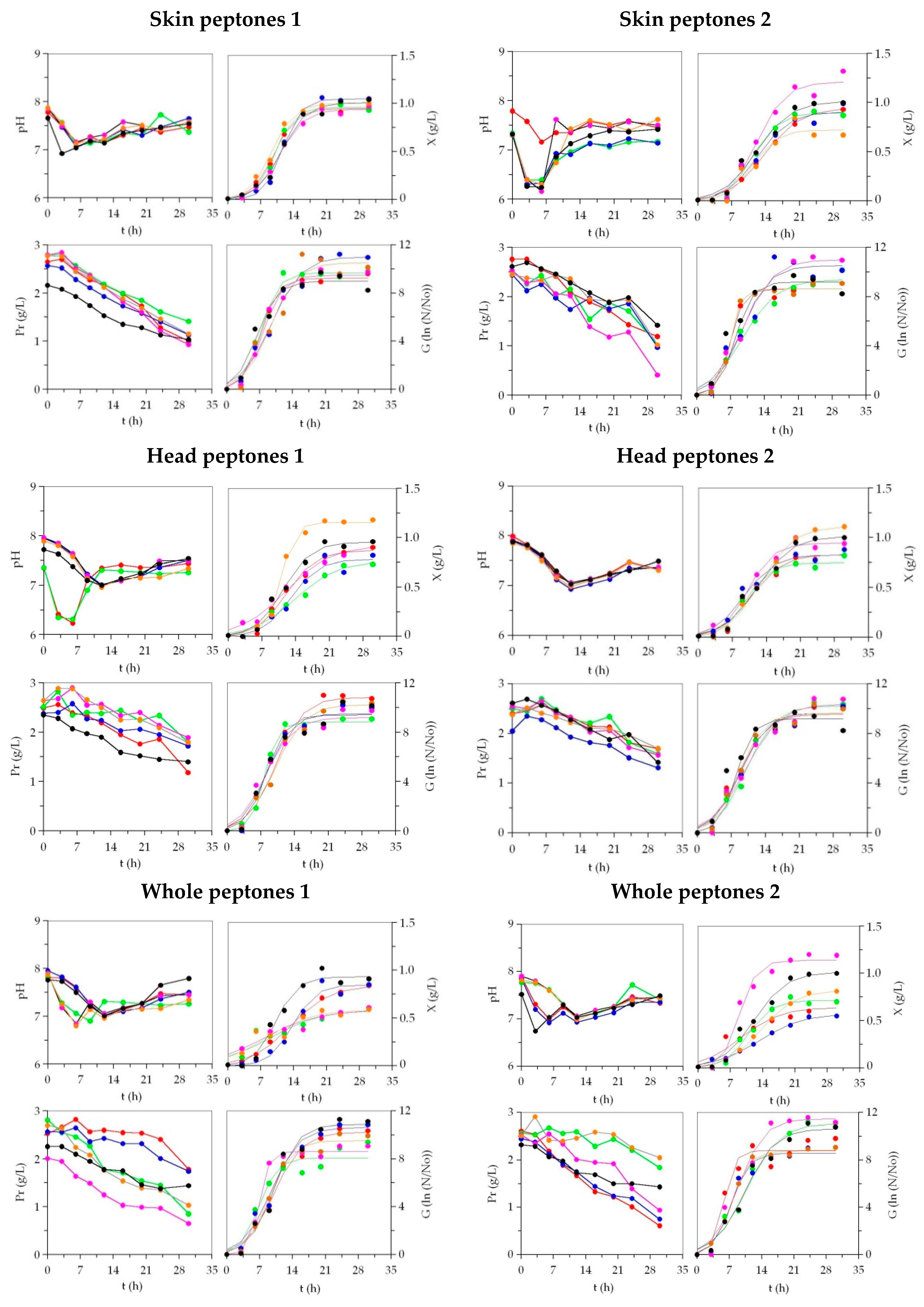

| Biomass (X) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sk_BW | Sk_RS | Sk_Ma | Sk_Po | Sk_Gu | Sk_Gr | Sk_Me | Sk_Ha | Sk_Bo | Sk_AHM | He_BW | He_RS | He_Ma | He_Po | He_Gu | MM1 | |
| Xm | 0.95 ± 0.04 | 0.94 ± 0.04 | 0.96 ± 0.04 | 0.95 ± 0.05 | 1.05 ± 0.04 | 1.00 ± 0.05 | 0.90 ± 0.08 | 0.91 ± 0.15 | 1.21 ± 0.15 | 0.72 ± 0.14 | 0.87 ± 0.09 | 0.77 ± 0.12 | 0.78 ± 0.13 | 0.92 ± 0.13 | 1.16 ± 0.06 | 1.01 ± 0.05 |
| vm | 0.10 ± 0.02 | 0.09 ± 0.02 | 0.12 ± 0.02 | 0.06 ± 0.01 | 0.13 ± 0.03 | 0.11 ± 0.03 | 0.08 ± 0.03 | 0.07 ± 0.05 | 0.10 ± 0.05 | 0.07 ± 0.06 | 0.07 ± 0.03 | 0.04 ± 0.01 | 0.07 ± 0.04 | 0.05 ± 0.01 | 0.19 ± 0.06 | 0.10 ± 0.02 |
| λX | 5.27 ± 0.99 | 5.83 ± 1.02 | 5.91 ± 0.89 | 6.43 ± 1.02 | 7.66 ± 0.82 | 4.61 ± 1.22 | 5.88 ± 1.98 | 5.79 ± 3.98 | 6.40 ± 2.67 | 6.16 ± 4.37 | 5.97 ± 2.27 | 5.03 ± 2.69 | 7.17 ± 3.29 | 2.50 (NS) | 7.81 ± 0.89 | 6.19 ± 1.73 |
| μX | 0.44 ± 0.09 | 0.38 ± 0.08 | 0.49 ± 0.10 | 0.27 ± 0.04 | 0.51 ± 0.11 | 0.42 ± 0.11 | 0.37 ± 0.14 | 0.30 ± 0.18 | 0.34 ± 0.17 | 0.39 ± 0.35 | 0.33 ± 0.13 | 0.23 ± 0.09 | 0.48 (NS) | 0.21 ± 0.08 | 0.65 ± 0.24 | 0.38 ± 0.13 |
| τX | 9.86 ± 0.56 | 11.0 ± 0.6 | 9.98 ± 0.49 | 13.8 ± 0.8 | 11.6 ± 0.5 | 9.37 ± 0.69 | 11.3 ± 1.3 | 12.4 ± 2.4 | 12.3 ± 1.8 | 11.3 ± 2.7 | 12.1 ± 1.5 | 13.9 ± 2.4 | 13.1 ± 3.5 | 11.9 ± 2.3 | 10.9 ± 0.4 | 11.1 ± 1.1 |
| tmX | 14.5 ± 1.3 | 16.3 ± 1.4 | 14.1 ± 1.1 | 21.1 ± 1.7 | 15.6 ± 1.1 | 14.1 ± 1.5 | 16.7 ± 2.8 | 18.9 ± 5.5 | 18.2 ± 4.0 | 16.4 ± 6.1 | 18.2 ± 3.4 | 22.8 ± 5.2 | 18.9 ± 4.9 | 21.4 ± 5.3 | 13.9 ± 1.1 | 16.7 ± 2.4 |
| YX/YPr | 0.564 | 0.522 | 0.674 | 0.592 | 0.729 | 0.615 | 0.582 | 0.669 | 0.616 | 0.468 | 0.682 | 1.130 | 1.228 | 1.273 | 1.362 | 0.920 |
| R2 | 0.996 | 0.997 | 0.997 | 0.998 | 0.988 | 0.995 | 0.989 | 0.970 | 0.981 | 0.943 | 0.987 | 0.984 | 0.968 | 0.983 | 0.998 | 0.990 |
| p-value | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 |
| Cells (G) | ||||||||||||||||
| Sk_BW | Sk_RS | Sk_Ma | Sk_Po | Sk_Gu | Sk_Gr | Sk_Me | Sk_Ha | Sk_Bo | Sk_AHM | He_BW | He_RS | He_Ma | He_Po | He_Gu | MM1 | |
| Gm | 9.23 ± 0.44 | 9.48 ± 0.69 | 9.68 ± 0.72 | 10.2 ± 1.2 | 11.1 ± 1.5 | 10.2 ± 0.8 | 9.40 ± 1.27 | 10.5 ± 1.3 | 11.0 ± 1.2 | 8.62 ± 0.39 | 10.8 ± 1.3 | 8.84 ± 0.34 | 9.40 ± 0.86 | 9.17 ± 1.09 | 10.4 ± 1.6 | 10.5 ± 2.2 |
| vG | 1.24 ± 0.33 | 1.13 ± 0.43 | 1.27 ± 0.52 | 1.14 ± 0.34 | 0.79 ± 0.33 | 1.49 ± 0.58 | 0.67 ± 0.30 | 0.94 ± 0.63 | 0.86 ± 0.35 | 2.00 ± 0.73 | 0.95 ± 0.43 | 1.61 ± 0.39 | 1.12 ± 0.53 | 0.82 ± 0.42 | 0.86 ± 0.41 | 1.03 ± 0.44 |
| λG | 3.54 ± 1.13 | 3.58 ± 1.77 | 3.51 ± 1.77 | 3.86 ± 1.62 | 3.78 ± 3.04 | 6.87 ± 1.57 | 2.54 (NS) | 3.72 (NS) | 3.17 ± 2.75 | 4.69 ± 0.79 | 4.41 ± 2.76 | 5.04 ± 0.78 | 3.96 ± 2.21 | 2.48 (NS) | 3.72 ± 3.30 | 3.37 ± 2.54 |
| μG | 0.54 ± 0.15 | 0.48 ± 0.19 | 0.53 ± 0.23 | 0.41 ± 0.13 | 0.29 ± 0.14 | 0.53 ± 0.22 | 0.28 ± 0.15 | 0.36 ± 0.26 | 0.31 ± 0.17 | 0.93 ± 0.35 | 0.35 ± 0.18 | 0.73 ± 0.18 | 0.48 ± 0.24 | 0.36 ± 0.20 | 0.30 ± 0.16 | 0.36 ± 0.17 |
| τG | 7.27 ± 0.62 | 7.76 ± 0.97 | 7.32 ± 0.96 | 8.79 ± 0.91 | 10.8 ± 2.0 | 10.6 ± 0.9 | 9.59 ± 2.12 | 9.35 ± 2.39 | 9.59 ± 1.69 | 6.84 ± 0.49 | 10.1 ± 1.7 | 7.79 ± 0.43 | 8.20 ± 1.21 | 8.07 ± 1.76 | 10.4 ± 2.1 | 8.93 ± 1.47 |
| tmG | 11.0 ± 1.3 | 12.0 ± 2.1 | 11.1 ± 2.0 | 13.7 ± 2.0 | 17.8 ± 4.6 | 14.4 ± 1.9 | 16.6 ± 4.9 | 15.1 ± 5.4 | 16.0 ± 3.9 | 9.00 ± 1.08 | 15.9 ± 5.2 | 10.5 ± 0.8 | 12.4 ± 2.6 | 13.7 ± 3.9 | 17.0 ± 4.9 | 14.5 ± 3.3 |
| YG/YPr | 4.71 | 4.83 | 3.70 | 5.989 | 24.13 | 8.449 | 6.81 | 5.989 | 14.54 | 6.37 | 14.74 | 12.80 | 15.13 | 13.05 | 6.500 | 13.63 |
| R2 | 0.994 | 0.988 | 0.985 | 0.991 | 0.977 | 0.990 | 0.973 | 0.949 | 0.979 | 0.994 | 0.977 | 0.996 | 0.981 | 0.971 | 0.969 | 0.981 |
| p-value | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 |
| Biomass (X) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| He_Gr | He_Bo | He_Ha | He_AHM | He_Me | Wh_BW | Wh_RS | Wh_Ma | Wh_Po | Wh_Gu | Wh_Gr | Wh_Bo | Wh_Ha | Wh_Me | Wh_AHM | MM2 | |
| Xm | 0.83 ± 0.11 | 0.94 ± 0.09 | 0.83 ± 0.10 | 1.11 ± 0.08 | 0.75 ± 0.07 | 0.85 ± 0.05 | 0.69 ± 0.20 | 0.85 ± 0.11 | 0.68 ± 0.19 | 0.69 ± 0.17 | 0.74 ± 0.16 | 1.14 ± 0.20 | 0.68 ± 0.05 | 0.71 ± 0.07 | 0.82 ± 0.04 | 0.95 ± 0.10 |
| vm | 0.07 ± 0.03 | 0.09 ± 0.04 | 0.07 ± 0.04 | 0.08 ± 0.02 | 0.08 ± 0.02 | 0.05 ± 0.01 | 0.04 ± 0.02 | 0.09 ± 0.04 | 0.04 ± 0.02 | 0.05 ± 0.02 | 0.06 ± 0.03 | 0.15 ± 0.11 | 0.05 ± 0.01 | 0.07 ± 0.03 | 0.06 ± 0.01 | 0.08 ± 0.03 |
| λX | 4.55 ± 3.07 | 4.89 ± 2.36 | 3.10 ± 3.09 | 6.04 ± 1.49 | 5.00 ± 2.11 | 4.90 ± 1.15 | −2.1 (NS) | 8.81 ± 2.54 | −3.9 (NS) | −1.6 (NS) | 0.75 (NS) | 5.27 ± 2.32 | 3.47 ± 1.63 | 5.80 ± 2.32 | 6.42 ± 1.01 | 5.72 ± 2.26 |
| μX | 0.31 ± 0.16 | 0.38 ± 0.17 | 0.32 ± 0.16 | 0.29 ± 0.07 | 0.42 ± 0.18 | 0.24 ± 0.04 | 0.18 ± 0.17 | 0.40 ± 0.21 | 0.17 ± 0.17 | 0.20 ± 0.19 | 0.24 ± 0.20 | 0.54 ± 0.19 | 0.21 ± 0.05 | 0.40 ± 0.19 | 0.27 ± 0.04 | 0.35 ± 0.14 |
| τX | 10.9 ± 2.0 | 10.2 ± 1.4 | 9.38 ± 1.87 | 12.9 ± 1.1 | 9.82 ± 1.20 | 13.4 ± 1.0 | 9.06 ± 6.16 | 13.8 ± 1.7 | 7.65 ± 6.00 | 8.23 ± 5.07 | 9.25 ± 4.19 | 8.99 ± 1.40 | 13.1 ± 1.5 | 10.8 ± 1.4 | 13.8 ± 0.8 | 11.5 ± 1.5 |
| tmX | 17.3 ± 4.5 | 15.5 ± 3.2 | 15.7 ± 4.3 | 20.1 ± 2.6 | 14.6 ± 2.7 | 21.9 ± 2.1 | 20.3 ± 15.3 | 18.8 ± 3.6 | 19.2 ± 15.7 | 18.0 ± 12.6 | 17.8 ± 9.9 | 12.7 ± 3.1 | 22.8 ± 3.3 | 15.9 ± 3.1 | 21.1 ± 1.7 | 17.3 ± 3.3 |
| YX/YPr | 1.225 | 0.970 | 1.196 | 1.639 | 0.884 | 1.103 | 0.463 | 1.009 | 0.588 | 0.653 | 0.547 | 0.899 | 0.463 | 0.955 | 1.749 | 1.006 |
| R2 | 0.975 | 0.981 | 0.973 | 0.995 | 0.985 | 0.997 | 0.924 | 0.979 | 0.925 | 0.924 | 0.917 | 0.993 | 0.994 | 0.983 | 0.998 | 0.986 |
| p-value | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.05 | <0.005 | <0.05 | <0.05 | <0.01 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 |
| Cells (G) | ||||||||||||||||
| He_Gr | He_Bo | He_Ha | He_AHM | He_Me | Wh_BW | Wh_RS | Wh_Ma | Wh_Po | Wh_Gu | Wh_Gr | Wh_Bo | Wh_Ha | Wh_Me | Wh_AHM | MM2 | |
| Gm | 9.56 ± 1.09 | 10.4 ± 1.5 | 9.67 ± 1.06 | 10.6 ± 0.9 | 8.62 ± 0.48 | 9.20 ± 1.00 | 8.09 ± 1.16 | 10.6 ± 1.1 | 8.82 ± 1.02 | 9.52 ± 0.73 | 10.1 ± 1.1 | 11.4 ± 1.3 | 8.57 ± 0.70 | 8.78 ± 0.33 | 9.39 ± 0.58 | 9.99 ± 2.18 |
| vG | 0.85 ± 0.41 | 0.74 ± 0.35 | 0.87 ± 0.40 | 0.94 ± 0.38 | 1.54 ± 0.58 | 0.86 ± 0.32 | 1.04 ± 0.82 | 0.90 ± 0.36 | 1.77 ± 1.77 | 0.99 ± 0.35 | 0.92 ± 0.36 | 0.81 ± 0.33 | 1.13 ± 0.51 | 1.13 ± 0.23 | 1.09 ± 0.34 | 0.94 ± 0.53 |
| λG | 3.00 ± 2.92 | 3.04 (NS) | 3.27 ± 2.77 | 3.49 ± 2.26 | 4.06 ± 1.16 | 4.11 ± 2.36 | 2.97 (NS) | 3.54 ± 2.55 | 3.31 ± 2.81 | 4.22 ± 1.87 | 4.43 ± 2.40 | 3.80 ± 2.80 | 3.63 ± 1.93 | 3.54 ± 0.89 | 3.04 ± 1.53 | 3.26 ± 3.13 |
| μG | 0.36 ± 0.19 | 0.29 ± 0.16 | 0.36 ± 0.18 | 0.39 ± 0.17 | 0.71 ± 0.28 | 0.33 ± 0.14 | 0.51 ± 0.43 | 0.34 ± 0.15 | 5.80 ± 1.27 | 0.42 ± 0.16 | 0.34 ± 0.15 | 0.30 ± 0.14 | 0.53 ± 0.25 | 0.52 ± 0.11 | 0.46 ± 0.15 | 0.39 ± 0.24 |
| τG | 8.61 ± 1.68 | 10.0 ± 2.2 | 8.83 ± 1.60 | 8.61 ± 1.27 | 6.86 ± 0.64 | 10.1 ± 1.5 | 6.88 ± 1.90 | 9.47 ± 1.52 | 11.0 (NS) | 9.02 ± 1.05 | 10.3 ± 1.5 | 10.4 ± 1.8 | 7.42 ± 1.05 | 7.41 ± 0.49 | 7.36 ± 0.84 | 8.33 ± 1.76 |
| tmG | 14.2 ± 3.8 | 17.0 ± 5.2 | 14.4 ± 3.6 | 13.8 ± 2.8 | 9.67 ± 1.36 | 16.1 ± 3.3 | 10.8 ± 4.0 | 15.4 ± 3.5 | 8.29 ± 2.90 | 13.8 ± 2.3 | 16.2 ± 3.3 | 17.0 ± 4.1 | 11.2 ± 2.2 | 11.3 ± 1.0 | 11.7 ± 1.8 | 13.4 ± 3.9 |
| YG/YPr | 14.12 | 11.21 | 13.91 | 14.85 | 4.27 | 11.31 | 5.10 | 13.00 | 4.44 | 11.71 | 5.273 | 6.77 | 6.20 | 6.95 | 4.67 | 10.67 |
| R2 | 0.975 | 0.971 | 0.976 | 0.983 | 0.991 | 0.984 | 0.946 | 0.981 | 0.956 | 0.988 | 0.982 | 0.979 | 0.983 | 0.996 | 0.990 | 0.968 |
| p-value | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 | <0.005 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vázquez, J.A.; Durán, A.; Nogueira, M.; Menduíña, A.; Antunes, J.; Freitas, A.C.; Gomes, A.M. Production of Marine Probiotic Bacteria in a Cost-Effective Marine Media Based on Peptones Obtained from Discarded Fish By-Products. Microorganisms 2020, 8, 1121. https://doi.org/10.3390/microorganisms8081121
Vázquez JA, Durán A, Nogueira M, Menduíña A, Antunes J, Freitas AC, Gomes AM. Production of Marine Probiotic Bacteria in a Cost-Effective Marine Media Based on Peptones Obtained from Discarded Fish By-Products. Microorganisms. 2020; 8(8):1121. https://doi.org/10.3390/microorganisms8081121
Chicago/Turabian StyleVázquez, José Antonio, Ana Durán, Margarita Nogueira, Araceli Menduíña, Joana Antunes, Ana Cristina Freitas, and Ana María Gomes. 2020. "Production of Marine Probiotic Bacteria in a Cost-Effective Marine Media Based on Peptones Obtained from Discarded Fish By-Products" Microorganisms 8, no. 8: 1121. https://doi.org/10.3390/microorganisms8081121
APA StyleVázquez, J. A., Durán, A., Nogueira, M., Menduíña, A., Antunes, J., Freitas, A. C., & Gomes, A. M. (2020). Production of Marine Probiotic Bacteria in a Cost-Effective Marine Media Based on Peptones Obtained from Discarded Fish By-Products. Microorganisms, 8(8), 1121. https://doi.org/10.3390/microorganisms8081121








