Genome Analysis of Endobacterium cerealis, a Novel Genus and Species Isolated from Zea mays Roots in North Spain
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain Isolation
2.2. 16S rRNA Gene Phylogenetic Analysis
2.3. Genome Sequencing, Assembling, Annotation and Analysis
2.4. Fatty Acid Analysis and Phenotypic Characterization
3. Results
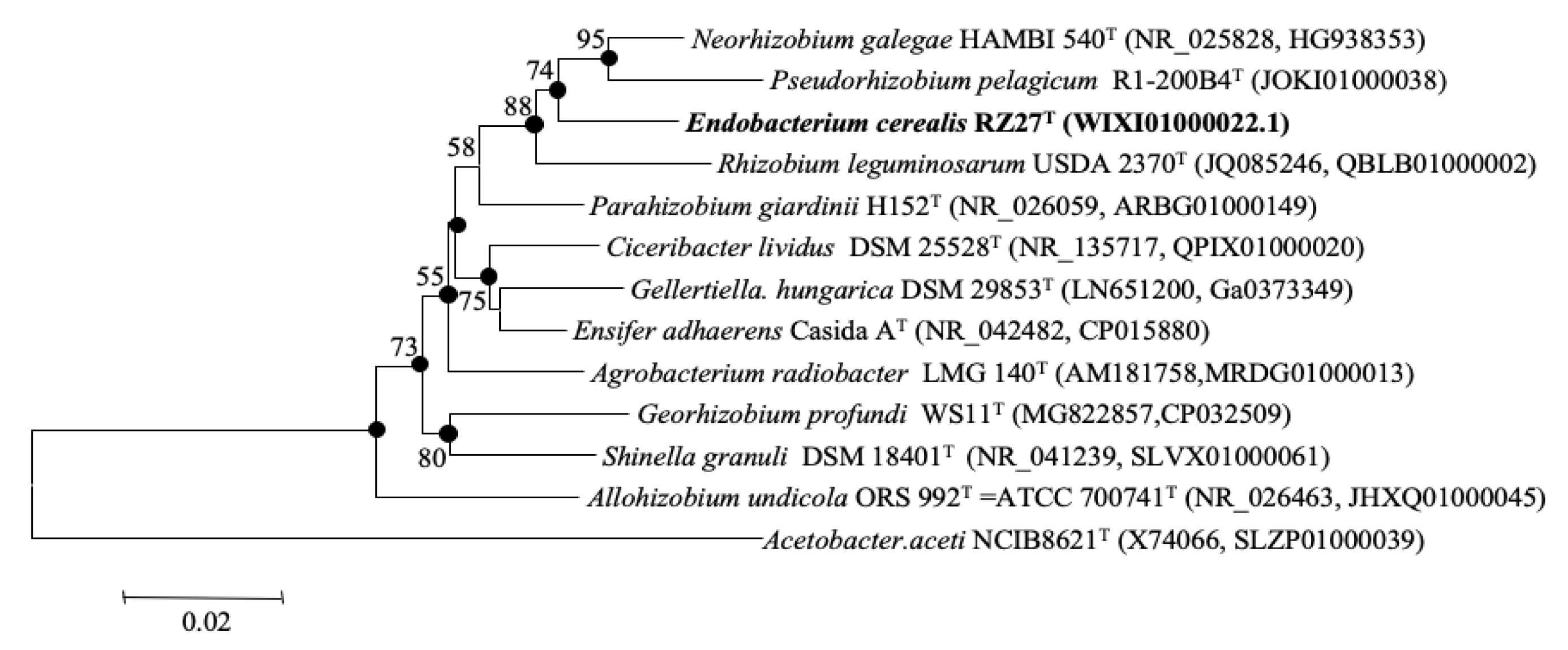
3.1. 16S rRNA Gene Phylogenetic Analysis
3.2. Genome Analyses
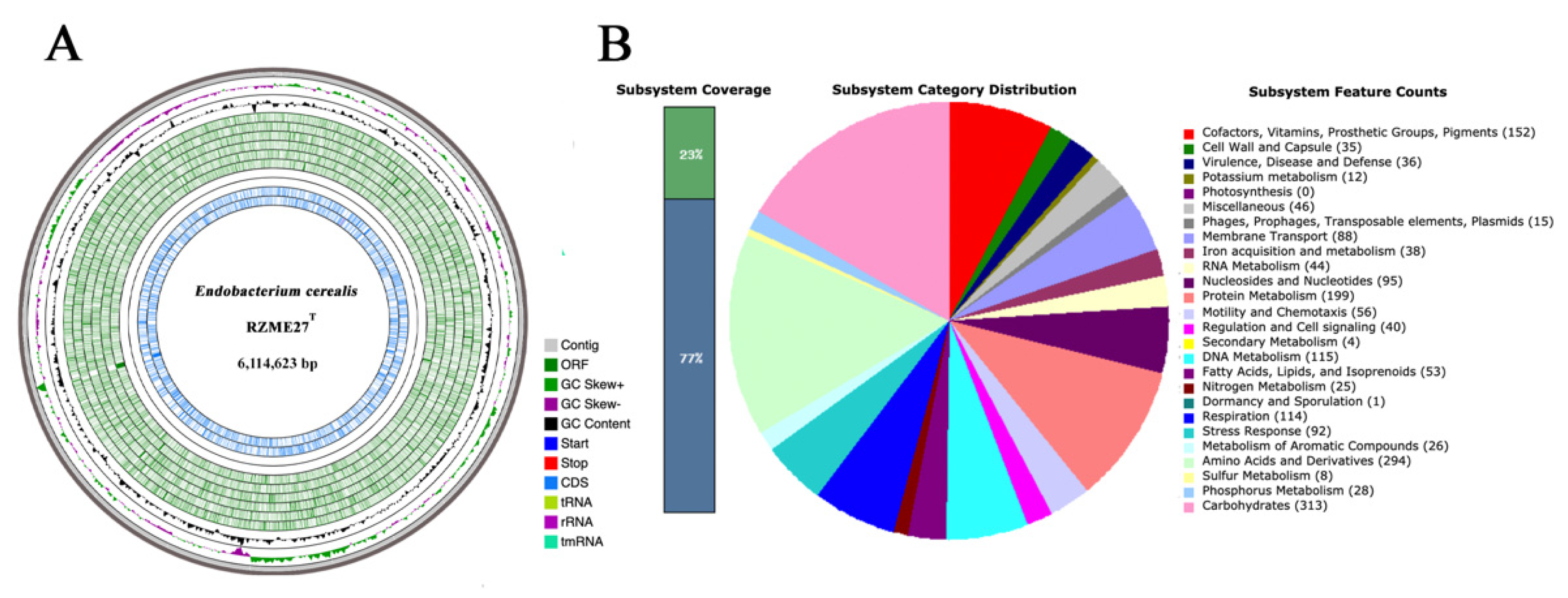
3.2.1. Genome Properties
3.2.2. Genome Mining
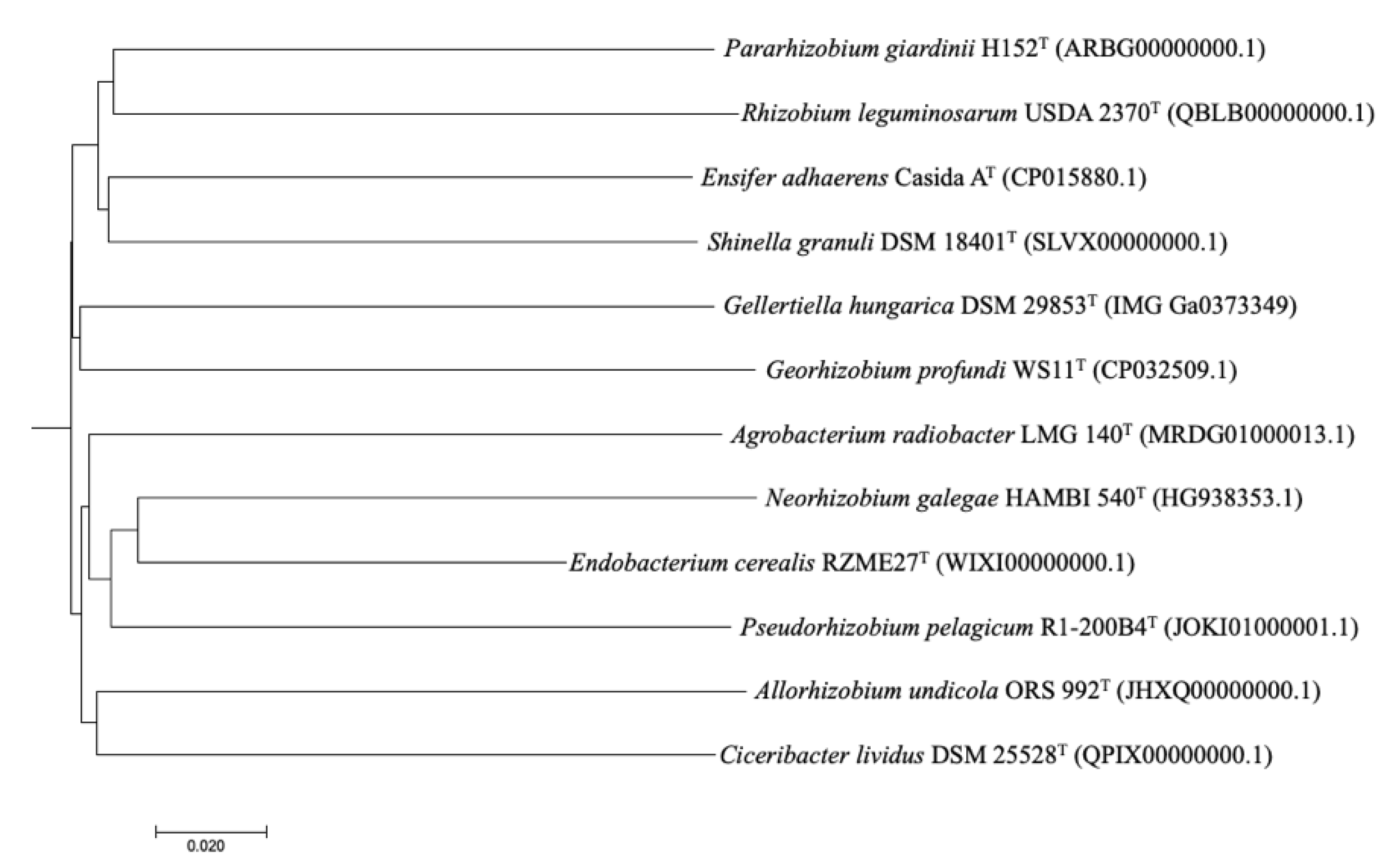
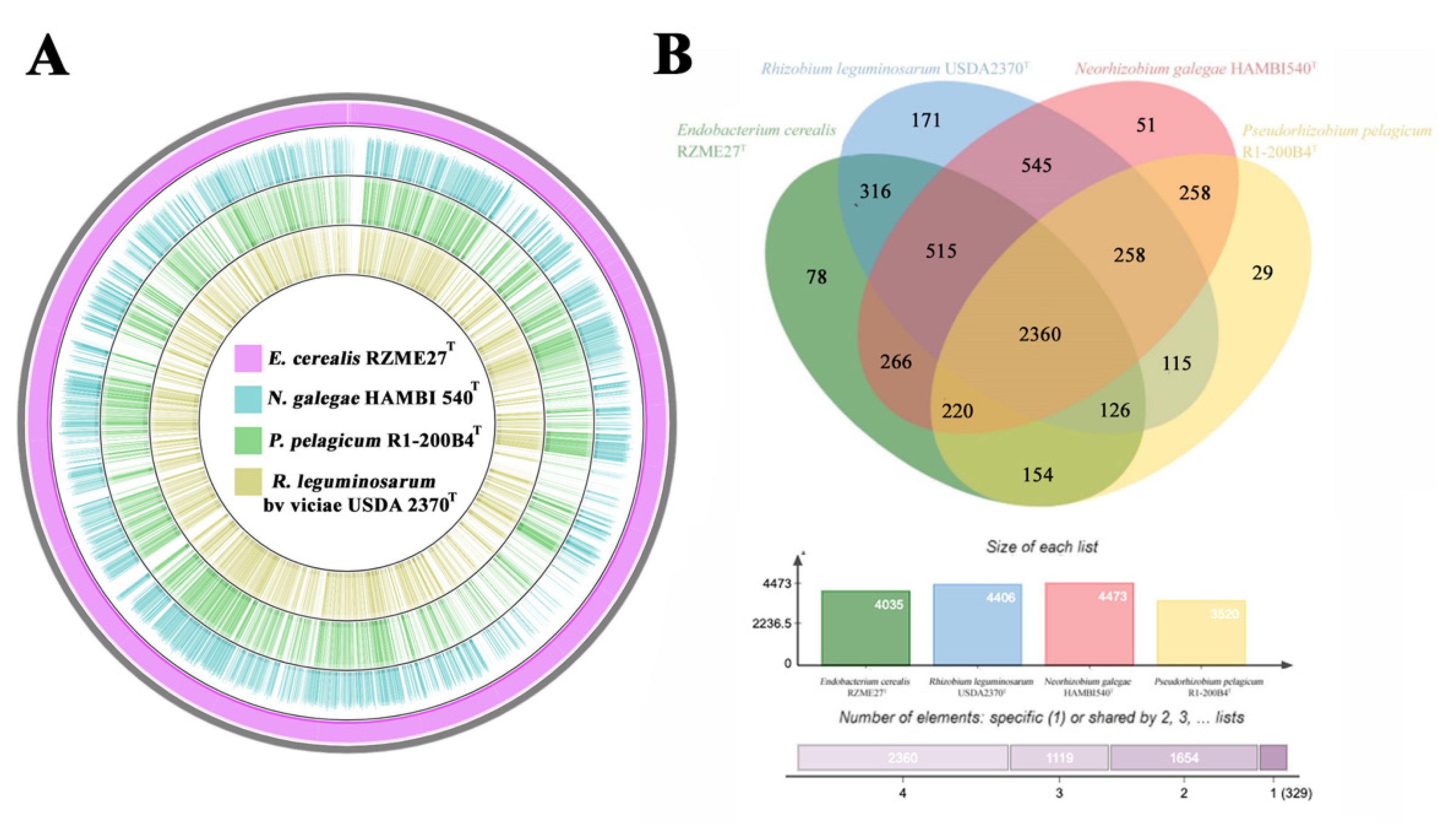
3.2.3. Genetic Relatedness and Pangenome Analysis
3.3. Fatty Acid Analysis and Phenotypic Characterization
4. Discussion
5. Conclusions
6. Description of Endobacterium gen. nov.
7. Description of Endobacterium cerealis sp. nov.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Conn, H.J. Taxonomic relationships of certain non-sporeforming rods in soil. J. Bacteriol. 1938, 36, 320–321. [Google Scholar]
- Frank, B. Ueber die pilzsymbiose der leguminosen. Bet. Dtsch. Bot. Ges. 1889, 7, 332–346. [Google Scholar]
- Conn, H.J. Validity of the genus Alcaligenes. J. Bacteriol. 1942, 44, 353–360. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.X.; Yan, G.H.; Li, J.L. Numerical taxonomic study of fast-growing soybean rhizobia and a proposal that Rhizobium fredii be assigned to Sinorhizobium gen. nov. Int. J. Syst. Bacteriol. 1988, 38, 392–397. [Google Scholar] [CrossRef]
- Casida, L.E., Jr. Ensifer adhaerens gen. nov., sp. nov.: A bacterial predator of bacteria in soil. Int. J. Syst. Bacteriol. 1982, 32, 339–345. [Google Scholar] [CrossRef]
- De Lajudie, P.; Laurent-Fulele, E.; Willems, A.; Torck, U.; Coopman, R.; Collins, M.D.; Kersters, K.; Dreyfus, B.; Gillis, M. Allorhizobium undicola gen. nov., sp. nov., nitrogen-fixing bacteria that efficiently nodulate Neptunia natans in Senegal. Int. J. Syst. Bacteriol. 1998, 48, 1277–1290. [Google Scholar] [CrossRef] [PubMed]
- An, D.S.; Im, W.T.; Yang, H.C.; Lee, S.T. Shinella granuli gen. nov., sp. nov., and proposal of the reclassification of Zoogloea ramigera ATCC 19623 as Shinella zoogloeoides sp. nov. Int. J. Syst. Evol. Microbiol. 2006, 56, 443–448. [Google Scholar] [CrossRef]
- Kathiravan, R.; Jegan, S.; Ganga, V.; Prabavathy, V.R.; Tushar, L.; Sasikala, C.; Ramana, C.V. Ciceribacter lividus gen. nov., sp. nov., isolated from rhizosphere soil of chickpea (Cicer arietinum L.). Int. J. Syst. Evol. Microbiol. 2013, 63, 4484–4488. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Österman, J.; Wahlberg, N.; Nesme, X.; Lavire, C.; Vial, L.; Paulin, L.; de Lajudie, P.; Lindström, K. Phylogeny of the Rhizobium-Allorhizobium-Agrobacterium clade supports the delineation of Neorhizobium gen. nov. Syst. Appl. Microbiol. 2014, 37, 208–215. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Willems, A.; Nesme, X.; de Lajudie, P.; Lindström, K. Revised phylogeny of Rhizobiaceae: Proposal of the delineation of Pararhizobium gen. nov., and 13 new species combinations. Syst. Appl. Microbiol. 2015, 38, 84–90. [Google Scholar] [CrossRef]
- Kimes, N.E.; López-Pérez, M.; Flores-Félix, J.D.; Ramírez-Bahena, M.H.; Igual, J.M.; Peix, A.; Rodriguez-Valera, F.; Velázquez, E. Pseudorhizobium pelagicum gen. nov., sp. nov. isolated from a pelagic Mediterranean zone. Syst. Appl. Microbiol. 2015, 38, 293–299. [Google Scholar] [CrossRef] [PubMed]
- Tóth, E.; Szuróczki, S.; Kéki, Z.; Bóka, K.; Szili-Kovács, T.; Schumann, P. Gellertiella hungarica gen. nov., sp. nov., a novel bacterium of the family Rhizobiaceae isolated from a spa in Budapest. Int. J. Syst. Evol. Microbiol. 2017, 67, 4565–4571. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Wei, Y.; Lai, Q.; Wu, Y.; Deng, J.; Li, J.; Liu, R.; Wang, L.; Fang, J. Georhizobium profundi gen. nov., sp. nov., a piezotolerant bacterium isolated from a deep-sea sediment sample of the New Britain Trench. Int. J. Syst. Evol. Microbiol. 2020, 70, 373–379. [Google Scholar] [CrossRef]
- Peix, A.; Ramírez-Bahena, M.H.; Velázquez, E.; Bedmar, E.J. Bacterial associations with legumes. Crit. Rev. Plant Sci. 2015, 34, 17–42. [Google Scholar] [CrossRef]
- Velázquez, E.; Carro, L.; Flores-Félix, J.D.; Martínez-Hidalgo, P.; Menéndez, E.; Ramírez-Bahena, M.H.; Mulas, R.; González-Andrés, P.; Martínez-Molina, E.; Peix, A. The legume nodule microbiome: A source of plant growth-promoting bacteria. In Probiotics and Plant Health; Kumar, V., Kumar, M., Sharma, S., Prasad, R., Eds.; Springer: Singapore, 2017; pp. 41–70. [Google Scholar]
- Velázquez, E.; García-Fraile, P.; Ramírez-Bahena, M.H.; Rivas, R.; Martínez-Molina, E. Current status of the taxonomy of bacteria able to establish nitrogen-fixing legume symbiosis. In Microbes for Legume Improvement; Zaidi, A., Khan, M., Musarrat, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 1–43. [Google Scholar]
- Flores-Félix, J.D.; Menéndez, E.; Peix, A.; García-Fraile, P.; Velázquez, E. History and current taxonomic status of genus Agrobacterium. Syst. Appl. Microbiol. 2020, 43, 126046. [Google Scholar] [CrossRef]
- Celador-Lera, L.; Menéndez, E.; Peix, A.; Igual, J.M.; Velázquez, E.; Rivas, R. Rhizobium zeae sp. nov., isolated from maize (Zea mays L.) roots. Int. J. Syst. Evol. Microbiol. 2017, 67, 2306–2311. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.L.; Sun, P.; Wang, X.M.; Lv, F.Y.; Mao, X.J.; Sun, J.G. Rhizobium wenxiniae sp. nov., an endophytic bacterium isolated from maize root. Int. J. Syst. Evol. Microbiol. 2017, 67, 2798–2803. [Google Scholar] [CrossRef]
- Chelius, M.K.; Triplett, E.W. Dyadobacter fermentans gen. nov., sp. nov., a novel gram-negative bacterium isolated from surface-sterilized Zea mays stems. Int. J. Syst. Evol. Microbiol. 2000, 50, 751–758. [Google Scholar] [CrossRef]
- Chelius, M.K.; Henn, J.A.; Triplett, E.W. Runella zeae sp. nov., a novel gram-negative bacterium from the stems of surface-sterilized Zea mays. Int. J. Syst. Evol. Microbiol. 2002, 52, 2061–2063. [Google Scholar]
- Rivas, R.; García-Fraile, P.; Mateos, P.F.; Martínez-Molina, E.; Velázquez, E. Characterization of xylanolytic bacteria present in the bract phyllosphere of the date palm Phoenix dactylifera. Lett. Appl. Microbiol. 2007, 44, 181–187. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The clustalX windows interface: Flexible strategies for multiple sequence alignement aided by quality analysis tools. Nucleic Acids Res. 1997, 24, 4876–4882. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. A neighbour-joining method: A new method for reconstructing phylogenetics trees. Mol. Biol. Evol. 1987, 44, 406–425. [Google Scholar]
- Rogers, J.S.; Swofford, D.L. A fast method for approximating maximum likelihoods of phylogenetic trees from nucleotide sequences. Syst. Biol. 1998, 47, 77–89. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 3, 1870–1874. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics 2008, 9, 75. [Google Scholar] [CrossRef]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Haft, D.H.; DiCuccio, M.; Badretdin, A.; Brover, V.; Chetvernin, V.; O’Neill, K.; Li, W.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; et al. RefSeq: An update on prokaryotic genome annotation and curation. Nucleic Acids Res. 2018, 46, D851–D860. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Stothard, P. The CGView server: A comparative genomics tool for circular genomes. Nucleic Acids Res. 2008, 36, W181–W184. [Google Scholar] [CrossRef] [PubMed]
- Petkau, A.; Stuart-Edwards, M.; Stothard, P.; van Domselaar, G. Interactive microbial genome visualization with GView. Bioinformatics 2010, 26, 3125–3126. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Mora, R.; Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef]
- Zhang, H.; Yohe, T.; Huang, L.; Entwistle, S.; Wu, P.; Yang, Z.; Busk, P.K.; Xu, Y.; Yin, Y. dbCAN2: A meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2018, 46, W95–W101. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Aramaki, T.; Blanc-Mathieu, R.; Endo, H.; Ohkubo, K.; Kanehisa, M.; Goto, S.; Ogata, H. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 2019, 19, 2251–2252. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Sasser, M. Identification of bacteria by gas chromatography of cellular fatty acids. USFCC Newsl. 1990, 20, 1–6. [Google Scholar]
- Beringer, J.E. R factors transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef]
- Doetsch, R.N. Determinative methods of light microscopy. In Manual of Methods for General Bacteriology; Gerdhardt, P., Murray, R.G.E., Costilow, R.N., Nester, E.W., Wood, W.A., Krieg, N.R., Phillips, G.B., Eds.; American Society for Microbiology: Washington, DC, USA, 1981; pp. 21–33. [Google Scholar]
- Vincent, J.M. A Manual for the Practical Study of the Root-nodule Bacteria. IBP Handbook 15; Black Well Scientific Publications: Oxford, UK, 1970. [Google Scholar]
- Felis, G.E.; Dellaglio, F. On species descriptions based on a single strain: Proposal to introduce the status species proponenda (sp. pr.). Int. J. Syst. Evol. Microbiol. 2007, 57, 2185–2187. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; de Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Stephan, R.; Grim, C.J.; Gopinath, G.R.; Mammel, M.K.; Sathyamoorthy, V.; Trach, L.H.; Chase, H.R.; Fanning, S.; Tall, B.D. Re-examination of the taxonomic status of Enterobacter helveticus, Enterobacter pulveris and Enterobacter turicensis as members of the genus Cronobacter and their reclassification in the genera Franconibacter gen. nov. and Siccibacter gen. nov. as Franconibacter helveticus comb. nov., Franconibacter pulveris comb. nov. and Siccibacter turicensis comb. nov., respectively. Int. J. Syst. Evol. Microbiol. 2019, 64, 3402–3410. [Google Scholar]
- Marathe, N.P.; Salvà-Serra, F.; Karlsson, R.; Larsson, D.G.K.; Moore, E.R.B.; Svensson-Stadler, L.; Jakobsson, H.E. Scandinavium goeteborgense gen. nov., sp. nov., a new member of the family Enterobacteriaceae isolated from a wound infection, carries a novel quinolone resistance gene variant. Front. Microbiol. 2019, 10, 2511. [Google Scholar] [CrossRef]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo-and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; Orozco-Mosqueda, M.C.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant–bacteria associations. Plant Mol. Biol. 2016, 90, 549–559. [Google Scholar] [CrossRef]
- Kumar, M.; Rajput, A. Phylogenomics and evolutionary perspective of quorum sensing regulators (LuxI/LuxR) in Prokaryotes. In Quorum Sensing and its Biotechnological Applications; Kalia, V.P., Ed.; Springer: Singapore, 2018; pp. 61–70. [Google Scholar]
- Menéndez, E.; Martínez-Hidalgo, P.; Silva, L.R.; Velázquez, E.; Mateos, P.F.; Peix, A. Recent advances in the active biomolecules involved in rhizobia-legume symbiosis. In Microbes for Legume Improvement; Zaidi, A., Khan, M., Musarrat, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 45–74. [Google Scholar]
- Kandel, S.L.; Joubert, P.M.; Doty, S.L. Bacterial endophyte colonization and distribution within plants. Microorganisms 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Robledo, M.; Rivera, L.; Jiménez-Zurdo, J.I.; Rivas, R.; Dazzo, F.; Velázquez, E.; Martínez-Molina, E.; Hirsch, A.M.; Mateos, P.F. Role of Rhizobium endoglucanase CelC2 in cellulose biosynthesis and biofilm formation on plant roots and abiotic surfaces. Microb. Cell Fact. 2012, 11, 125. [Google Scholar] [CrossRef]
- Boch, J.; Kempf, B.; Bremer, E. Osmoregulation in Bacillus subtilis: Synthesis of the osmoprotectant glycine betaine from exogenously provided choline. J. Bacteriol. 1994, 176, 5364–5371. [Google Scholar] [CrossRef] [PubMed]
- Scott, R.A.; Thilmony, R.; Harden, L.A.; Zhou, Y.; Brandl, M.T. Escherichia coli O157: H7 converts plant-derived choline to glycine betaine for osmoprotection during pre- and post-harvest colonization of injured lettuce leaves. Front. Microbiol. 2017, 8, 2436. [Google Scholar] [CrossRef]
- Boncompagni, E.; Osteras, M.; Poggi, M.C.; le Rudulier, D. Occurrence of choline and glycine betaine uptake and metabolism in the family Rhizobiaceae and their roles in osmoprotection. Appl. Environ. Microbiol. 1999, 65, 2072–2077. [Google Scholar] [CrossRef]
- McInerney, J.O.; McNally, A.; O’Connell, M.J. Why prokaryotes have pangenomes. Nat. Microbiol. 2017, 2, 17040. [Google Scholar] [CrossRef]
- Brockhurst, M.A.; Harrison, E.; Hall, J.P.J.; Richards, T.; McNally, A.; MacLean, C. The ecology and evolution of pangenomes. Curr. Biol. 2019, 29, R1094–R1103. [Google Scholar] [CrossRef]
| Attributes | E. cerealis RZME27T | P. pelagicum RI-200B4T | N. galegae HAMBI 540T | R. leguminosarum USDA 2370T |
|---|---|---|---|---|
| Sequence size (bp) | 6,114,623 | 5,134,606 | 6,455,027 | 7,851,935 |
| Number of contigs | 51 | 62 | 2 | 108 |
| GC content (%) | 58.8 | 62.8 | 61.3 | 60.6 |
| Longest contig size | 649,048 | 392,382 | 4,647,962 | 1,820,721 |
| Shortest contig size | 285 | 4118 | 1,807,065 | 502 |
| N50 value | 345,671 | 184,300 | 4,647,962 | 413,138 |
| L50 value | 7 | 11 | 1 | 5 |
| Total number of genes | 5893 | 4509 | 6248 | 7753 |
| Total CDSs | 5836 | 4458 | 6,181 | 7699 |
| Number of coding sequences/CDSs (with protein) | 5705 | 4417 | 6014 | 7312 |
| Pseudogenes | 131 | 41 | 167 | 387 |
| Number of RNAs (tRNAs, rRNAs, ncRNAs) | 57 (49, 4, 4) | 51 (47, 3, 1) | 67 (51, 9, 7) | 54 (46, 4, 4) |
| Species | RZME27T | HAMBI 540T | R1-200B4T | USDA 2370T | WS11T | DSM 29853T | DSM 25528T | DSM 18401T | Casida AT | ORS 992T | LMG 140T | H152T |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RZME17T | 100 | |||||||||||
| HAMBI 540T | 75.61 | 100 | ||||||||||
| R1-200B4T | 75.09 | 75.40 | 100 | |||||||||
| USDA 2370T | 73.34 | 74.49 | 73.03 | 100 | ||||||||
| WS11T | 70.91 | 70.23 | 70.69 | 70.24 | 100 | |||||||
| DSM 29853T | 72.78 | 72.34 | 73.02 | 72.56 | 70.75 | 100 | ||||||
| DSM 25528T | 73.82 | 73.47 | 74.19 | 73.15 | 70.87 | 73.33 | 100 | |||||
| DSM 18401T | 73.14 | 73.53 | 73.40 | 73.92 | 71.08 | 73.55 | 73.70 | 100 | ||||
| Casida AT | 72.75 | 73.20 | 73.03 | 73.64 | 70.87 | 72.82 | 73.04 | 76.03 | 100 | |||
| ORS 992T | 72.97 | 72.70 | 72.65 | 72.81 | 69.94 | 72.50 | 73.27 | 72.75 | 72.23 | 100 | ||
| DSM 30147T | 74.30 | 73.77 | 73.39 | 73.29 | 70.10 | 72.26 | 73.11 | 73.20 | 72.63 | 72.76 | 100 | |
| H152T | 72.90 | 73.17 | 72.73 | 74.57 | 70.70 | 72.57 | 72.97 | 75.43 | 75.74 | 71.96 | 72.52 | 100 |
| Fatty Acid * | E. cerealis RZME27T | N. galegae HAMBI 540T | P. pelagicum R1-200B4T | R. leguminosarum USDA 2370T |
|---|---|---|---|---|
| C16:0 | 8.86 | 11.58 | 7.69 | 7.18 |
| C17:0 | 0.10 | 1.91 | nd | nd |
| C18:0 | 2.08 | 2.28 | 2.26 | 12.71 |
| C16:0 3OH | 2.14 | 2.54 | nd | nd |
| C18:0 3OH | 1.30 | 1.60 | 1.49 | 0.99 |
| C18:1ω7c 11-methyl | 0.38 | 0.55 | 3.04 | 7.06 |
| C19:0 cyclo ω8c | nd | 10.76 | 4.37 | 3.74 |
| Summed feature 2 1 | 5.18 | 4.63 | 4.41 | 3.59 |
| Summed feature 3 2 | 1.78 | 0.63 | 1.49 | 1.19 |
| Summed feature 8 3 | 77.70 | 61.14 | 73.84 | 62.77 |
| Characteristics | E. cerealis RZME27T | N. galegae HAMBI 540T | P. pelagicum R1-200B4T | R. leguminosarum USDA 2370T |
|---|---|---|---|---|
| Colony colour (on YMA) | white | white-pink | white | white-cream |
| Nitrate reduction | + | − | + | − |
| Growth in 1% NaCl | + | + | + | − |
| Growth in 4% NaCl | + | − | + | − |
| Growth in 7% NaCl | − | − | + | − |
| Growth at 37 °C | w | + | + | − |
| Assimilation of (API 20NE): | ||||
| Malate | + | + | − | w |
| Assimilation of (API 32GN): | ||||
| l-rhamnose | + | − | + | + |
| d-ribose | w | + | − | + |
| Inositol | + | + | − | + |
| Melibiose | + | + | − | + |
| 5-keto-gluconate | + | + | + | − |
| 2-keto-gluconate | − | − | w | − |
| Propionate | + | − | − | − |
| 3-hydroxybutyrate | − | − | − | + |
| l-histidine | + | + | − | + |
| l-alanine | w | + | w | − |
| l-serine | − | − | + | − |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Menéndez, E.; Flores-Félix, J.D.; Ramírez-Bahena, M.H.; Igual, J.M.; García-Fraile, P.; Peix, A.; Velázquez, E. Genome Analysis of Endobacterium cerealis, a Novel Genus and Species Isolated from Zea mays Roots in North Spain. Microorganisms 2020, 8, 939. https://doi.org/10.3390/microorganisms8060939
Menéndez E, Flores-Félix JD, Ramírez-Bahena MH, Igual JM, García-Fraile P, Peix A, Velázquez E. Genome Analysis of Endobacterium cerealis, a Novel Genus and Species Isolated from Zea mays Roots in North Spain. Microorganisms. 2020; 8(6):939. https://doi.org/10.3390/microorganisms8060939
Chicago/Turabian StyleMenéndez, Esther, Jose David Flores-Félix, Martha Helena Ramírez-Bahena, Jose M. Igual, Paula García-Fraile, Alvaro Peix, and Encarna Velázquez. 2020. "Genome Analysis of Endobacterium cerealis, a Novel Genus and Species Isolated from Zea mays Roots in North Spain" Microorganisms 8, no. 6: 939. https://doi.org/10.3390/microorganisms8060939
APA StyleMenéndez, E., Flores-Félix, J. D., Ramírez-Bahena, M. H., Igual, J. M., García-Fraile, P., Peix, A., & Velázquez, E. (2020). Genome Analysis of Endobacterium cerealis, a Novel Genus and Species Isolated from Zea mays Roots in North Spain. Microorganisms, 8(6), 939. https://doi.org/10.3390/microorganisms8060939







