Metabarcoding Analyses of Gut Microbiota of Nile Tilapia (Oreochromis niloticus) from Lake Awassa and Lake Chamo, Ethiopia
Abstract
1. Introduction
2. Materials and Methods
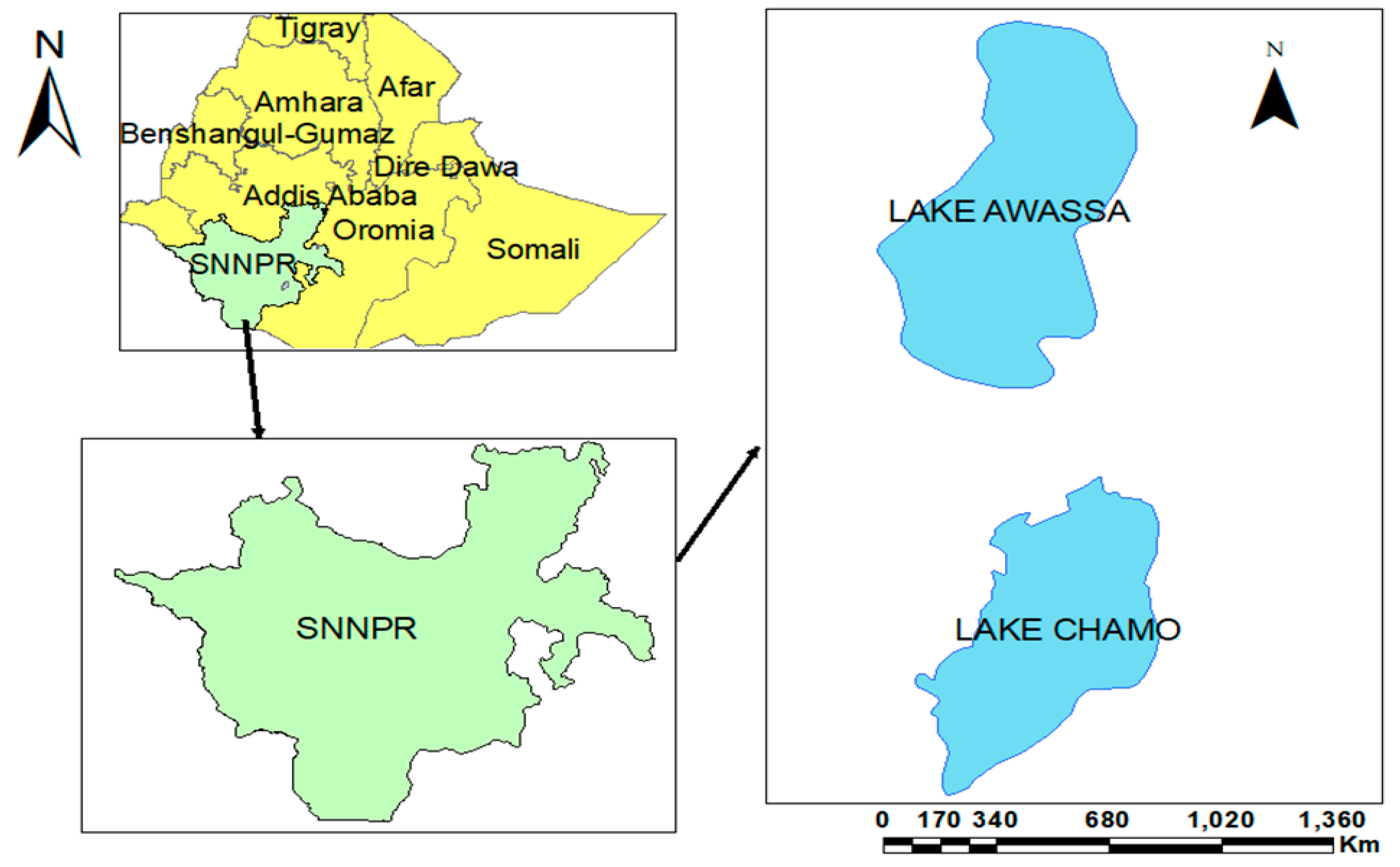
2.1. Description of the Specimen Collection Sites
2.2. Fish Sampling and Processing
2.3. DNA Extraction
2.4. PCR Amplification and Sequencing
2.5. Sequencing Data Analysis
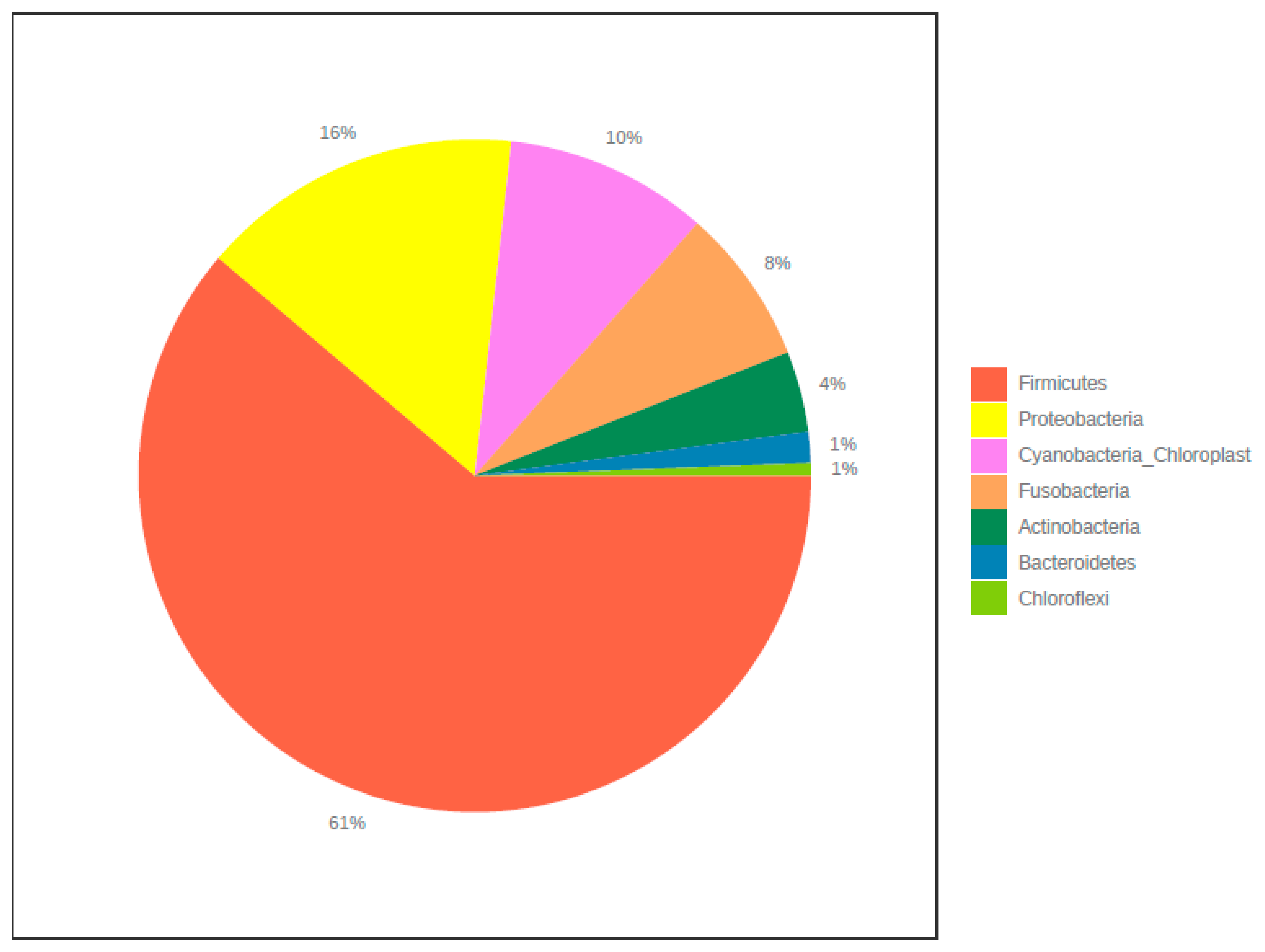
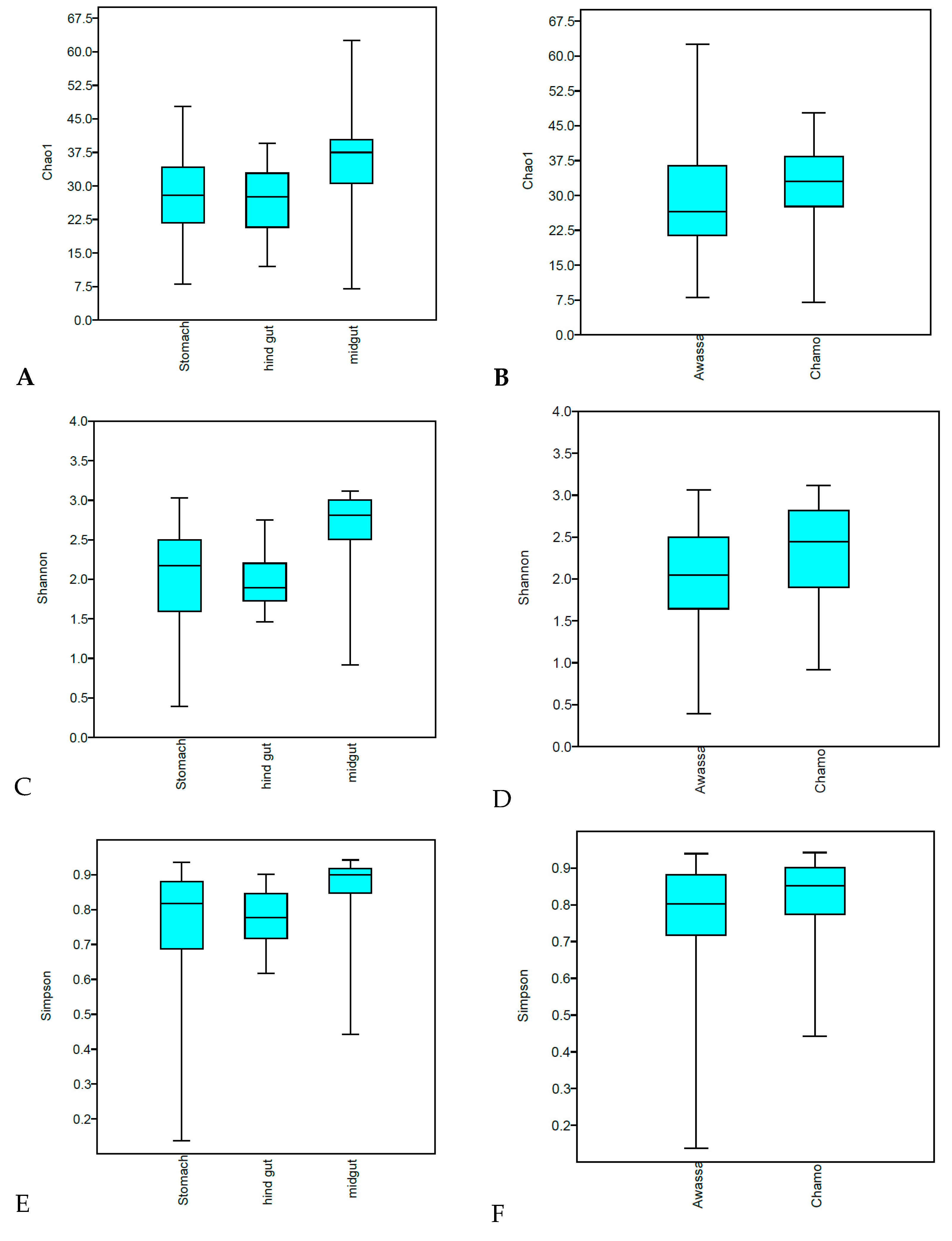
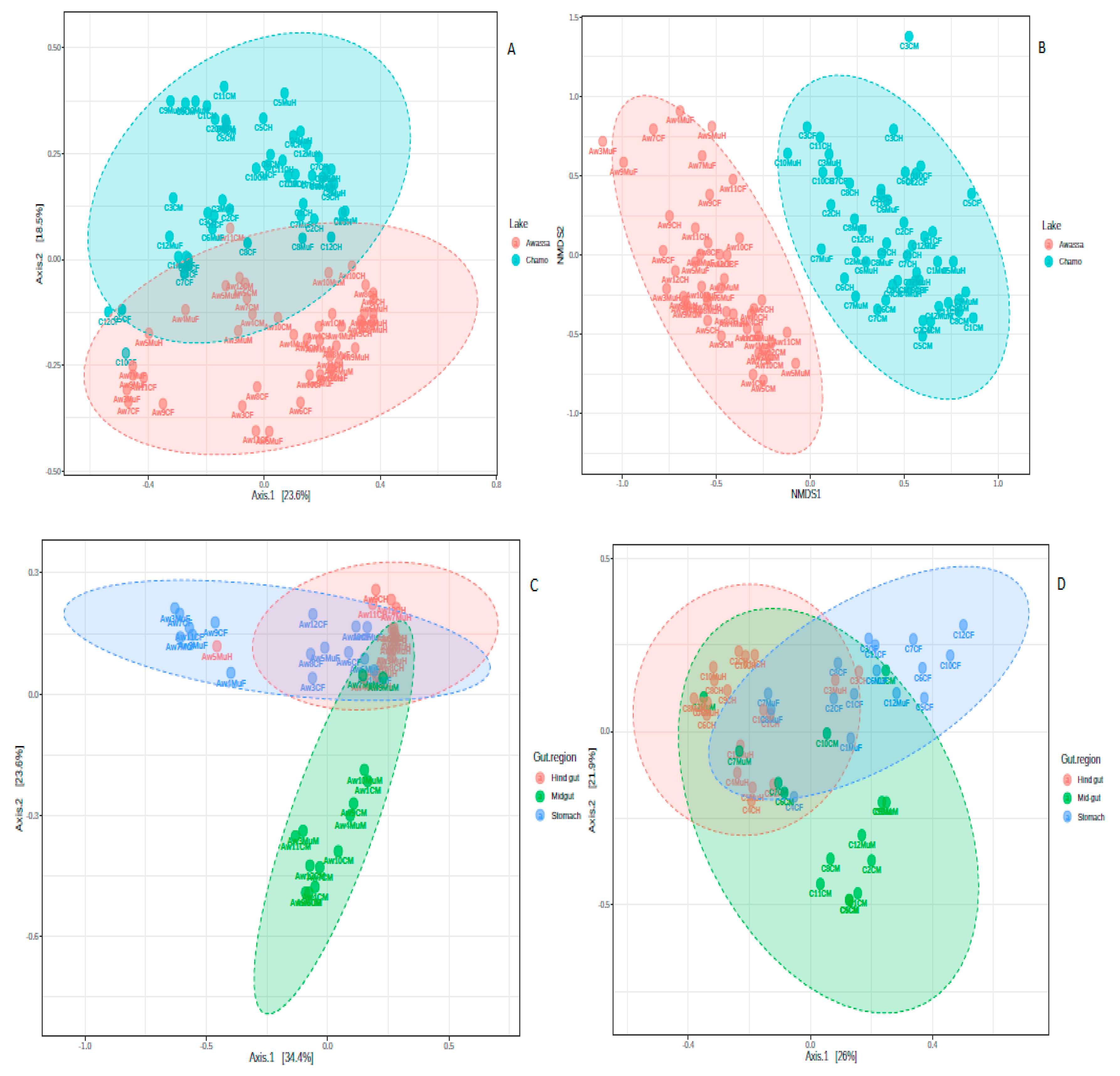
3. Results
3.1. Diversity Measures
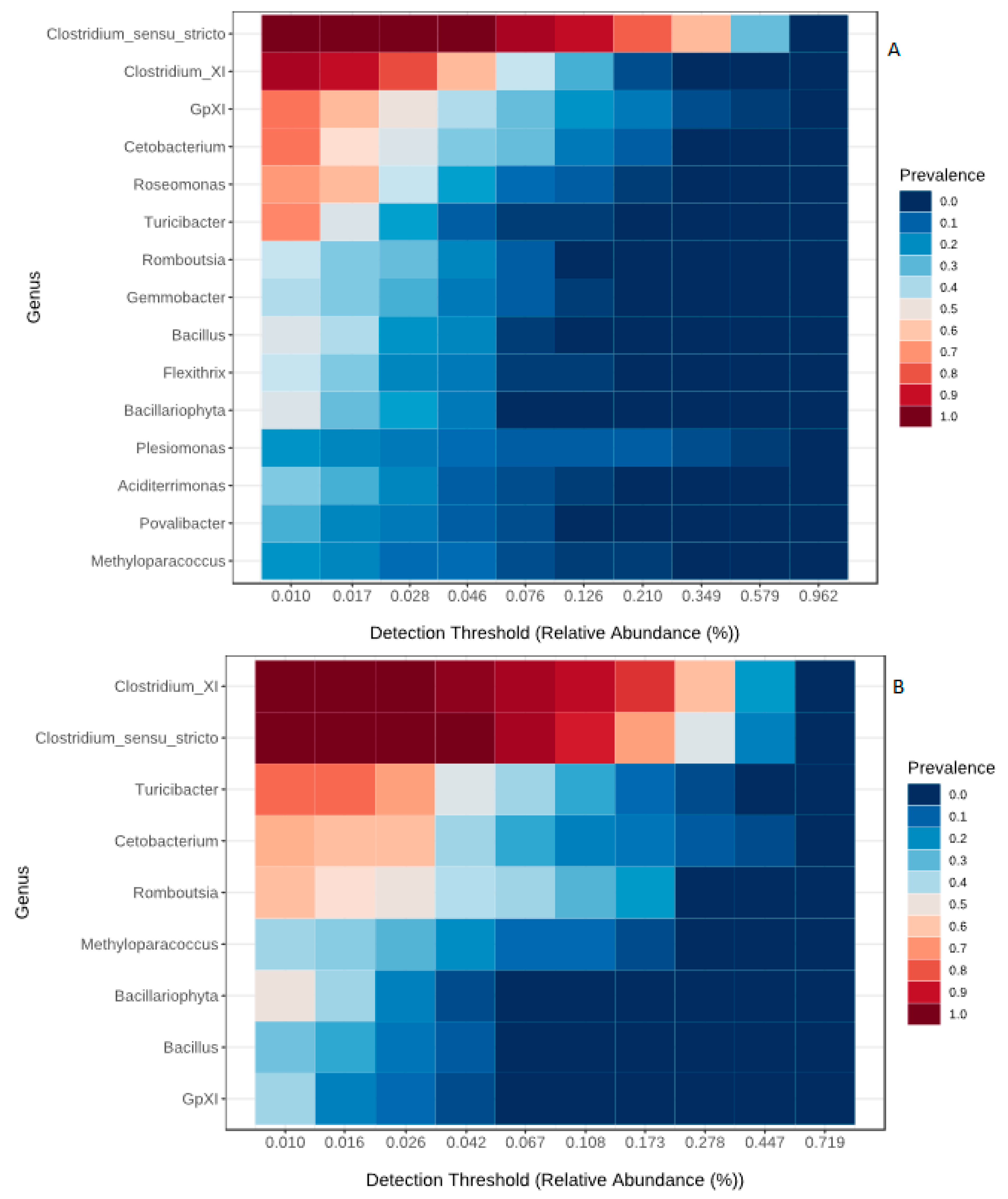
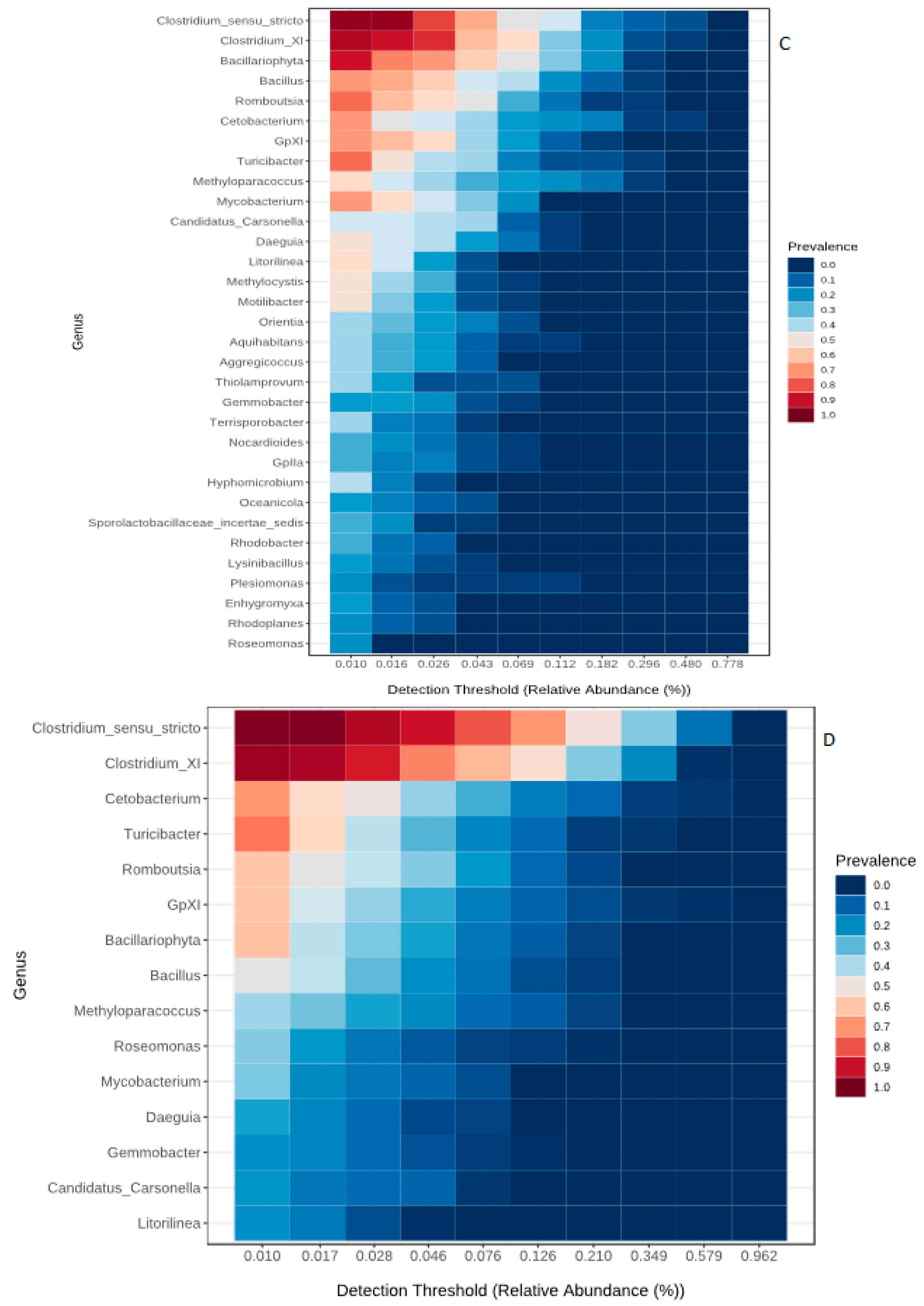
3.2. Core Microbiome
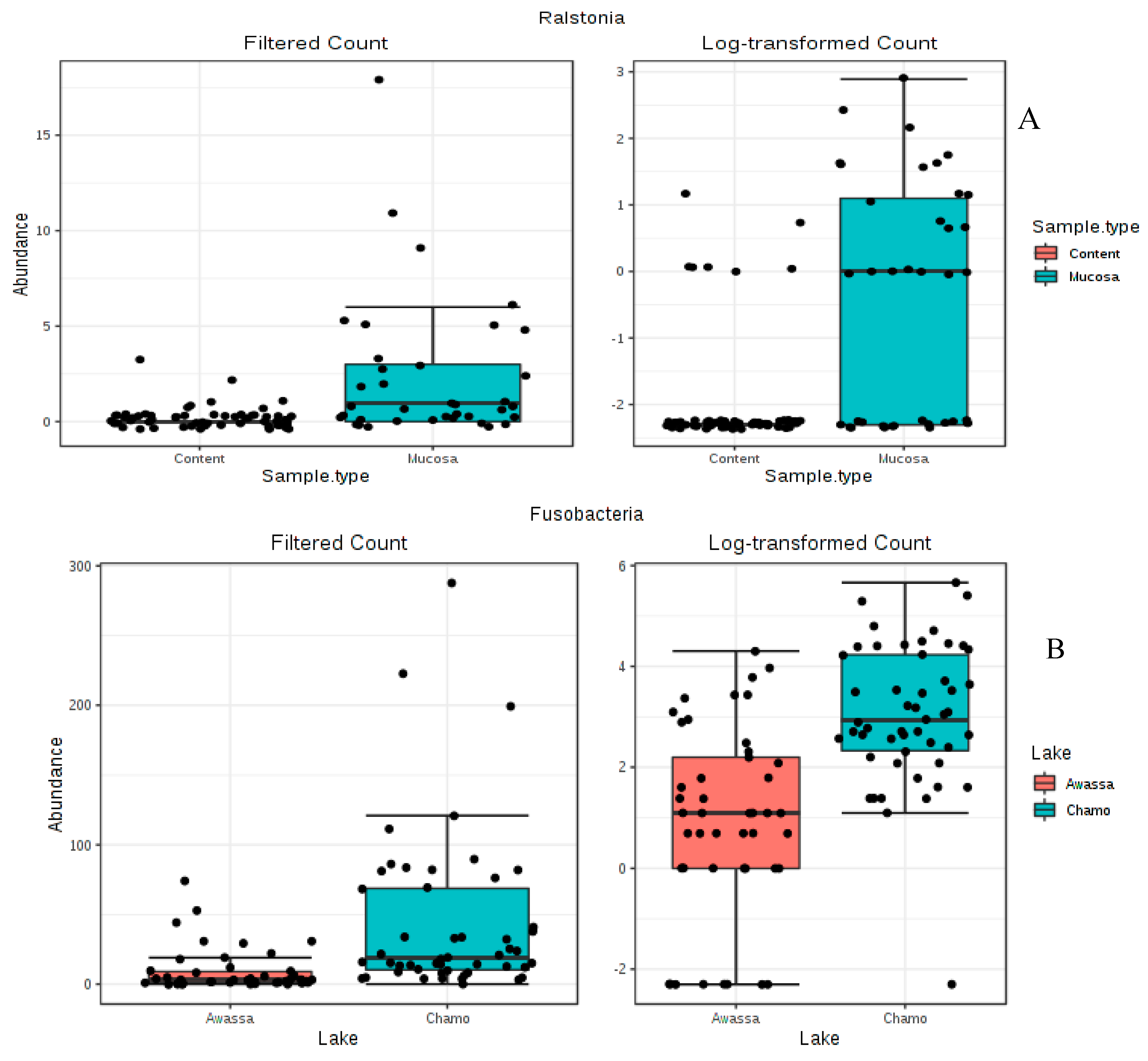
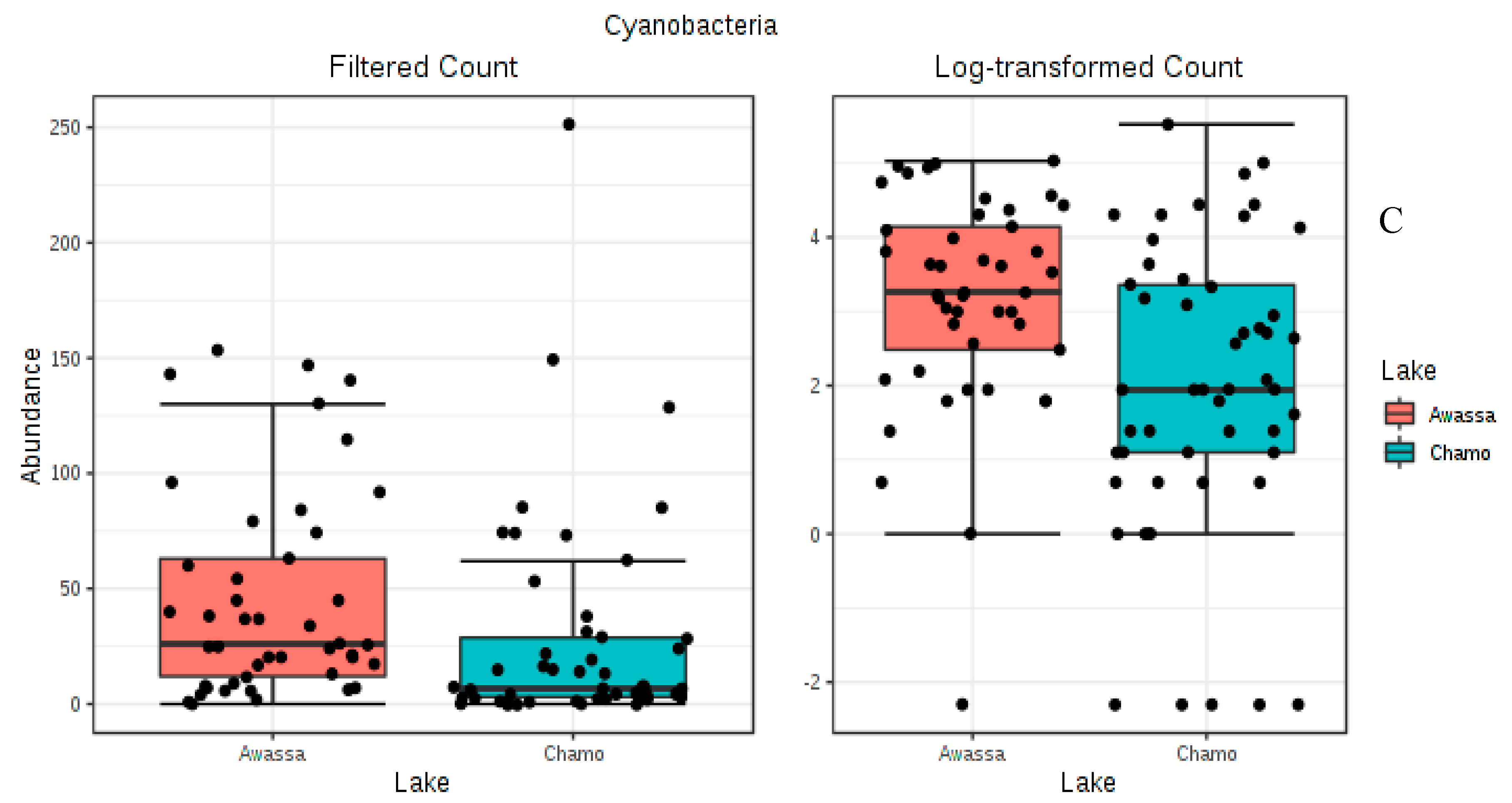
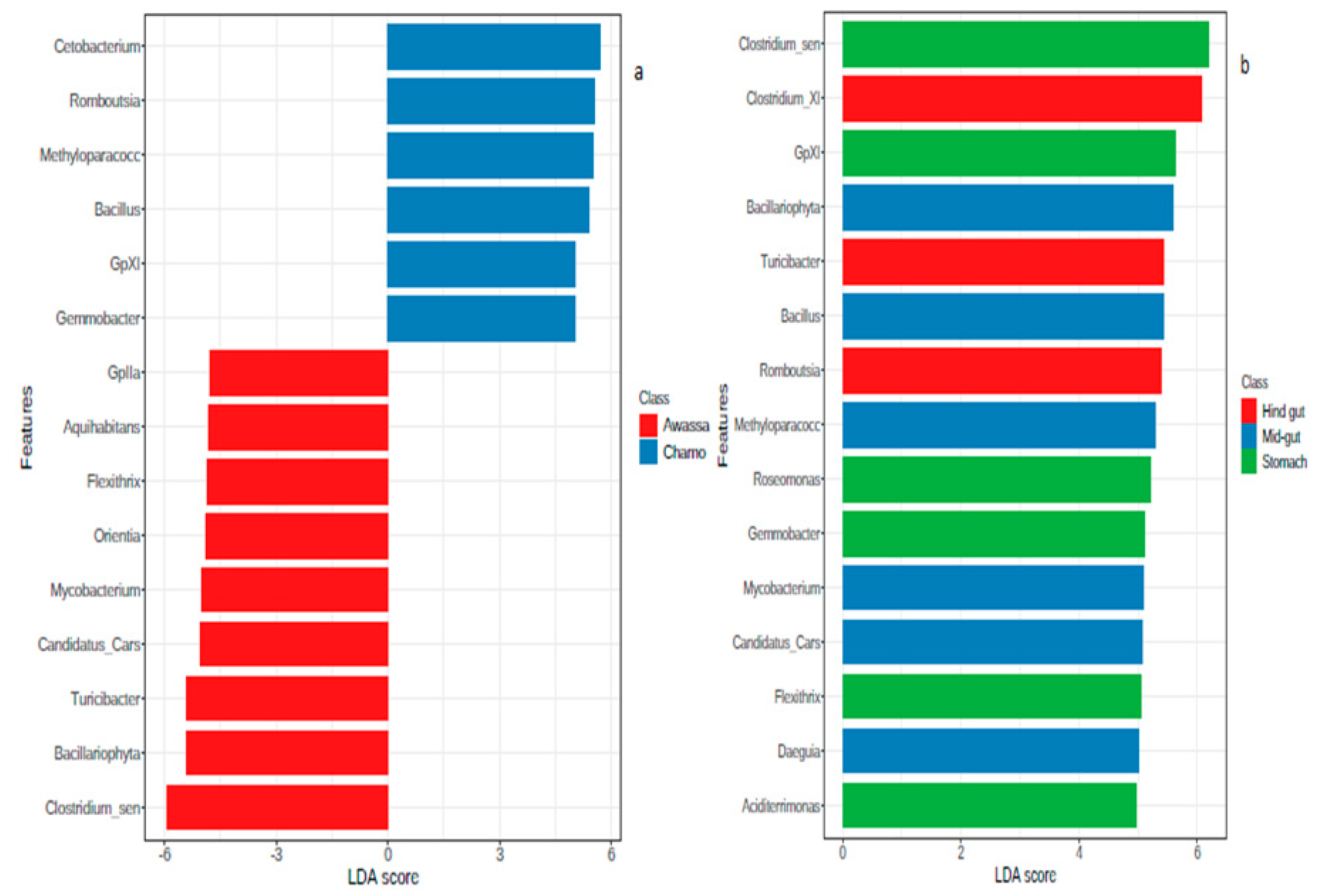
3.3. Differential Abundance Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schoeni, J.L.; Wong, A.C. Inhibition of Campylobacter jejuni colonization in chicks by defined competitive exclusion bacteria. Appl. Environ. Microbiol. 1994, 60, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Tlaskalová-Hogenová, H.; Stěpánková, R.; Kozáková, H.; Hudcovic, T.; Vannucci, L.; Tučková, L.; Rossmann, P.; Hrnčíř, T.; Kverka, M.; Zákostelská, Z.; et al. The role of gut microbiota (commensal bacteria) and the mucosal barrier in the pathogenesis of inflammatory and autoimmune diseases and cancer: Contribution of germ-free and gnotobiotic animal models of human diseases. Cell. Mol. Immunol. 2011, 8, 110–120. [Google Scholar] [CrossRef]
- Guarner, F.; Malagelada, J.-R. Gut flora in health and disease. Lancet 2003, 361, 512–519. [Google Scholar] [CrossRef]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Navarrete, P.; Magne, F.; Araneda, C.; Fuentes, P.; Barros, L.; Opazo, R.; Espejo, R.; Romero, J. PCR-TTGE analysis of 16S rRNA from rainbow trout (Oncorhynchus mykiss) gut microbiota reveals host-specific communities of active bacteria. PLoS ONE 2012, 7, e31335. [Google Scholar] [CrossRef]
- Prabu, E.; Rajagopalsamy CB, T.; Ahilan, B.; Jeevagan IJ, M.A.; Renuhadevi, M. Tilapia—An Excellent Candidate Species for World Aquaculture: A Review. ARRB 2019, 31, 1–14. [Google Scholar] [CrossRef]
- Charo-Karisa, H.; Rezk, M.A.; Bovenhuis, H.; Komen, H. Heritability of cold tolerance in Nile tilapia, Oreochromis niloticus, juveniles. Aquaculture 2005, 249, 115–123. [Google Scholar] [CrossRef]
- Song, W.; Li, L.; Huang, H.; Jiang, K.; Zhang, F.; Chen, X.; Zhao, M.; Ma, L. The Gut Microbial Community of Antarctic Fish Detected by 16S rRNA Gene Sequence Analysis. BioMed Res. Int. 2016, 2016, 3241529. [Google Scholar] [CrossRef]
- Maiwore, J.; Tatsadjieu, N.L.; Montet, D.; Loiseau, G.; Mbofung, C.M.F. Comparison of bacterial communities of tilapia fish from Cameroon and Vietnam using PCR-DGGE (polymerase chain reaction-denaturing gradient gel electrophoresis). Afr. J. Biotechnol. 2009, 8, 24. [Google Scholar]
- Molinari, L.M.; Oliveira SD de Pedroso, R.B.; De Lucas Rodrigues Bittencourt, N.; Nakamura, C.V.; Ueda-Nakamura, T.; Abreu, B.A.; Dias, B.P. Bacterial microflora in the gastrointestinal tract of Nile tilapia, Oreochromis niloticus, cultured in a semi-intensive system. Acta Sci. Biol. Sci. 2003, 25, 267–271. [Google Scholar]
- Thillaimaharani, K.A.; Logesh, A.R.; Sharmila, K.; Magdoom, B.K.; Kalaiselvam, M. Studies on the intestinal bacterial flora of tilapia Oreochromis mossambicus (Peters, 1852) and optimization of alkaline protease by Virgibacillus pantothenticus. J. Microbiol. Antimicrob. 2012, 4, 79–87. [Google Scholar] [CrossRef][Green Version]
- Françoise, L. Occurrence and role of lactic acid bacteria in seafood products. Food Microbiol. 2010, 27, 698–709. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Rawls, J.F. Intestinal microbiota composition in fishes is influenced by host ecology and environment. Mol. Ecol. 2012, 21, 3100–3102. [Google Scholar] [CrossRef] [PubMed]
- Le Nguyen, D.D.; Ngoc, H.H.; Dijoux, D.; Loiseau, G.; Montet, D. Determination of fish origin by using 16S rDNA fingerprinting of bacterial communities by PCR-DGGE: An application on Pangasius fish from Viet Nam. Food Cont. 2008, 19, 454–460. [Google Scholar] [CrossRef]
- Vaughan, E.E.; Schut, F.; Heilig, H.; Zoetendal, E.G.; de Vos, W.M.; Akkermans, A.D.L. A molecular view of the intestinal ecosystem. Curr. Issues Intest. Microbiol. 2000, 1, 1–12. [Google Scholar]
- Bates, J.M.; Mittge, E.; Kuhlman, J.; Baden, K.N.; Cheesman, S.E.; Guillemin, K. Distinct signals from the microbiota promote different aspects of zebrafish gut differentiation. Dev. Biol. 2006, 297, 374–386. [Google Scholar] [CrossRef]
- Romero, J.; Navarrete, P. 16S rDNA-based analysis of dominant bacterial populations associated with early life stages of coho salmon (Oncorhynchus kisutch). Microb. Ecol. 2006, 51, 422–430. [Google Scholar] [CrossRef] [PubMed]
- Baldo, L.; Riera, J.L.; Tooming-Klunderud, A.; Albà, M.M.; Salzburger, W. Gut Microbiota Dynamics during Dietary Shift in Eastern African Cichlid Fishes. PLoS ONE 2015, 10, e0127462. [Google Scholar] [CrossRef]
- Wassel, M.A.I.; Elsaied, H.; Rashed, M. Biodiversity of gut microflora of Oreochromis niloticus based on culture-independent rRNA gene analyses at Lake Nasser, Egypt. Egypt J. Genet. Cytol. 2016, 45, 215–233. [Google Scholar] [CrossRef]
- Gebretsadik, T.; Mereke, K. Threats and Opportunities to Major Rift Valley Lakes Wetlands of Ethiopia. Agric. Res. Technol. 2017, 9, 1–6. [Google Scholar] [CrossRef]
- Haile, M.Z.; Mohammed, E.T. Evaluation of the current water quality of Lake Hawassa, Ethiopia. Int. J. Water Res. Environ. Eng. 2019, 11, 120–128. [Google Scholar] [CrossRef]
- Pattnaik, D.B. Species Diversity of Lake Hawassa, Ethiopia. Int. J. Sci. Res. 2014, 3, 33–35. [Google Scholar]
- Vijverberg, J.; Dejen, E.; Getahun, A.; Nagelkerke, L.A.J. The composition of fish communities of nine Ethiopian lakes along a north-south gradient: Threats and possible solutions. Anim. Biol. 2012, 62, 315–335. [Google Scholar] [CrossRef]
- Anteneh, W.; Getahun, A.; Dejen, E. The lacustrine species of Labeobarbus of Lake Tana (Ethiopia) spawning at Megech and Dirma tributary rivers. SINET: Ethiop. J. Sci. 2008, 31, 21–28. [Google Scholar] [CrossRef]
- de Graaf, M.; Palstra, A.; Sibbing, F. Riverine spawning and reproductive segregation in a lacustrine cyprinid species flock, facilitated by homing? Anim. Biol. 2004, 54, 393–415. [Google Scholar] [CrossRef]
- Nagelkerke, L.A.J.; Sibbing, F.A. Reproductive segregation among the Barbus intermedius complex of Lake Tana, Ethiopia. An example of intralacustrine speciation? J. Fish Biol. 1996, 49, 1244–1266. [Google Scholar] [CrossRef]
- Tafa, B.; Assefa, E. Detection of Copper and Zinc (Heavy Metals) in Water of Lake Chamo, Arbaminch Ethiopia. World J. Chem. Educ. 2014, 2, 42–47. [Google Scholar]
- Deriggi, G.F.; Inoue, L.A.; Moraes, G. Stress responses to handling in Nile tilapia (Oreochromis niloticus Linnaeus): Assessment of eugenol as an alternative anesthetic. Acta Sci. Biol. Sci. 2006, 28, 269–274. [Google Scholar]
- Ghanbari, M.; Kneifel, W.; Domig, K.J. A new view of the fish gut microbiome: Advances from next-generation sequencing. Aquaculture 2015, 448, 464–475. [Google Scholar] [CrossRef]
- Gong, J.; Forster, R.J.; Yu, H.; Chambers, J.R.; Sabour, P.M.; Wheatcroft, R.; Chen, S. Diversity and phylogenetic analysis of bacteria in the mucosa of chicken ceca and comparison with bacteria in the cecal lumen. FEMS Microbiol. Lett. 2002, 208, 1–7. [Google Scholar] [CrossRef]
- Gong, J.; Forster, R.J.; Yu, H.; Chambers, J.R.; Wheatcroft, R.; Sabour, P.M.; Chen, S. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microb. Ecol. 2002, 41, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Shokralla, S.; Hellberg, R.S.; Handy, S.M.; King, I.; Hajibabaei, M. A DNA Mini-Barcoding System for Authentication of Processed Fish Products. Sci. Rep. 2015, 5, 15894. [Google Scholar] [CrossRef] [PubMed]
- Nossa, C.W.; Oberdorf, W.E.; Yang, L.; Aas, J.A.; Paster, B.J.; Desantis, T.Z.; Brodie, E.L.; Malamud, D.; Poles, M.A.; Pei, Z. Design of 16S rRNA gene primers for 454 pyrosequencing of the human foregut microbiome. World J. Gastroenterol. 2010, 16, 4135–4144. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Zhang, J.; Kobert, K.; Flouri, T.; Stamatakis, A. PEAR: A fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 2014, 30, 614–620. [Google Scholar] [CrossRef]
- Curto, M.; Winter, S.; Seiter, A.; Schmid, L.; Scheicher, K.; Barthel LM, F.; Plass, J.; Meimberg, H. Application of a SSR-GBS marker system on investigation of European Hedgehog species and their hybrid zone dynamics. Ecol. Evol. 2019, 9, 2814–2832. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Dhariwal, A.; Chong, J.; Habib, S.; King, I.L.; Agellon, L.B.; Xia, J. MicrobiomeAnalyst: A web-based tool for comprehensive statistical, visual and meta-analysis of microbiome data. Nucleic Acids Res. 2017, 45, W180–W188. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Hamady, M.; Fraser-Liggett, C.M.; Knight, R.; Gordon, J.I. The human microbiome project. Nature 2007, 449, 804–810. [Google Scholar] [CrossRef]
- Yohannes, Y.B.; Ikenaka, Y.; Saengtienchai, A.; Watanabe, K.P.; Nakayama SM, M.; Ishizuka, M. Occurrence, distribution, and ecological risk assessment of DDTs and heavy metals in surface sediments from Lake Awassa–Ethiopian Rift Valley Lake. Environ. Sci. Pollut. Res. Int. 2013, 20, 8663–8671. [Google Scholar] [CrossRef]
- Jin, Y.; Xia, J.; Pan, Z.; Yang, J.; Wang, W.; Fu, Z. Polystyrene microplastics induce microbiota dysbiosis and inflammation in the gut of adult zebrafish. Environ. Pollut. 2018, 235, 322–329. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.-L.; Li, S.; Qin, C.-B.; Zhu, Z.-X.; Hu, W.-P.; Yang, L.-P.; Lu, R.-H.; Li, W.-J.; Nie, G.-X. Intestinal microbiota and lipid metabolism responses in the common carp (Cyprinus carpio L.) following copper exposure. Ecotoxicol. Environ. Saf. 2018, 160, 257–264. [Google Scholar] [CrossRef]
- Miyake, S.; Ngugi, D.K.; Stingl, U. Diet strongly influences the gut microbiota of surgeon fishes. Mol. Ecol. 2015, 24, 656–672. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Wang, Y.; Liu, S.; Huang, J.; Zhai, Z.; He, C.; Ding, J.; Wang, J.; Wang, H.; Fan, W.; et al. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS ONE 2015, 10, e0117441. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Qiao, N.; Li, T.; Yu, R.; Zhai, Q.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Dietary supplementation with probiotics regulates gut microbiota structure and function in Nile tilapia exposed to aluminum. PeerJ 2019, 7, e6963. [Google Scholar] [CrossRef]
- Egerton, S.; Culloty, S.; Whooley, J.; Stanton, C.; Ross, R.P. The Gut Microbiota of Marine Fish. Front. Microbiol. 2018, 9, 873. [Google Scholar] [CrossRef]
- Ghanbari, M.; Shahraki, H.; Kneifel, W.; Domig, K.J. A first insight into the intestinal microbiota of snow trout (Schizothorax zarudnyi). Symbiosis 2017, 72, 183–193. [Google Scholar] [CrossRef]
- Ringel, Y.; Maharshak, N.; Ringel-Kulka, T.; Wolber, E.A.; Sartor, R.B.; Carroll, I.M. High throughput sequencing reveals distinct microbial populations within the mucosal and luminal niches in healthy individuals. Gut Microbes 2015, 6, 173–181. [Google Scholar] [CrossRef]
- Zheng, Y.; Wu, W.; Hu, G.; Qiu, L.; Meng, S.; Song, C.; Fan, L.; Zhao, Z.; Bing, X.; Chen, J. Gut microbiota analysis of juvenile genetically improved farmed tilapia (Oreochromis niloticus) by dietary supplementation of different resveratrol concentrations. Fish Shellfish Immun. 2018, 77, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Ran, C.; Huang, L.; Liu, Z.; Xu, L.; Yang, Y.; Tacon, P.; Auclair, E.; Zhou, Z. A Comparison of the Beneficial Effects of Live and Heat-Inactivated Baker’s Yeast on Nile Tilapia: Suggestions on the Role and Function of the Secretory Metabolites Released from the Yeast. PLoS ONE 2015, 10, e0145448. [Google Scholar] [CrossRef]
- Ray, C.; Bujan, N.; Tarnecki, A.; Davis, A.D.; Browdy, C.; Arias, C.R. Analysis of the Gut Microbiome of Nile Tilapia Oreochromis Niloticus L. Fed Diets Supplemented with Previda® and Saponin. J. Fish. Sci. 2017, 11, 36–45. [Google Scholar] [CrossRef][Green Version]
- Shade, A.; Handelsman, J. Beyond the Venn diagram: The hunt for a core microbiome. Environ. microbiol. 2012, 14, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Wong, S.; Waldrop, T.; Summerfelt, S.; Davidson, J.; Barrows, F.; Kenney, P.B.; Welch, T.; Wiens, G.D.; Snekvik, K.; Rawls, J.F.; et al. Aquacultured rainbow trout (Oncorhynchus mykiss) possess a large core intestinal microbiota that is resistant to variation in diet and rearing density. Appl. Environ. Microbiol. 2013, 79, 4974–4984. [Google Scholar] [CrossRef]
- Riiser, E.S.; Haverkamp TH, A.; Borgan, Ø.; Jakobsen, K.S.; Jentoft, S.; Star, B. A Single Vibrionales 16S rRNA Oligotype Dominates the Intestinal Microbiome in Two Geographically Separated Atlantic cod Populations. Front. Microbiol. 2018, 9, 1561. [Google Scholar] [CrossRef] [PubMed]
- Dehler, C.E.; Secombes, C.J.; Martin, S.A.M. Environmental and physiological factors shape the gut microbiota of Atlantic salmon parr (Salmo salar L.). Aquaculture 2017, 467, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Roeselers, G.; Mittge, E.K.; Stephens, W.Z.; Parichy, D.M.; Cavanaugh, C.M.; Guillemin, K.; Rawls, J.F. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011, 5, 1595–1608. [Google Scholar] [CrossRef]
- Liu, H.; Guo, X.; Gooneratne, R.; Lai, R.; Zeng, C.; Zhan, F.; Wang, W. The gut microbiome and degradation enzyme activity of wild freshwater fishes influenced by their trophic levels. Sci. Rep. 2016, 6, 24340. [Google Scholar] [CrossRef]
- Koo, H.; Hakim, J.A.; Powell, M.L.; Kumar, R.; Eipers, P.G.; Morrow, C.D.; Crowley, M.; Lefkowitz, E.J.; Watts, S.A.; Bej, A.K. Metagenomics approach to the study of the gut microbiome structure and function in zebrafish Danio rerio fed with gluten formulated diet. J. Microbiol. Meth. 2017, 135, 69–76. [Google Scholar] [CrossRef]
- Tsuchiya, C.; Sakata, T.; Sugita, H. Novel ecological niche of Cetobacterium somerae, an anaerobic bacterium in the intestinal tracts of freshwater fish. Lett. Appl. Microbiol. 2008, 46, 43–48. [Google Scholar] [CrossRef]
- Degnan, P.H.; Taga, M.E.; Goodman, A.L. Vitamin B12 as a modulator of gut microbial ecology. Cell Metab. 2014, 20, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Borsodi, A.K.; Szabó, A.; Krett, G.; Felföldi, T.; Specziár, A.; Boros, G. Gut content microbiota of introduced bigheaded carps (Hypophthalmichthys spp.) inhabiting the largest shallow lake in Central Europe. Microbiol. Res. 2017, 195, 40–50. [Google Scholar] [CrossRef]
- Elsaied, H.E.; Soliman, T.; Abu-Taleb, H.T.; Goto, H.; Jenke-Kodam, H. Phylogenetic characterization of eukaryotic and prokaryotic gut flora of Nile tilapia, Oreochromis niloticus, along niches of Lake Nasser, Egypt, based on rRNA gene high-throughput sequences. Ecol. Genet. Genom. 2019, 11, 100037. [Google Scholar] [CrossRef]
- Estruch, G.; Collado, M.C.; Peñaranda, D.S.; Tomás Vidal, A.; Jover Cerdá, M.; Pérez Martínez, G.; Martinez-Llorens, S. Impact of Fishmeal Replacement in Diets for Gilthead Sea Bream (Sparus aurata) on the Gastrointestinal Microbiota Determined by Pyrosequencing the 16S rRNA Gene. PLoS ONE 2015, 10, e0136389. [Google Scholar] [CrossRef] [PubMed]
- Semyalo, R.; Rohrlack, T.; Kayiira, D.; Kizito, Y.S.; Byarujali, S.; Nyakairu, G.; Larsson, P. On the diet of Nile tilapia in two eutrophic tropical lakes containing toxin producing cyanobacteria. Limnologica 2011, 41, 30–36. [Google Scholar] [CrossRef]
- Clements, K.D.; Pasch, I.B.Y.; Moran, D.; Turner, S.J. Clostridia dominate 16S rRNA gene libraries prepared from the hindgut of temperate marine herbivorous fishes. Mar. Biol. 2007, 150, 1431–1440. [Google Scholar] [CrossRef]
- Sarkar, B.; Ghosh, K. Gastrointestinal microbiota in Oreochromis mossambicus (Peters) and Oreochromis niloticus (Linnaeus): Scanning electron microscopy and microbiological study. Int. J. Fish Aquat. Stud. 2014, 2, 78–88. [Google Scholar]
- Poletto, T.V.; Vieira CR, W.; Silva, C.P.; Fracalossi, D.M. Isolation and Identification of a Potential Amylolytic Probiotic Bacterium from the Gut of Jundiá Catfish, Rhamdia quelen. Braz. Arch. Biol. Technol. 2018, 61, e18161205. [Google Scholar] [CrossRef]
- Carda-Diéguez, M.; Mira, A.; Fouz, B. Pyrosequencing survey of intestinal microbiota diversity in cultured sea bass (Dicentrarchus labrax) fed functional diets. FEMS Microbiol. Ecol. 2014, 87, 451–459. [Google Scholar] [CrossRef]
- Zhou, Z.; Liu, Y.; Shi, P.; He, S.; Yao, B.; Ringø, E. Molecular characterization of the autochthonous microbiota in the gastrointestinal tract of adult yellow grouper (Epinephelus awoara) cultured in cages. Aquaculture 2009, 286, 184–189. [Google Scholar] [CrossRef]
| Phylum | Class | Order | Family | Genus |
|---|---|---|---|---|
| Firmicutes | Clostridia | Clostridiales | Clostridiaceae_1 | Clostridium_sensu_stricto |
| Peptostreptococcaceae | Clostridium_XI | |||
| Romboutsia | ||||
| Bacilli | Bacillales | Bacillaceae 1 | Bacillus | |
| Erysipelotrichia | Erysipelotrichales | Erysipelotrichaceae | Turicibacter | |
| Cyanobacteria | Cyanobacteria | Family XI | Family XI | GpXI |
| Chloroplast | Chloroplast | Chloroplast | Bacillariophyta | |
| Fusobacteria | Fusobacteriia | Fusobacteriales | Fusobacteriaceae | Cetobacterium |
| Proteobacteria | Gammaproteobacteria | Methylococcales | Methylococcaceae | Methyloparacoccus |
| Gammaproteobacteria_incertae_sedis | Candidatus Carsonella | Candidatus Carsonella | ||
| Alphaproteobacteria | Rhodospirillales | Acetobacteraceae | Roseomonas | |
| Rhizobiales | Brucellaceae | Daeguia | ||
| Rhodobacterales | Rhodobacteraceae | Gemmobacter | ||
| Deltaproteobacteria | Myxococcales | |||
| Actinobacteria | Actinobacteria | Actinomycetales | Mycobacteriaceae | Mycobacterium |
| Acidimicrobiales | ||||
| Chloroflexi | Caldilineae | Caldilineales | Caldineaceae | Litorilinea |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bereded, N.K.; Curto, M.; Domig, K.J.; Abebe, G.B.; Fanta, S.W.; Waidbacher, H.; Meimberg, H. Metabarcoding Analyses of Gut Microbiota of Nile Tilapia (Oreochromis niloticus) from Lake Awassa and Lake Chamo, Ethiopia. Microorganisms 2020, 8, 1040. https://doi.org/10.3390/microorganisms8071040
Bereded NK, Curto M, Domig KJ, Abebe GB, Fanta SW, Waidbacher H, Meimberg H. Metabarcoding Analyses of Gut Microbiota of Nile Tilapia (Oreochromis niloticus) from Lake Awassa and Lake Chamo, Ethiopia. Microorganisms. 2020; 8(7):1040. https://doi.org/10.3390/microorganisms8071040
Chicago/Turabian StyleBereded, Negash Kabtimer, Manuel Curto, Konrad J. Domig, Getachew Beneberu Abebe, Solomon Workneh Fanta, Herwig Waidbacher, and Harald Meimberg. 2020. "Metabarcoding Analyses of Gut Microbiota of Nile Tilapia (Oreochromis niloticus) from Lake Awassa and Lake Chamo, Ethiopia" Microorganisms 8, no. 7: 1040. https://doi.org/10.3390/microorganisms8071040
APA StyleBereded, N. K., Curto, M., Domig, K. J., Abebe, G. B., Fanta, S. W., Waidbacher, H., & Meimberg, H. (2020). Metabarcoding Analyses of Gut Microbiota of Nile Tilapia (Oreochromis niloticus) from Lake Awassa and Lake Chamo, Ethiopia. Microorganisms, 8(7), 1040. https://doi.org/10.3390/microorganisms8071040






