The Differential Phosphorylation-Dependent Signaling and Glucose Immunometabolic Responses Induced during Infection by Salmonella Enteritidis and Salmonella Heidelberg in Chicken Macrophage-like cells
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line and Maintenance
2.2. Bacteria Serovars
2.3. Infection of Cells with Salmonella
2.4. Gentamicin Protection Assay
2.5. Kinome Peptide Array Analysis
2.6. Seahorse XFp Metabolic Assay
2.7. Statistics
3. Results
3.1. Salmonella Alters Host (HD11 Cells) Immunometabolism
3.2. S. Enteritidis and S. Heidelberg Induce Differential Phosphorylation of Cytoskeletal Proteins
3.3. S. Enteritidis and S. Heidelberg Induce Differential Phosphorylation of Metabolic Proteins to Promote Their Survival
3.4. S. Enteritidis Infection Induces an Early Increase in Glucose Metabolism, and Early S. Heidelberg Infection Dampens Glucose Metabolism
3.5. S. Enteritidis and S. Heidelberg Induce Differential Phosphorylation of Inflammatory Proteins
3.6. S. Enteritidis Is More Invasive Than S. Heidelberg
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Protein Name | Human UniProt Accession | Human Site | Chicken Site |
|---|---|---|---|
| Transforming protein RhoA (RhoA) | P61586 | S188 | S199 |
| Rho-associated, coiled-coil-containing protein kinase 2 (ROCK2) | O75116 | Y722 | Y507 |
| Rho GTPase-activating protein 6 | O43182 | Y407 | Y211 |
| Rho GTPase-activating protein 17 | Q68EM7 | S484 | S479 |
| Caspase-1 | P29466 | S227 | S106 |
| NACHT, LRR, and PYD domains-containing protein 3 (NLRP3) | Q96P20 | T233 | T24 |
| Interferon regulatory factor 1 (IRF-1) | P10914 | Y109 | Y109 |
| Caspase recruitment domain-containing protein 11 (CARD11) | Q9BXL7 | S116 | S118 |
| Toll-like receptor 5 (TLR5) | O60602 | Y798 | Y800 |
| Toll-like receptor 3 (TLR3) | O15455 | Y858 | Y854 |
| Interleukin-6 receptor subunit beta (IL-6R) | P40189 | S782 | S757 |
| Caspase-3 | P42574 | S150 | S158 |
| Caspase-8 | Q14790 | S347 | S350 |
| MAP kinase-interacting protein kinase 1 (MNK1) | Q9BUB5 | T255 | T199 |
| Jun N-terminal kinase 1 (JNK1) | P45983 | T183 | T183 |
| NF-kappa-B inhibitor alpha/I-kappa-B-alpha (IkB-α) | P25963 | Y42 | Y46 |
| Serine/threonine-protein kinase mTOR (mTOR) | P42345 | S2448 | S2352 |
| 6-Phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFK2) | Q16875 | S461 | S462 |
| Glucose-6-phosphate isomerase (GPI) | P06744 | S185 | S184 |
| 5′-AMP-activated protein kinase catalytic subunit α-1 (AMPK) | Q13131 | T183 | T185 |
| Hypoxia-inducible factor 1-alpha (HIF-1α) | Q16665 | S247 | S247 |
References
- Andino, A.; Hanning, I. Salmonella enterica: Survival, Colonization, and Virulence Differences among Serovars. Sci. World J. 2015, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Swaggerty, C.; Kogut, M.H.; He, H.; Genovese, K.J.; Johnson, C.; Arsenault, R.J. Differential Levels of Cecal Colonization by Salmonella Enteritidis in Chickens Triggers Distinct Immune Kinome Profiles. Front. Vet. Sci. 2017, 4, 4. [Google Scholar] [CrossRef] [PubMed]
- Majowicz, S.E.; Musto, J.; Scallan, E.; Angulo, F.J.; Kirk, M.D.; O’Brien, S.; Jones, T.F.; Fazil, A.; Hoekstra, R.M. The Global Burden of Nontyphoidal Salmonella Gastroenteritis. Clin. Infect. Dis. 2010, 50, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Gal-Mor, O.; Boyle, E.C.; Grassl, G.A. Same species, different diseases: How and why typhoidal and non-typhoidal Salmonella enterica serovars differ. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef]
- Feasey, N.A.; Dougan, G.; Kingsley, R.A.; Heyderman, R.S.; Gordon, M.A. Invasive non-typhoidal salmonella disease: An emerging and neglected tropical disease in Africa. Lancet 2012, 379, 2489–2499. [Google Scholar] [CrossRef]
- Porwollik, S.; Boyd, E.F.; Choy, C.; Cheng, P.; Florea, L.; Proctor, E.; McClelland, M. Characterization of Salmonella enterica Subspecies I Genovars by Use of Microarrays. J. Bacteriol. 2004, 186, 5883–5898. [Google Scholar] [CrossRef] [PubMed]
- Demczuk, W.; Soule, G.; Clark, C.; Ackermann, H.-W.; Easy, R.; Khakhria, R.; Rodgers, F.; Ahmed, R. Phage-Based Typing Scheme for Salmonella enterica Serovar Heidelberg, a Causative Agent of Food Poisonings in Canada. J. Clin. Microbiol. 2003, 41, 4279–4284. [Google Scholar] [CrossRef]
- Haeusler, G.M.; Curtis, N. Non-typhoidal Salmonella in children: Microbiology, epidemiology, and treatment. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2013; Volume 764, pp. 13–26. [Google Scholar]
- Nair, D.V.T.; Venkitanarayanan, K.; Johny, A.K. Antibiotic-Resistant Salmonella in the Food Supply and the Potential Role of Antibiotic Alternatives for Control. Foods 2018, 7, 167. [Google Scholar] [CrossRef]
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.-A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne Illness Acquired in the United States—Major Pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef]
- Gordon, M.A. Salmonella infections in immunocompromised adults. J. Infect. 2008, 56, 413–422. [Google Scholar] [CrossRef]
- Shimoni, Z.; Pitlik, S.; Leibovici, L.; Samra, Z.; Konigsberger, H.; Drucker, M.; Agmon, V.; Ashkenazi, S.; Weinberger, M. Nontyphoid Salmonella Bacteremia: Age-Related Differences in Clinical Presentation, Bacteriology, and Outcome. Clin. Infect. Dis. 1999, 28, 822–827. [Google Scholar] [CrossRef]
- Zhao, X.; Gao, Y.; Ye, C.; Yang, L.; Wang, T.; Chang, W. Prevalence and Characteristics of Salmonella Isolated from Free-Range Chickens in Shandong Province, China. BioMed Res. Int. 2016, 1–6. [Google Scholar] [CrossRef]
- Andres, V.M.; Davies, R.H. Biosecurity Measures to Control Salmonella and Other Infectious Agents in Pig Farms: A Review. Compr. Rev. Food Sci. Food Saf. 2015, 14, 317–335. [Google Scholar] [CrossRef]
- Shah, D.H.; Paul, N.C.; Sischo, W.C.; Crespo, R.; Guard, J. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult. Sci. 2017, 96, 687–702. [Google Scholar] [CrossRef]
- Olsen, S.J.; Bishop, R.; Brenner, F.W.; Roels, T.H.; Bean, N.; Tauxe, R.V.; Slutsker, L. The Changing Epidemiology of Salmonella: Trends in Serotypes Isolated from Humans in the United States, 1987–1997. J. Infect. Dis. 2001, 183, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Djeffal, S.; Mamache, B.; Elgroud, R.; Hireche, S.; Bouaziz, O. Prevalence and risk factors for Salmonella spp. contamination in broiler chicken farms and slaughterhouses in the northeast of Algeria. Vet. World 2018, 11, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Arsenault, R.J.; Genovese, K.J.; Johnson, C.; Kogut, M.H. Chicken macrophages infected with Salmonella (S.) Enteritidis or S. Heidelberg produce differential responses in immune and metabolic signaling pathways. Vet. Immunol. Immunopathol. 2018, 195, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Ciraci, C.; Tuggle, C.K.; Wannemuehler, M.J.; Nettleton, D.S.; Lamont, S.J. Unique genome-wide transcriptome profiles of chicken macrophages exposed to Salmonella-derived endotoxin. BMC Genom. 2010, 11, 545. [Google Scholar] [CrossRef]
- Jarvis, N.A.; Donaldson, J.R.; O’Bryan, C.A.; Ricke, S.C.; Crandall, P.G. Listeria monocytogenes infection of HD11, chicken macrophage-like cells. Poult. Sci. 2017, 96, 950–956. [Google Scholar] [CrossRef]
- Beug, H.; von Kirchbach, A.; Doderlein, G.; Conscience, J.-F.; Graf, T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell 1979, 18, 375–390. [Google Scholar] [CrossRef]
- Wisner, A.L.S.; Potter, A.A.; Köster, W. Effect of the Salmonella Pathogenicity Island 2 Type III Secretion System on Salmonella Survival in Activated Chicken Macrophage-Like HD11 Cells. PLoS ONE 2011, 6, e29787. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 19, 92. [Google Scholar] [CrossRef] [PubMed]
- Langston, P.K.; Shibata, M.; Horng, T. Metabolism Supports Macrophage Activation. Front. Immunol. 2017, 8, 723. [Google Scholar] [CrossRef] [PubMed]
- Gog, J.R.; Murcia, A.; Osterman, N.; Restif, O.; McKinley, T.J.; Sheppard, M.; Achouri, S.; Wei, B.; Mastroeni, P.; Wood, J.L.N.; et al. Dynamics of Salmonella infection of macrophages at the single cell level. J. R. Soc. Interface 2012, 9, 2696–2707. [Google Scholar] [CrossRef] [PubMed]
- Gorbach, S.L. Microbiology of the Gastrointestinal Tract. In Medical Microbiology; Baron, S., Ed.; University of Texas Medical Branch at Galveston: Galveston, TX, USA, 1996; ISBN 978-0-9631172-1-2. [Google Scholar]
- Yang, M.; Xu, J.; Wang, Q.; Zhang, A.; Wang, K. An obligatory anaerobic Salmonella typhimurium strain redirects M2 macrophage to the M1 phenotype. Oncol. Lett. 2018, 15, 3918–3922. [Google Scholar] [CrossRef]
- Brundu, S.F.A. Polarization and Repolarization of Macrophages. J. Clin. Cell. Immunol. 2015, 6, 1–10. [Google Scholar] [CrossRef]
- Arsenault, R.J.; Kogut, M.H. Immunometabolism and the Kinome Peptide Array: A New Perspective and Tool for the Study of Gut Health. Front. Vet. Sci. 2015, 2, 633. [Google Scholar] [CrossRef]
- Li, Y.; Arsenault, R.J.; Trost, B.; Slind, J.; Griebel, P.J.; Napper, S.; Kusalik, A.J. A Systematic Approach for Analysis of Peptide Array Kinome Data. Sci. Signal. 2012, 5, pl2. [Google Scholar] [CrossRef]
- Arsenault, R.J.; Kogut, M.H. Chicken-Specific Peptide Arrays for Kinome Analysis: Flight for the Flightless. Curr. Top. Biotechnol. 2012, 7, 79–89. [Google Scholar]
- Jalal, S.; Arsenault, R.; Potter, A.; Babiuk, L.A.; Griebel, P.J.; Napper, S. Genome to Kinome: Species-Specific Peptide Arrays for Kinome Analysis. Sci. Signal. 2009, 2, pl1. [Google Scholar] [CrossRef]
- Parikh, K.; Peppelenbosch, M.P.; Ritsema, T. Kinome Profiling Using Peptide Arrays in Eukaryotic Cells. In Advanced Structural Safety Studies; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2009; Volume 527, pp. 269–280. [Google Scholar]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Muzio, L.L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Borsoi, A.; Santos, L.R.D.; Rodrigues, L.B.; Moraes, H.L.D.S.; Salle, C.T.P.; Nascimento, V.P.D. Behavior of Salmonella Heidelberg and Salmonella Enteritidis Strains Following Broiler Chick Inoculation: Evaluation of Cecal Morphometry, Liver and Cecum Bacterial Counts And Fecal Excretion Patterns. Braz. J. Microbiol. 2011, 42, 266–273. [Google Scholar] [CrossRef]
- Kogut, M.H.; Genovese, K.J.; He, H.; Arsenault, R.J. AMPK and mTOR: Sensors and regulators of immunometabolic changes during Salmonella infection in the chicken. Poult. Sci. 2016, 95, 345–353. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Genovese, K.J.; Swaggerty, C.; Nisbet, D.J.; Kogut, M.H. A Comparative Study on Invasion, Survival, Modulation of Oxidative Burst, and Nitric Oxide Responses of Macrophages (HD11), and Systemic Infection in Chickens by Prevalent Poultry Salmonella Serovars. Foodborne Pathog. Dis. 2012, 9, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Pugh, R.; Laughlin, R.C.; Andrews-Polymenis, H.; McClelland, M.; Bäumler, A.J.; Adams, L.G. High-throughput Assay to Phenotype Salmonella enterica Typhimurium Association, Invasion, and Replication in Macrophages. J. Vis. Exp. 2014, e51759. [Google Scholar] [CrossRef] [PubMed]
- Arsenault, R.; Lee, J.T.; Latham, R.; Carter, B.; Kogut, M.H. Changes in immune and metabolic gut response in broilers fed β-mannanase in β-mannan-containing diets. Poult. Sci. 2017, 96, 4307–4316. [Google Scholar] [CrossRef]
- Trost, B.; Kindrachuk, J.; Maattanen, P.; Napper, S.; Kusalik, A. PIIKA 2: An Expanded, Web-Based Platform for Analysis of Kinome Microarray Data. PLoS ONE 2013, 8, e80837. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.V.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2016, 45, D362–D368. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2015, 44, D457–D462. [Google Scholar] [CrossRef]
- Boutet, E.; Lieberherr, D.; Tognolli, M.; Schneider, M.; Bansal, P.; Bridge, A.; Poux, S.; Bougueleret, L.; Xenarios, I. UniProtKB/Swiss-Prot, the Manually Annotated Section of the UniProt KnowledgeBase: How to Use the Entry View. Adv. Struct. Saf. Stud. 2016, 1374, 23–54. [Google Scholar] [CrossRef]
- Zaru, R.; Magrane, M.; O’Donovan, C. The UniProt Consortium From the research laboratory to the database: The Caenorhabditis elegans kinome in UniProtKB. Biochem. J. 2017, 474, 493–515. [Google Scholar] [CrossRef] [PubMed]
- Pundir, S.; Martin, M.J.; O’Donovan, C. Chapter 2. Protein Knowledgebase. Breast Cancer 2017, 1558, 41–55. [Google Scholar] [CrossRef]
- Hornbeck, P.; Zhang, B.; Murray, B.; Kornhauser, J.M.; Latham, V.; Skrzypek, E. PhosphoSitePlus, 2014: Mutations, PTMs and recalibrations. Nucleic Acids Res. 2014, 43, D512–D520. [Google Scholar] [CrossRef]
- Mookerjee, S.A.; Brand, M.D. Measurement and Analysis of Extracellular Acid Production to Determine Glycolytic Rate. J. Vis. Exp. 2015, e53464. [Google Scholar] [CrossRef]
- Seahorse XFp Analyzer/Agilent. Available online: https://www.agilent.com/en/products/cell-analysis/seahorse-analyzers/seahorse-xfp-analyzer (accessed on 28 February 2019).
- Cascales, E. Inside the Chamber of Secrets of the Type III Secretion System. Cell 2017, 168, 949–951. [Google Scholar] [CrossRef] [PubMed]
- Jennings, E.; Thurston, T.L.; Holden, D.W. Salmonella SPI-2 Type III Secretion System Effectors: Molecular Mechanisms and Physiological Consequences. Cell Host Microbe 2017, 22, 217–231. [Google Scholar] [CrossRef]
- Crhanova, M.; Hradecka, H.; Faldynova, M.; Matulova, M.; Havlickova, H.; Sisak, F.; Rychlik, I. Immune Response of Chicken Gut to Natural Colonization by Gut Microflora and to Salmonella enterica Serovar Enteritidis Infection. Infect. Immun. 2011, 79, 2755–2763. [Google Scholar] [CrossRef]
- Patel, J.C.; Galán, J.E. Differential activation and function of Rho GTPases during Salmonella–host cell interactions. J. Cell Boil. 2006, 175, 453–463. [Google Scholar] [CrossRef]
- Quilliam, L.A.; Lambert, Q.T.; Mickelson-Young, L.A.; Westwick, J.K.; Sparks, A.B.; Kay, B.K.; Jenkins, N.A.; Gilbert, D.J.; Copeland, N.G.; Der, C.J. Isolation of a NCK-associated Kinase, PRK2, an SH3-binding Protein and Potential Effector of Rho Protein Signaling. J. Boil. Chem. 1996, 271, 28772–28776. [Google Scholar] [CrossRef]
- Matsuzawa, T.; Kuwae, A.; Yoshida, S.; Sasakawa, C.; Abe, A. Enteropathogenic Escherichia coli activates the RhoA signaling pathway via the stimulation of GEF-H1. EMBO J. 2004, 23, 3570–3582. [Google Scholar] [CrossRef]
- Feng, J.; Ito, M.; Ichikawa, K.; Isaka, N.; Nishikawa, M.; Hartshorne, D.J.; Nakano, T. Inhibitory Phosphorylation Site for Rho-associated Kinase on Smooth Muscle Myosin Phosphatase. J. Boil. Chem. 1999, 274, 37385–37390. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.; Kendall, M.M. Salmonella enterica Serovar Typhimurium Strategies for Host Adaptation. Front. Microbiol. 2017, 8. [Google Scholar] [CrossRef]
- Lopez, C.A.; Winter, S.E.; Rivera-Chávez, F.; Xavier, M.N.; Poon, V.; Nuccio, S.-P.; Tsolis, R.M.; Bäumler, A.J. Phage-Mediated Acquisition of a Type III Secreted Effector Protein Boosts Growth of Salmonella by Nitrate Respiration. mBio 2012, 3, e00143-12. [Google Scholar] [CrossRef] [PubMed]
- Wigley, P. Salmonella enterica in the Chicken: How it has Helped Our Understanding of Immunology in a Non-Biomedical Model Species. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Bratburd, J.R.; Keller, C.; Vivas, E.; Gemperline, E.; Li, L.; Rey, F.E.; Currie, C.R.; Hsiao, A.; Whiteson, K. Gut Microbial and Metabolic Responses to Salmonella enterica Serovar Typhimurium and Candida albicans. mBio 2018, 9, e02032-18. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G. AMP-activated protein kinase—An energy sensor that regulates all aspects of cell function. Genes Dev. 2011, 25, 1895–1908. [Google Scholar] [CrossRef]
- Berg, J.M.; Tymoczko, J.L.; Stryer, L. 20.3 the Pentose Phosphate Pathway Generates NADPH and Synthesizes Five-Carbon Sugars. Biochemistry 2002, 5. [Google Scholar]
- Poulsen, B.R.; Nøhr, J.; Douthwaite, S.; Hansen, L.V.; Iversen, J.J.L.; Visser, J.; Ruijter, G.J.G. Increased NADPH concentration obtained by metabolic engineering of the pentose phosphate pathway in Aspergillus niger. FEBS J. 2005, 272, 1313–1325. [Google Scholar] [CrossRef]
- Stunault, M.I.; Bories, G.; Guinamard, R.R.; Ivanov, S. Metabolism Plays a Key Role during Macrophage Activation. Available online: https://www.hindawi.com/journals/mi/2018/2426138/ (accessed on 17 May 2019).
- Müller, A.J.; Hoffmann, C.; Galle, M.; Broeke, A.V.D.; Heikenwalder, M.; Falter, L.; Misselwitz, B.; Kremer, M.; Beyaert, R.; Hardt, W.-D. The S. Typhimurium Effector SopE Induces Caspase-1 Activation in Stromal Cells to Initiate Gut Inflammation. Cell Host Microbe 2009, 6, 125–136. [Google Scholar] [CrossRef]
- Brint, E.K.; Fitzgerald, K.A.; Smith, P.; Coyle, A.J.; Gutierrez-Ramos, J.-C.; Fallon, P.G.; O’Neill, L.A.J. Characterization of Signaling Pathways Activated by the Interleukin 1 (IL-1) Receptor Homologue T1/ST2. J. Boil. Chem. 2002, 277, 49205–49211. [Google Scholar] [CrossRef]
- Dolniak, B.; Katsoulidis, E.; Carayol, N.; Altman, J.K.; Redig, A.J.; Tallman, M.S.; Ueda, T.; Watanabe-Fukunaga, R.; Fukunaga, R.; Platanias, L.C. Regulation of Arsenic Trioxide-induced Cellular Responses by Mnk1 and Mnk2. J. Boil. Chem. 2008, 283, 12034–12042. [Google Scholar] [CrossRef]
- Fan, C.; Yang, J.; Engelhardt, J.F. Temporal pattern of NF? B activation influences apoptotic cell fate in a stimuli-dependent fashion. J. Cell Sci. 2002, 115, 4843–4853. [Google Scholar] [CrossRef] [PubMed]
- Hahn-Windgassen, A.; Nogueira, V.; Chen, C.-C.; Skeen, J.E.; Sonenberg, N.; Hay, N. Akt Activates the Mammalian Target of Rapamycin by Regulating Cellular ATP Level and AMPK Activity. J. Boil. Chem. 2005, 280, 32081–32089. [Google Scholar] [CrossRef]
- Buerger, C.; Shirsath, N.; Lang, V.; Berard, A.; Diehl, S.; Kaufmann, R.; Boehncke, W.-H.; Wolf, P. Inflammation dependent mTORC1 signaling interferes with the switch from keratinocyte proliferation to differentiation. PLoS ONE 2017, 12, e0180853. [Google Scholar] [CrossRef] [PubMed]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Nakayama, M.; Kaibuchi, K. Rho-kinase/ROCK: A key regulator of the cytoskeleton and cell polarity. Cytoskeleton 2010, 67, 545–554. [Google Scholar] [CrossRef]
- Prakash, S.K.; Paylor, R.; Jenna, S.; Lamarche-Vane, N.; Armstrong, D.L.; Xu, B.; Mancini, M.A.; Zoghbi, H.Y. Functional analysis of ARHGAP6, a novel GTPase-activating protein for RhoA. Hum. Mol. Genet. 2000, 9, 477–488. [Google Scholar] [CrossRef]
- Behnsen, J.; Pérez-López, A.; Nuccio, S.-P.; Raffatellu, M. Exploiting host immunity: The Salmonella paradigm. Trends Immunol. 2015, 36, 112–120. [Google Scholar] [CrossRef]
- Schikora, A.; Virlogeux-Payant, I.; Bueso, E.; García, A.V.; Nilau, T.; Charrier, A.; Pelletier, S.; Menanteau, P.; Baccarini, M.; Velge, P.; et al. Conservation of Salmonella Infection Mechanisms in Plants and Animals. PLoS ONE 2011, 6, e24112. [Google Scholar] [CrossRef]
- Alvarado-Kristensson, M.; Melander, F.; Leandersson, K.; Rönnstrand, L.; Wernstedt, C.; Andersson, T. p38-MAPK Signals Survival by Phosphorylation of Caspase-8 and Caspase-3 in Human Neutrophils. J. Exp. Med. 2004, 199, 449–458. [Google Scholar] [CrossRef]
- Arpaia, N.; Godec, J.; Lau, L.; Sivick, K.E.; McLaughlin, L.M.; Jones, M.B.; Dracheva, T.; Peterson, S.N.; Monack, D.M.; Barton, G.M. TLR Signaling Is Required for Salmonella typhimurium Virulence. Cell 2011, 144, 675–688. [Google Scholar] [CrossRef] [PubMed]
- Elsheimer-Matulova, M.; Varmuzova, K.; Sisak, F.; Havlickova, H.; Babak, V.; Stejskal, K.; Zdráhal, Z.; Rychlik, I. Chicken innate immune response to oral infection with Salmonella enterica serovar Enteritidis. Vet. Res. 2013, 44, 37. [Google Scholar] [CrossRef] [PubMed]
- Venkitanarayanan, K.; Thakur, S.; Ricke, S.C. Food Safety in Poultry Meat Production; Springer: Berlin/Heidelberg, Germany, 2019; ISBN 978-3-030-05011-5. [Google Scholar]
- Vazquez-Torres, A.; Fang, F.C. Salmonella evasion of the NADPH phagocyte oxidase. Microbes Infect. 2001, 3, 1313–1320. [Google Scholar] [CrossRef]
- Choi, Y.J.; Park, Y.J.; Jeong, H.O.; Kim, D.H.; Ha, Y.M.; Kim, J.M.; Song, Y.M.; Heo, H.-S.; Yu, B.P. Inhibitory Effect of mTOR Activator MHY1485 on Autophagy: Suppression of Lysosomal Fusion. PLoS ONE 2012, 7, e43418. [Google Scholar] [CrossRef]
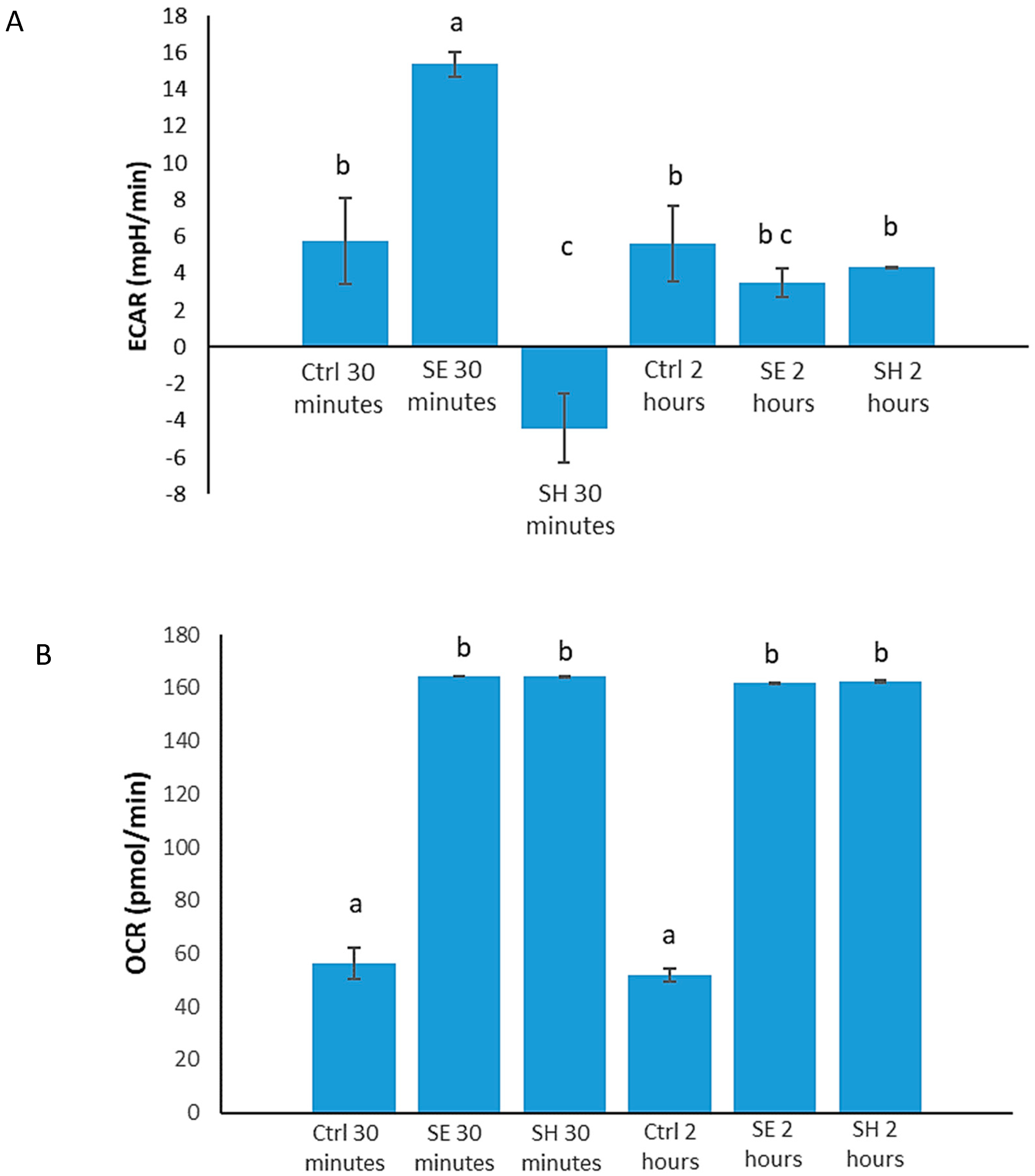
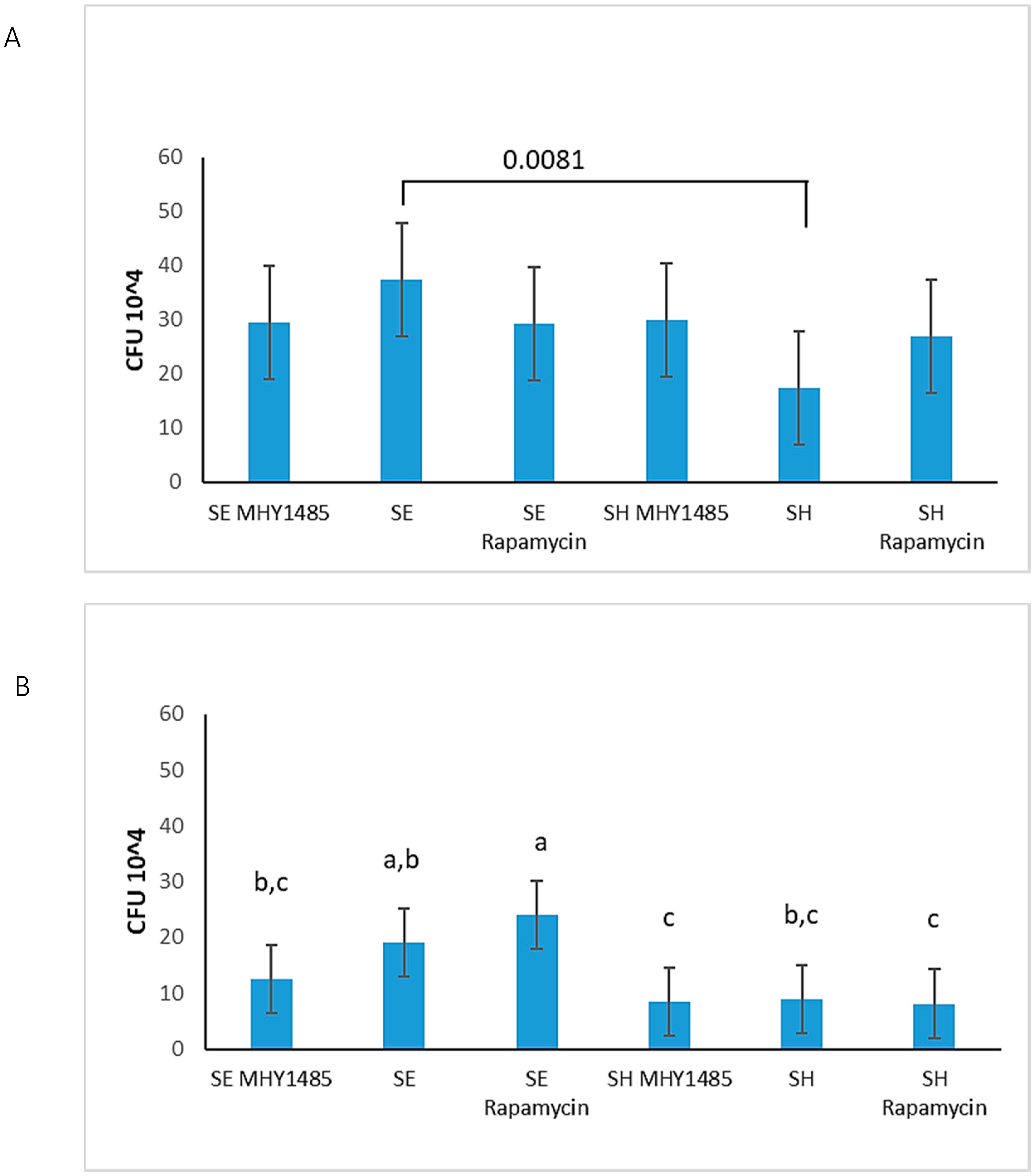
| Signal Transduction Pathways | Proteins altered 30 min p.i. | Proteins altered 2 h p.i. |
|---|---|---|
| Insulin signaling | 17 | 23 |
| AMPK signaling | 21 | 19 |
| mTOR signaling | 13 | 15 |
| HIF-1α signaling | 16 | 21 |
| UniProt Accession | Protein Name | Site/Effect of Phosphorylation | Phosphorylation Change 30 Min | Phosphorylation Change 2 h | ||
|---|---|---|---|---|---|---|
| S. Enteritidis | S. Heidelberg | S. Enteritidis | S. Heidelberg | |||
| P61586 | RhoA | S188/activity inhibited | ↑ | ↓ | ↑ | ↓ |
| O75116 | ROCK2 | Y722/activity inhibited | ↓ | ∅ | ↓ | ↓ |
| O43182 | RGH06 | Y407/unspecified | ↓ | ∅ | ∅ | ↓ |
| Q68EM7 | RGH17 | S484/unspecified | ↑ | ↑ | ↑ | ↑ |
| Protein/Peptide | Phosphorylation Change 30 Min | Phosphorylation Change 2 h | ||
|---|---|---|---|---|
| S. Enteritidis | S. Heidelberg | S. Enteritidis | S. Heidelberg | |
| PFK1 | ↑↓ | ↓ | ↑↑↑↓ | ↑↑↑↓ |
| PFK2 | ↑↑(S461) | ∅ | ↑↑(S461) | ↑↑(S461) |
| GPI | ↓(S185) | ↓(S185) | ∅ | ↑↑ |
| PhK | ↑↓ | ↑ | ↓↓↓ | ↑↓ |
| GAPDH | ↑↑ | ↑ | ∅ | ↑↓ |
| PGK | ↑ | ↑ | ↓ | ↓ |
| PGM | ↓ | ↓ | ↑↓↓ | ↓↓ |
| PKM | ↑ | ↑↑↑ | ↑↑ | ↑↑↓ |
| AMPK | ↑↑↑(T183)↓↓ | ↑↑↑(S496)↓↓↓ | ↓↓ | ↑↑↑↑(S496)↓↓ |
| HIF-1α | ↑↓(S247) | ↑↑(S247) | ↑(S247) | ↑(S247)↓ |
| S6K | ↓↓ | ↓ | ↑↓ | ↑ |
| 4EBP1 | ↑ | ↓↓ | ↓↓↓ | ↓↓ |
| UniProt Accession | Protein Name | Site/Effect of Phosphorylation | Phosphorylation Change 30 Min | Phosphorylation Change 2 h | ||
|---|---|---|---|---|---|---|
| S. Enteritidis | S. Heidelberg | S. Enteritidis | S. Heidelberg | |||
| P29466 | Caspase-1 | S227/unspecified | ↑ | ∅ | ↓ | ∅ |
| Q96P20 | NLRP3 | T233/unspecified | ↓ | ↑ | ∅ | ↓ |
| P10914 | IRF1 | Y109/unspecified | ∅ | ∅ | ↑ | ↑ |
| Q9BXL7 | CARD11 | S116/unspecified | ↑ | ↑ | ↓ | ↓ |
| O60602 | TLR5 | Y798/unspecified | ∅ | ↑ | ↓ | ↓ |
| O15455 | TLR3 | Y858/unspecified | ↓ | ∅ | ↑ | ↓ |
| P40189 | IL-6R | S782/unspecified | ↑ | ↑ | ↓ | ∅ |
| P42574 | Caspase-3 | S150/activity inhibited | ↓ | ↓ | ∅ | ↓ |
| Q14790 | Caspase-8 | S347/activity inhibited | ∅ | ∅ | ↓ | ↓ |
| Q9BUB5 | MNK | T255/activity induced | ∅ | ↓ | ∅ | ↓ |
| P45983 | JNK1 | T183/unspecified | ↑ | ∅ | ∅ | ↑ |
| P25963 | IkB-alpha | Y42/activity induced | ∅ | ↑ | ↓ | ∅ |
| P42345 | mTOR | S2448/activity induced | ∅ | ∅ | ↑ | ↑ |
| S. Enteritidis 30 min p.i. | S. Heidelberg 30 min p.i. | S. Enteritidis 2 h p.i. | S. Heidelberg 2 h p.i. |
|---|---|---|---|
| Increased rate of glycolysis and no significant change in pentose phosphate pathway activity | Decreased rate of glycolysis and increased pentose phosphate pathway activity | Decreased rate of glycolysis and no significant change in pentose phosphate pathway activity | Increased rate of glycolysis and maintained increased pentose phosphate pathway activity |
| Increased invasiveness and increased cell death * | No change in invasiveness and cell death | No change in invasiveness and decreased cell death | No changes in invasiveness and increased cell death |
| Increased rate of oxygen consumption | Increased rate of oxygen consumption | Increased rate of oxygen consumption | Increased rate of oxygen consumption |
| No response to mTOR treatments | No response to mTOR treatments | Response to mTOR treatments | No response to mTOR treatments |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Perry, F.; Johnson, C.; Aylward, B.; Arsenault, R.J. The Differential Phosphorylation-Dependent Signaling and Glucose Immunometabolic Responses Induced during Infection by Salmonella Enteritidis and Salmonella Heidelberg in Chicken Macrophage-like cells. Microorganisms 2020, 8, 1041. https://doi.org/10.3390/microorganisms8071041
Perry F, Johnson C, Aylward B, Arsenault RJ. The Differential Phosphorylation-Dependent Signaling and Glucose Immunometabolic Responses Induced during Infection by Salmonella Enteritidis and Salmonella Heidelberg in Chicken Macrophage-like cells. Microorganisms. 2020; 8(7):1041. https://doi.org/10.3390/microorganisms8071041
Chicago/Turabian StylePerry, Famatta, Casey Johnson, Bridget Aylward, and Ryan J. Arsenault. 2020. "The Differential Phosphorylation-Dependent Signaling and Glucose Immunometabolic Responses Induced during Infection by Salmonella Enteritidis and Salmonella Heidelberg in Chicken Macrophage-like cells" Microorganisms 8, no. 7: 1041. https://doi.org/10.3390/microorganisms8071041
APA StylePerry, F., Johnson, C., Aylward, B., & Arsenault, R. J. (2020). The Differential Phosphorylation-Dependent Signaling and Glucose Immunometabolic Responses Induced during Infection by Salmonella Enteritidis and Salmonella Heidelberg in Chicken Macrophage-like cells. Microorganisms, 8(7), 1041. https://doi.org/10.3390/microorganisms8071041





